Is the Secret of VDAC Isoforms in Their Gene Regulation? Characterization of Human VDAC Genes Expression Profile, Promoter Activity, and Transcriptional Regulators
Abstract
1. Introduction
2. Results
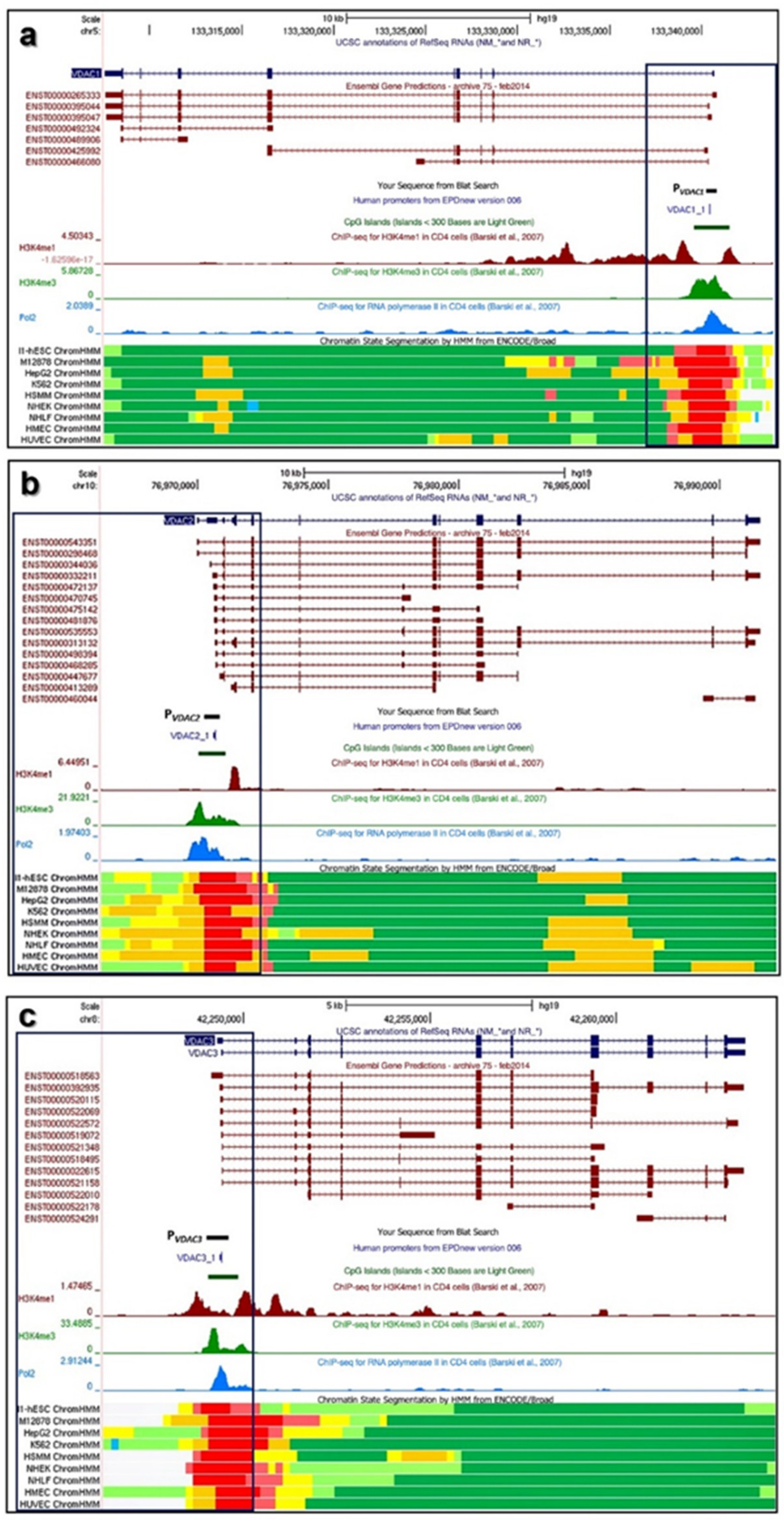
2.1. Structure of Human VDAC1, VDAC2, and VDAC3 Genes: Transcripts Variants and Promoters
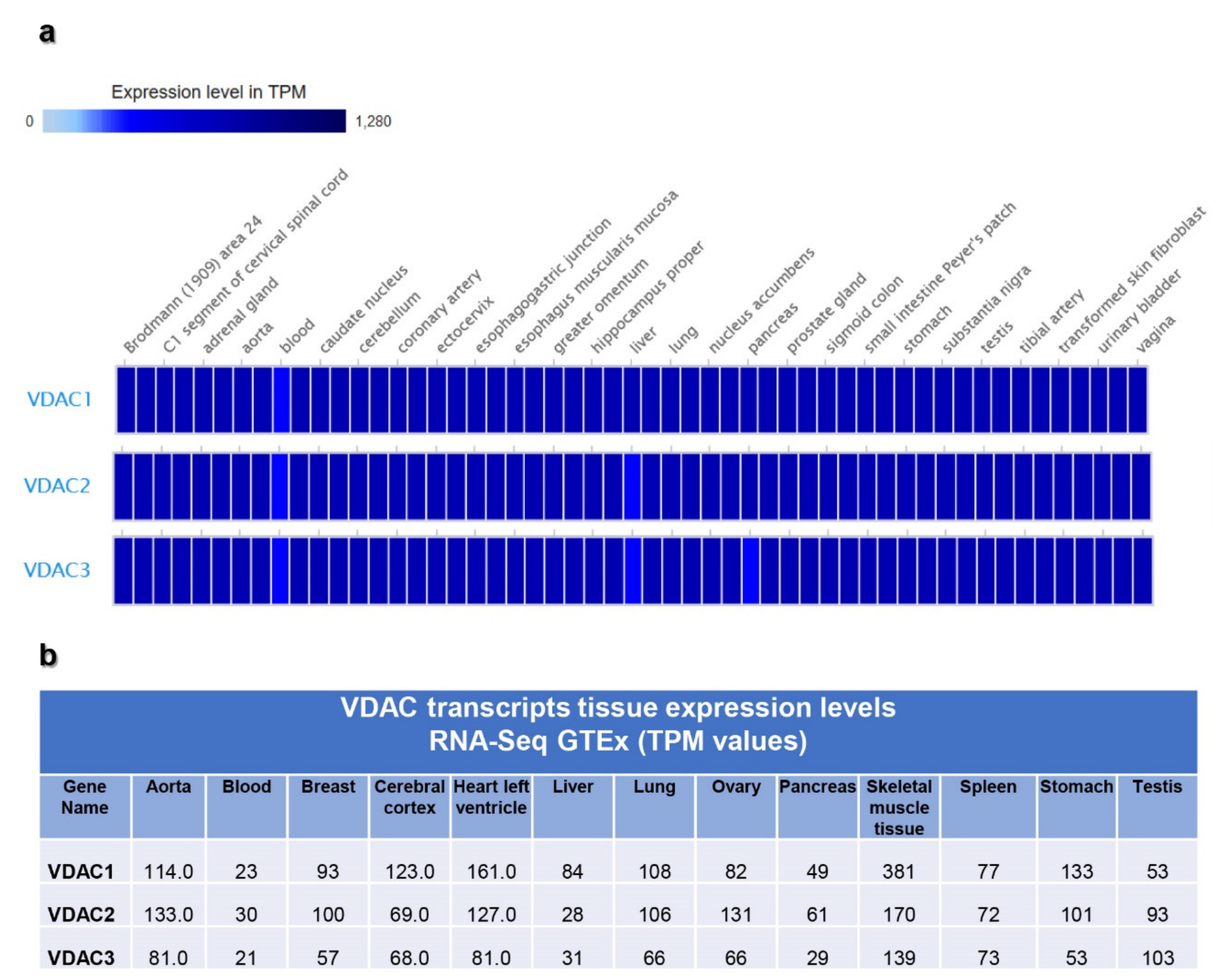
2.2. VDAC Gene Transcription by Expression Atlas Resources
