LINC00885 a Novel Oncogenic Long Non-Coding RNA Associated with Early Stage Breast Cancer Progression
Abstract
1. Introduction
2. Results and Discussion
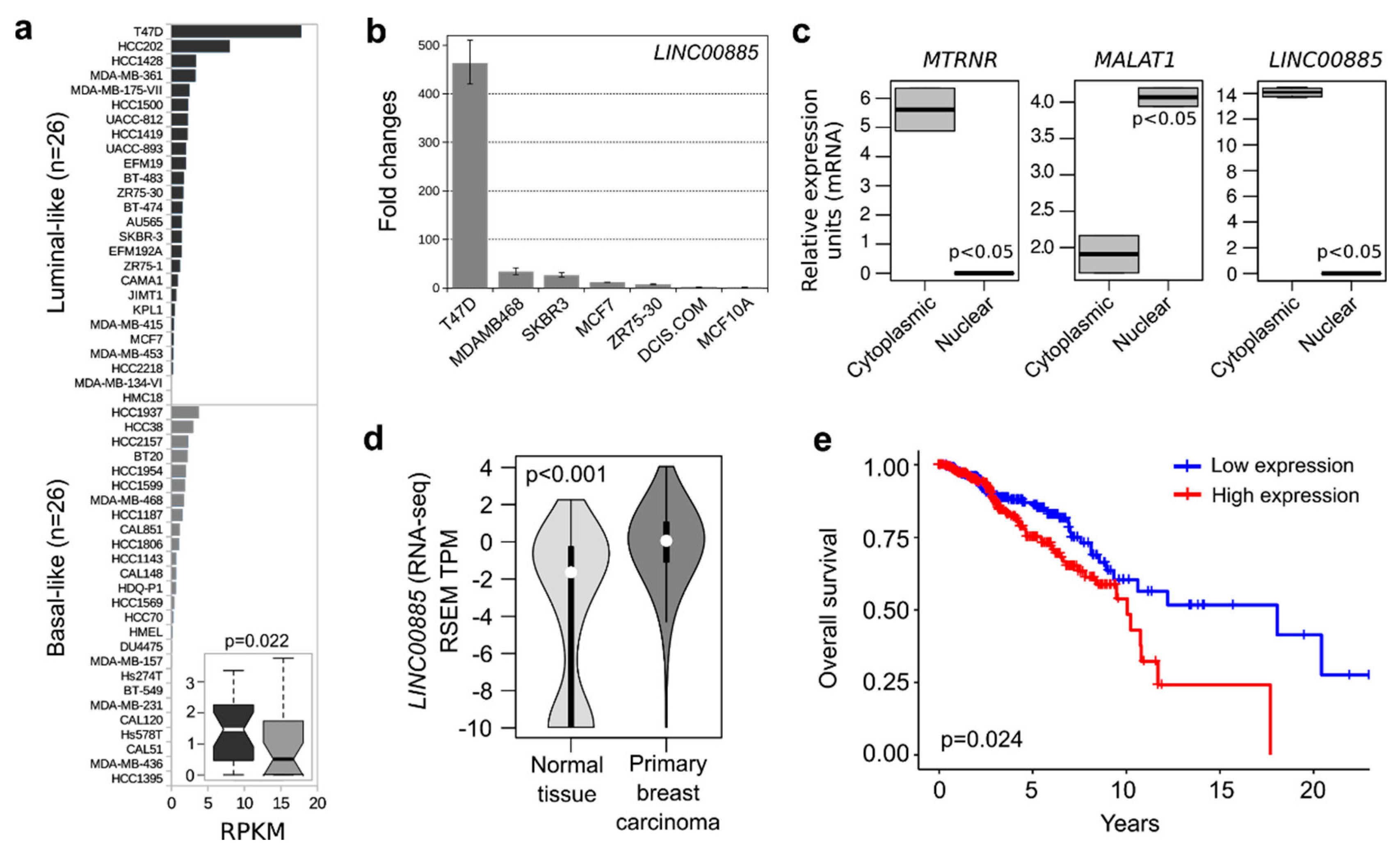
2.1. LINC00885 Expression and Subcellular Localization in Breast Cancer Lines
2.2. LINC00885 Overexpression Correlates with Poorer Breast Cancer Outcome
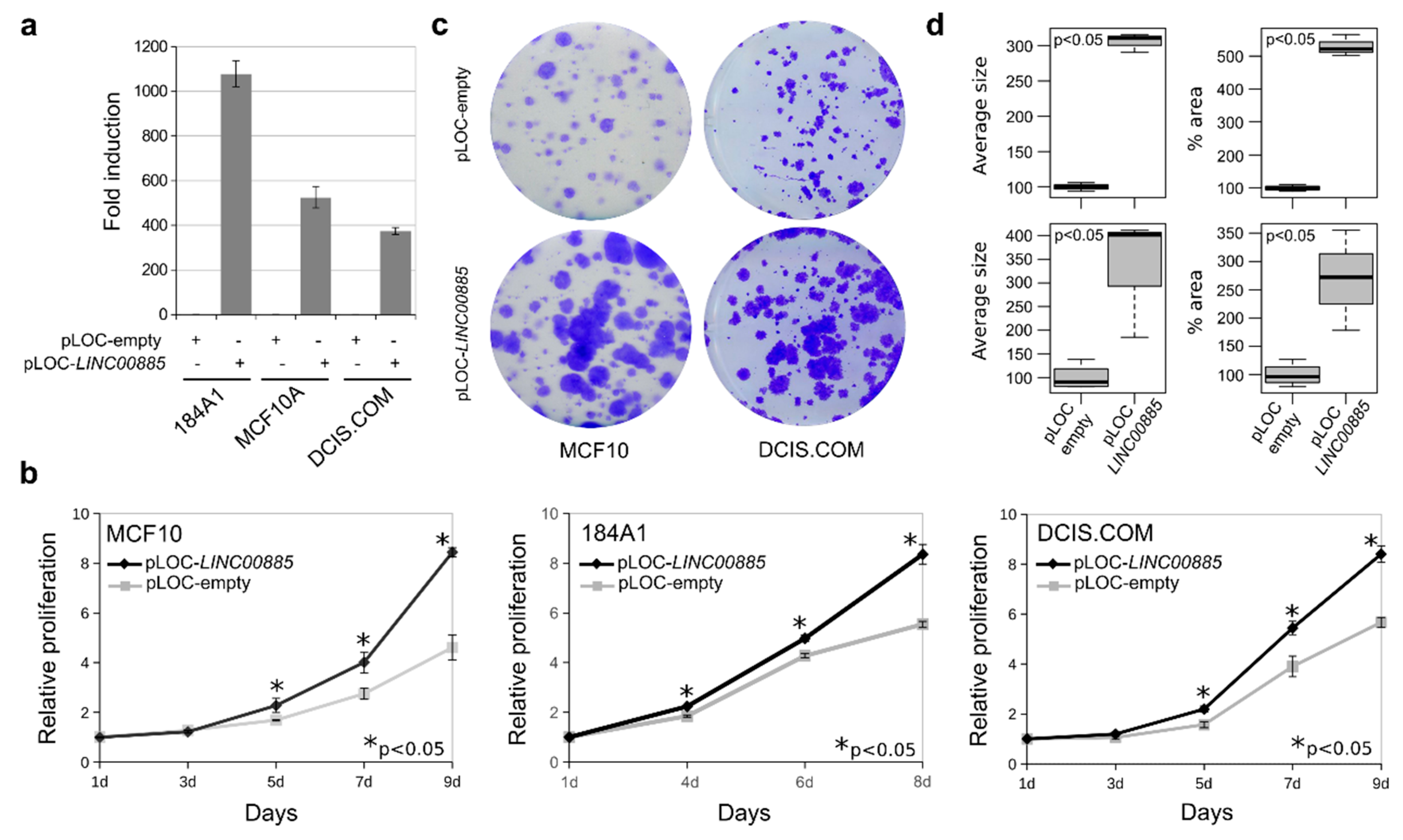
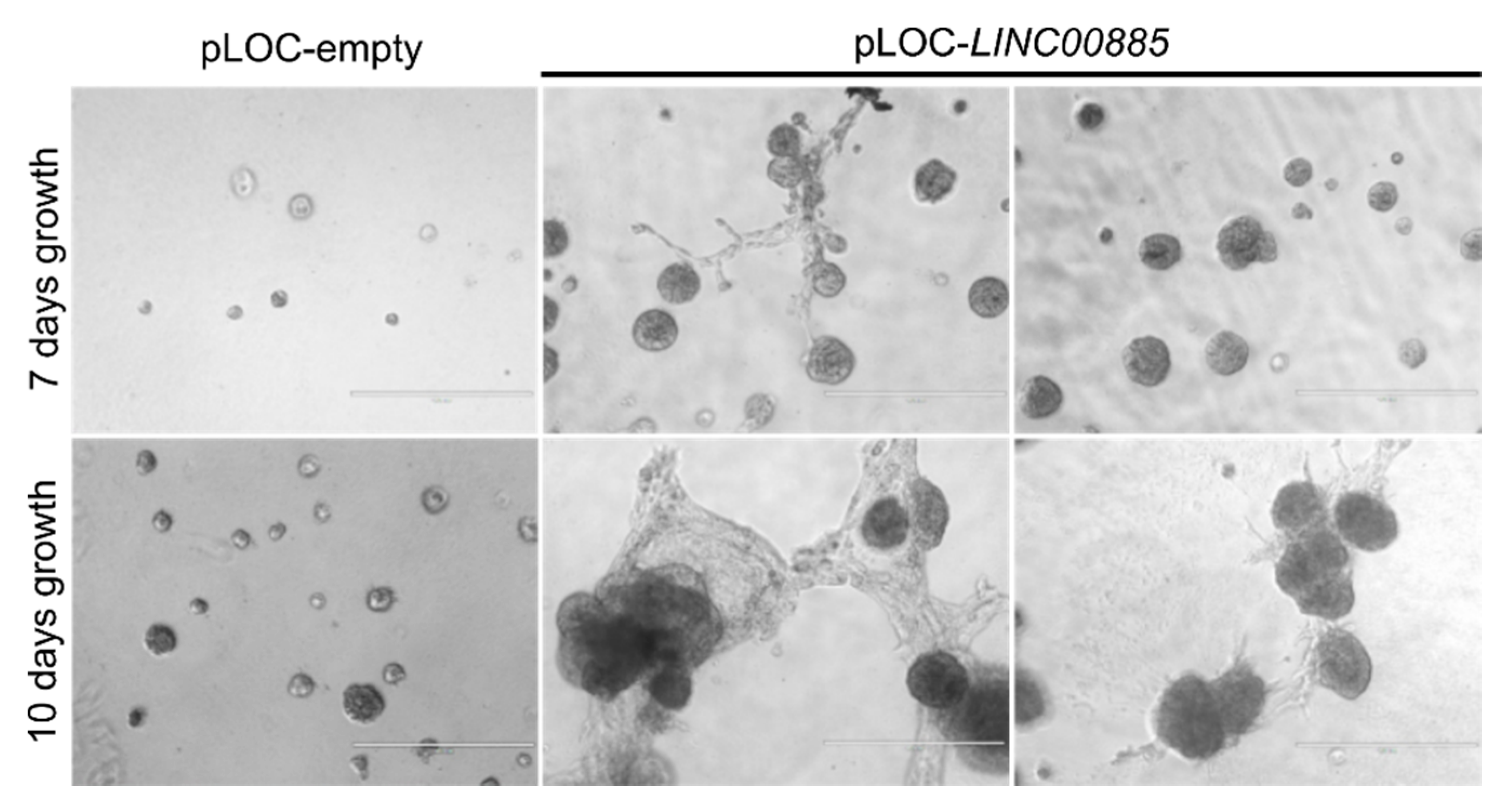
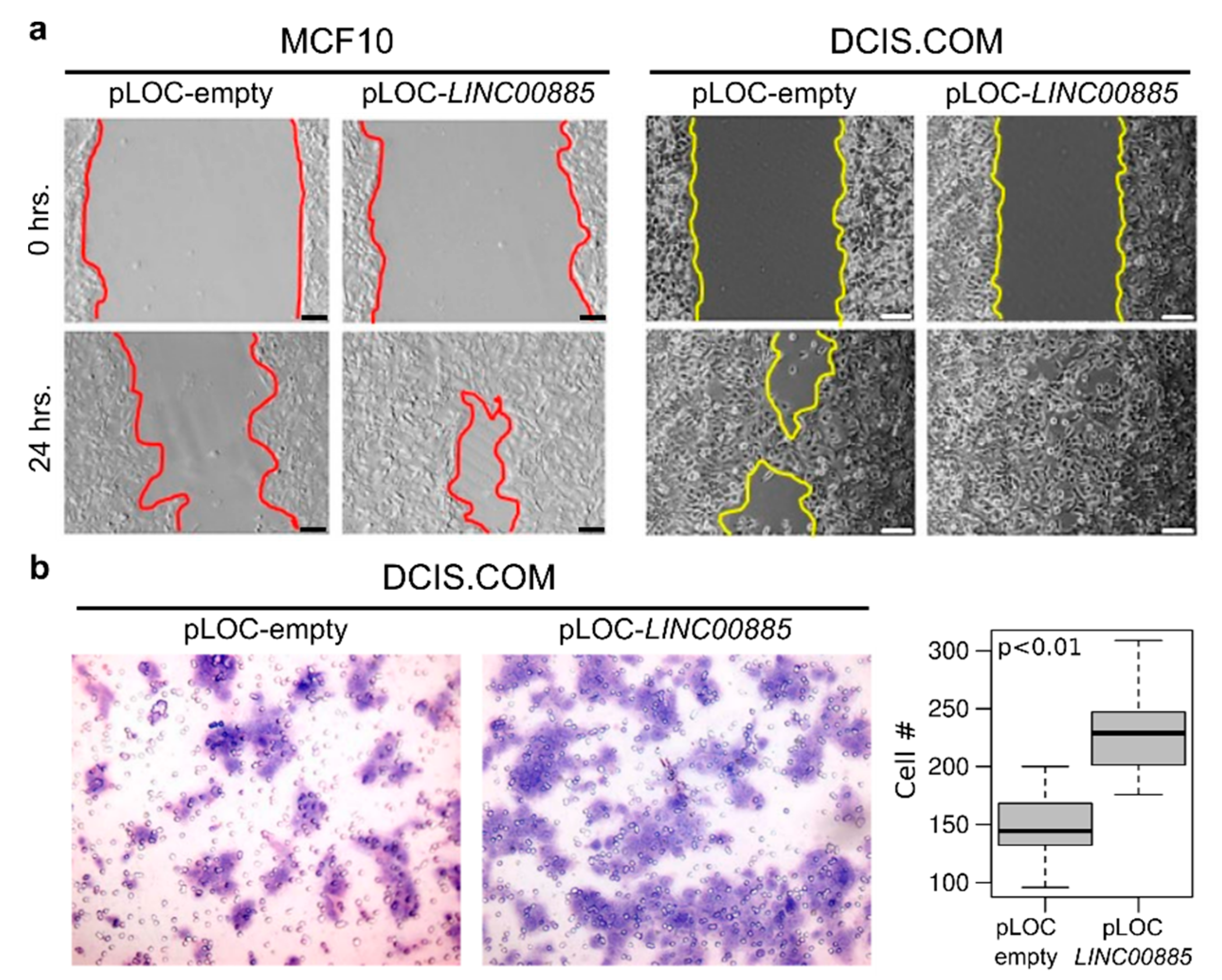
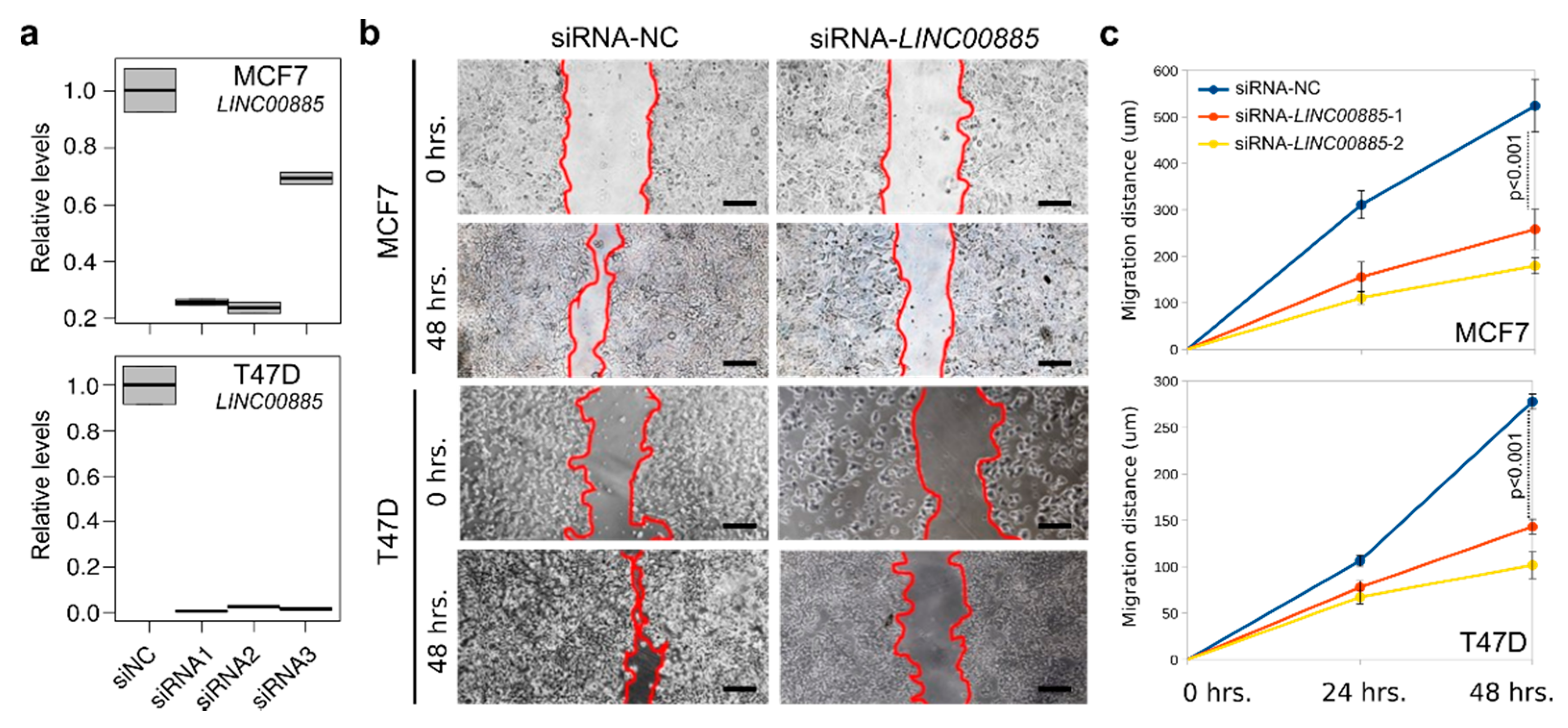
2.3. LINC00885 Overexpression Promotes In Vitro Proliferation, Migration, and Invasion of DCIS and Normal Breast Cancer Cells
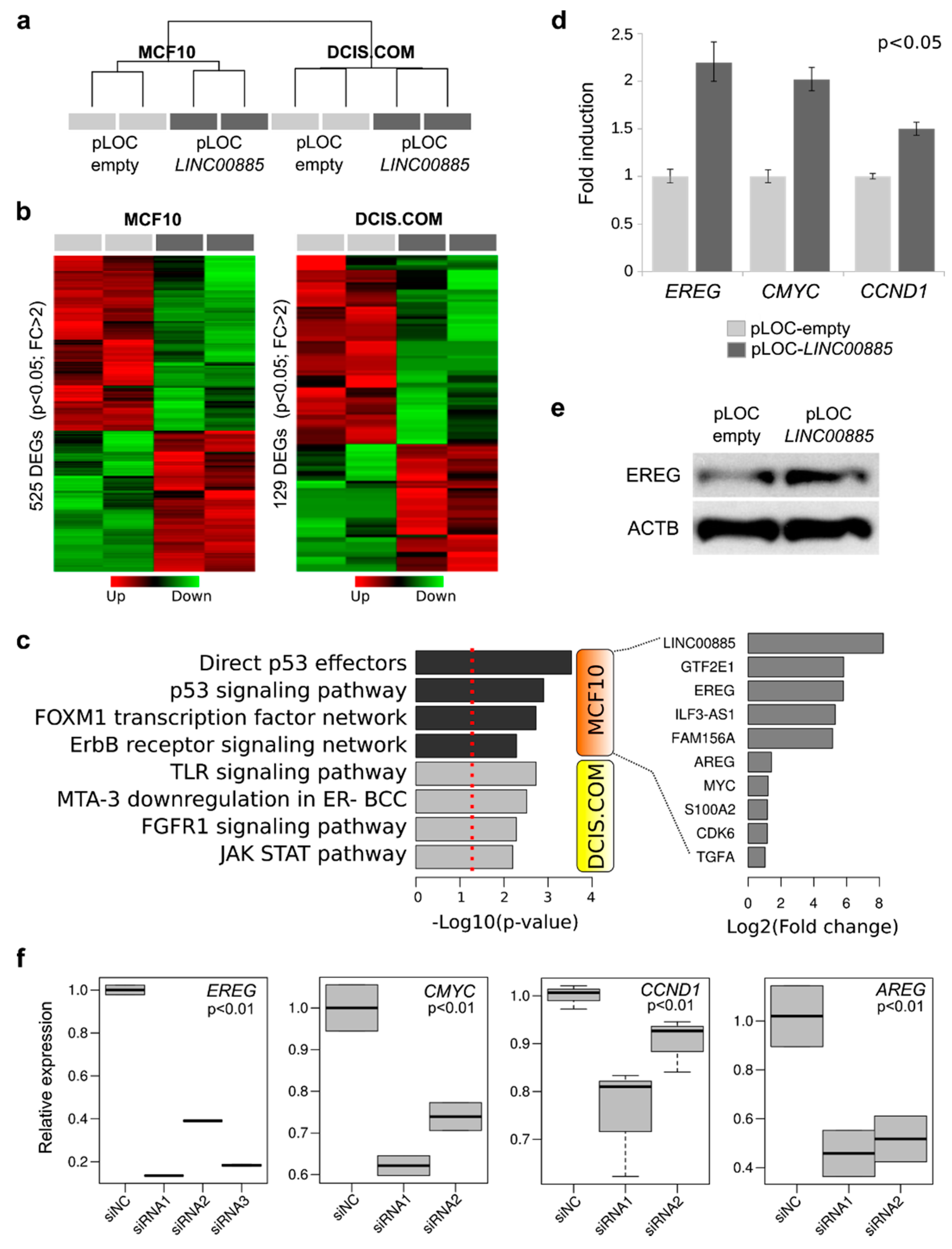
2.4. Transcriptome Analysis and RNA-Protein Interaction of LINC00885 Overexpressing Cells
3. Materials and Methods
3.1. Cell Lines, Cell Culture
3.2. LncRNA Subcellular Localization Methods
3.3. Stable LINC00885 Expressing Cells
3.4. RNA Interference (siRNA) Assay
3.5. RNA Isolation and Real Time Quantitative PCR (RT-qPCR)
3.6. Cell Proliferation, Clonal Growth, Cell Motility and Migration Assays
3.7. Tumorigenicity Assay
3.8. RNA-Seq Data Analysis
3.9. RNA-Pulldown and Mass Spectrometry Analysis
3.10. Western Blot Analysis
3.11. In Silico Analysis of LINC00885 in Breast Cancer
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DCIS | Ductal carcinoma in situ |
| lincRNA | Long-intergenic RNA |
| CNA | Copy number alteration |
| DEG | Differentially expressed genes |
References
- Allred, D.C. Ductal carcinoma in situ: Terminology, classification, and natural history. J. Natl. Cancer Inst. Monogr. 2010, 41, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Abba, M.C.; Gong, T.; Lu, Y.; Lee, J.; Zhong, Y.; Lacunza, E.; Butti, M.; Takata, Y.; Gaddis, S.; Shen, J.; et al. A molecular portrait of high-grade ductal carcinoma in situ. Cancer Res. 2015, 75, 3980–3990. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Bernassola, F.; Candi, E. Emerging roles of long non-coding RNAs in breast cancer biology and management. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, J.K. The emerging role of long noncoding RNAs in human disease. Methods Mol. Biol. 2018, 1706, 91–110. [Google Scholar]
- Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martín, D.; Merkel, A.; Lagarde, J.; Veeravalli, L.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Ma, M.-H.; An, J.-X.; Zhang, C.; Liu, J.; Liang, Y.; Zhang, C.-D.; Zhang, Z.; Dai, D.-Q. ZEB1-AS1 initiates a miRNA-mediated ceRNA network to facilitate gastric cancer progression. Cancer Cell Int. 2019, 19, 27. [Google Scholar] [CrossRef]
- Hah, N.; Danko, C.G.; Core, L.; Waterfall, J.J.; Siepel, A.; Lis, J.T.; Kraus, W.L. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 2011, 145, 622–634. [Google Scholar] [CrossRef]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef]
- Perez, D.S.; Hoage, T.R.; Pritchett, J.R.; Ducharme-Smith, A.L.; Halling, M.L.; Ganapathiraju, S.C.; Streng, P.S.; Smith, D.I. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum. Mol. Genet. 2008, 17, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.; Perez, D.S.; Pritchett, J.R.; Halling, M.L.; Tang, H.; Smith, D.I. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics 2010, 95, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Zaheed, O.; Samson, J.; Dean, K. A bioinformatics approach to identify novel long, non-coding RNAs in breast cancer cell lines from an existing RNA-sequencing dataset. Non-Cod. RNA Res. 2020, 5, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Malouf, G.G.; Chen, Y.; Zhang, J.; Yao, H.; Valero, V.; Weinstein, J.N.; Spano, J.P.; Meric-Bernstam, F.; Khayat, D.; et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget 2014, 5, 9864. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, Z.; Yang, Y.; Xiang, T.; Song, W.; Liu, S. Microarray expression profiling of dysregulated long non-coding RNAs in triple-negative breast cancer. Cancer Biol. Ther. 2015, 16, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Xu, P.; Wu, Y.; Huang, L.; Li, W.; Lv, S.; Wu, X.; Zeng, X.; Shen, R.; Jia, X.; et al. LncRNAs as new biomarkers to differentiate triple negative breast cancer from non-triple negative breast cancer. Oncotarget 2016, 7, 13047. [Google Scholar] [CrossRef]
- Deng, Y.; Luo, S.; Zhang, X.; Zou, C.; Yuan, H.; Liao, G.; Xu, L.; Deng, C.; Lan, Y.; Zhao, T.; et al. A pan-cancer atlas of cancer hallmark-associated candidate driver lncRNA s. Mol. Oncol. 2018, 12, 1980–2005. [Google Scholar] [CrossRef]
- Zhang, K.; Shi, Z.M.; Chang, Y.N.; Hu, Z.M.; Qi, H.X.; Hong, W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene 2014, 547, 1–9. [Google Scholar] [CrossRef]
- Farooqui, M.; Bohrer, L.R.; Brady, N.J.; Chuntova, P.; Kemp, S.E.; Wardwell, C.T.; Nelson, A.C.; Schwertfeger, K.L. Epiregulin contributes to breast tumorigenesis through regulating matrix metalloproteinase 1 and promoting cell survival. Mol. Cancer 2015, 14, 138. [Google Scholar] [CrossRef]
- Amsterdam, A.; Shezen, E.; Raanan, C.; Slilat, Y.; Ben-Arie, A.; Prus, D.; Schreiber, L. Epiregulin as a marker for the initial steps of ovarian cancer development. Int. J. Oncol. 2011, 39, 1165–1172. [Google Scholar] [CrossRef]
- Schneider, M.R.; Wolf, E. The epidermal growth factor receptor ligands at a glance. J. Cell Physiol. 2009, 218, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Seger, R. The ERK signaling cascade—Views from different subcellular compartments. Biofactors 2009, 35, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Keshet, Y.; Seger, R. The MAP kinase signaling cascades: A system of hundreds of components regulates a diverse array of physiological functions. Methods Mol. Biol. 2010, 661, 3–38. [Google Scholar] [PubMed]
- Li, M.; Liu, Y.; Zhang, X.; Liu, J.; Wang, P. Transcriptomic analysis of high-throughput sequencing about circRNA, lncRNA and mRNA in bladder cancer. Gene 2018, 677, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Degli Esposti, D.; Hernandez-Vargas, H.; Voegele, C.; Fernandez-Jimenez, N.; Forey, N.; Bancel, B.; Le Calvez-Kelm, F.; McKay, J.; Merle, P.; Herceg, Z. Identification of novel long non-coding RNAs deregulated in hepatocellular carcinoma using RNA-sequencing. Oncotarget 2016, 7, 31862. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, Y.; Yang, J. LncRNAs are altered in lung squamous cell carcinoma and lung adenocarcinoma. Oncotarget 2017, 8, 24275. [Google Scholar] [CrossRef] [PubMed]
- Hmeljak, J.; Sanchez-Vega, F.; Hoadley, K.A.; Shih, J.; Stewart, C.; Heiman, D.; Tarpey, P.; Danilova, L.; Drill, E.; Gibb, E.A.; et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018, 8, 1548–1565. [Google Scholar] [CrossRef] [PubMed]
- Brenner, A.J.; Stampfer, M.R.; Aldaz, C.M. Increased p16 expression with first senescence arrest in human mammary epithelial cells and extended growth capacity with p16 inactivation. Oncogene 1998, 17, 199–205. [Google Scholar] [CrossRef]
- Behbod, F.; Kittrell, F.S.; LaMarca, H.; Edwards, D.; Kerbawy, S.; Heestand, J.C.; Young, E.; Mukhopadhyay, P.; Yeh, H.W.; Allred, D.C.; et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009, 11, R66. [Google Scholar] [CrossRef]
- Miller, F.R.; Santner, S.J.; Tait, L.; Dawson, P.J. MCF10DCIS. com xenograft model of human comedo ductal carcinoma in situ. J. Natl. Cancer Inst. 2000, 92, 1185a–1186. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abba, M.C.; Canzoneri, R.; Gurruchaga, A.; Lee, J.; Tatineni, P.; Kil, H.; Lacunza, E.; Aldaz, C.M. LINC00885 a Novel Oncogenic Long Non-Coding RNA Associated with Early Stage Breast Cancer Progression. Int. J. Mol. Sci. 2020, 21, 7407. https://doi.org/10.3390/ijms21197407
Abba MC, Canzoneri R, Gurruchaga A, Lee J, Tatineni P, Kil H, Lacunza E, Aldaz CM. LINC00885 a Novel Oncogenic Long Non-Coding RNA Associated with Early Stage Breast Cancer Progression. International Journal of Molecular Sciences. 2020; 21(19):7407. https://doi.org/10.3390/ijms21197407
Chicago/Turabian StyleAbba, Martin C., Romina Canzoneri, Agustina Gurruchaga, Jaeho Lee, Pradeep Tatineni, Hyunsuk Kil, Ezequiel Lacunza, and C. Marcelo Aldaz. 2020. "LINC00885 a Novel Oncogenic Long Non-Coding RNA Associated with Early Stage Breast Cancer Progression" International Journal of Molecular Sciences 21, no. 19: 7407. https://doi.org/10.3390/ijms21197407
APA StyleAbba, M. C., Canzoneri, R., Gurruchaga, A., Lee, J., Tatineni, P., Kil, H., Lacunza, E., & Aldaz, C. M. (2020). LINC00885 a Novel Oncogenic Long Non-Coding RNA Associated with Early Stage Breast Cancer Progression. International Journal of Molecular Sciences, 21(19), 7407. https://doi.org/10.3390/ijms21197407