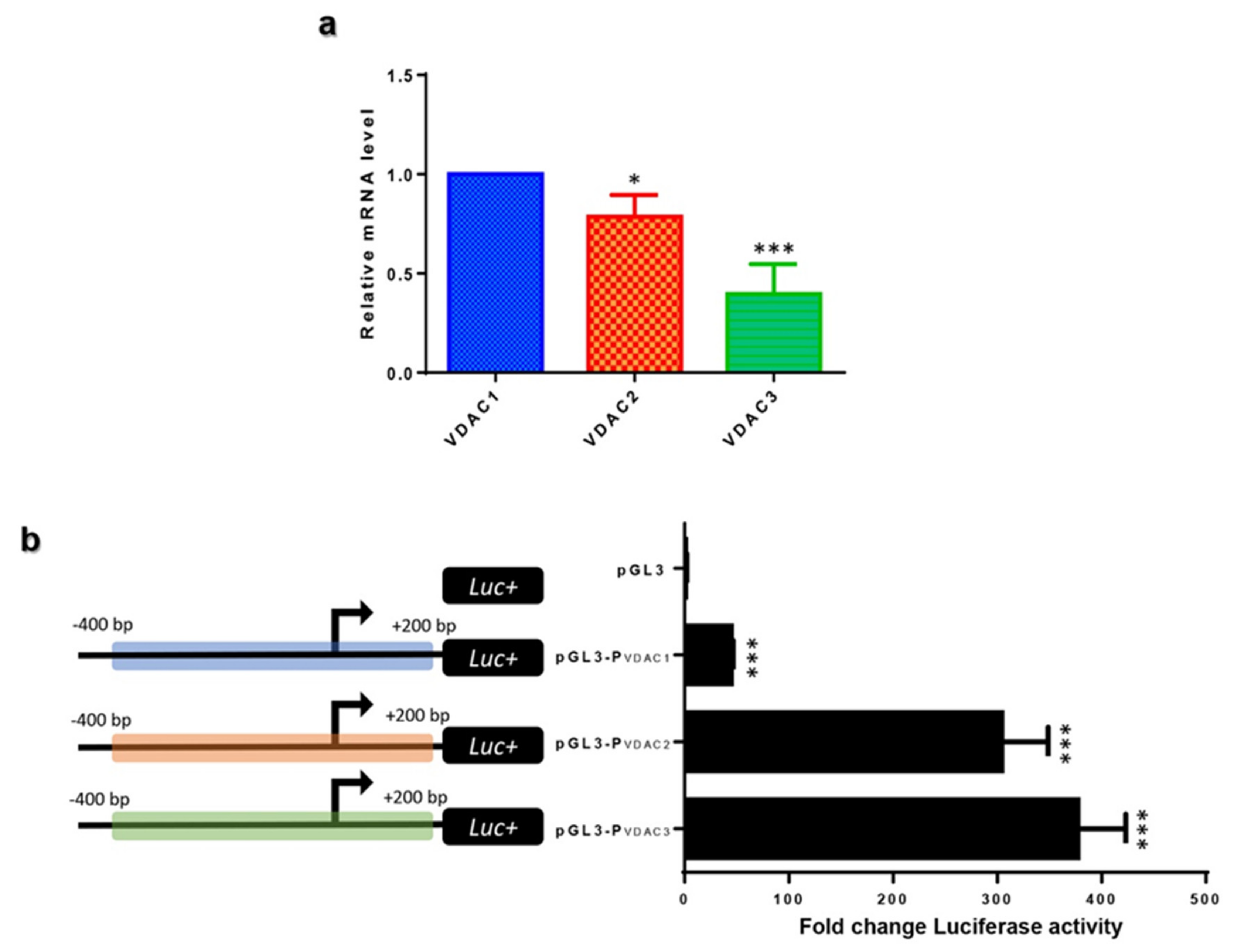
2.3. VDAC Isoforms Comparative Expression in HeLa Cells
2.4. VDAC Genes Transcriptional Promoter Activity
2.5. Characterization of VDAC Genes Core Promoters
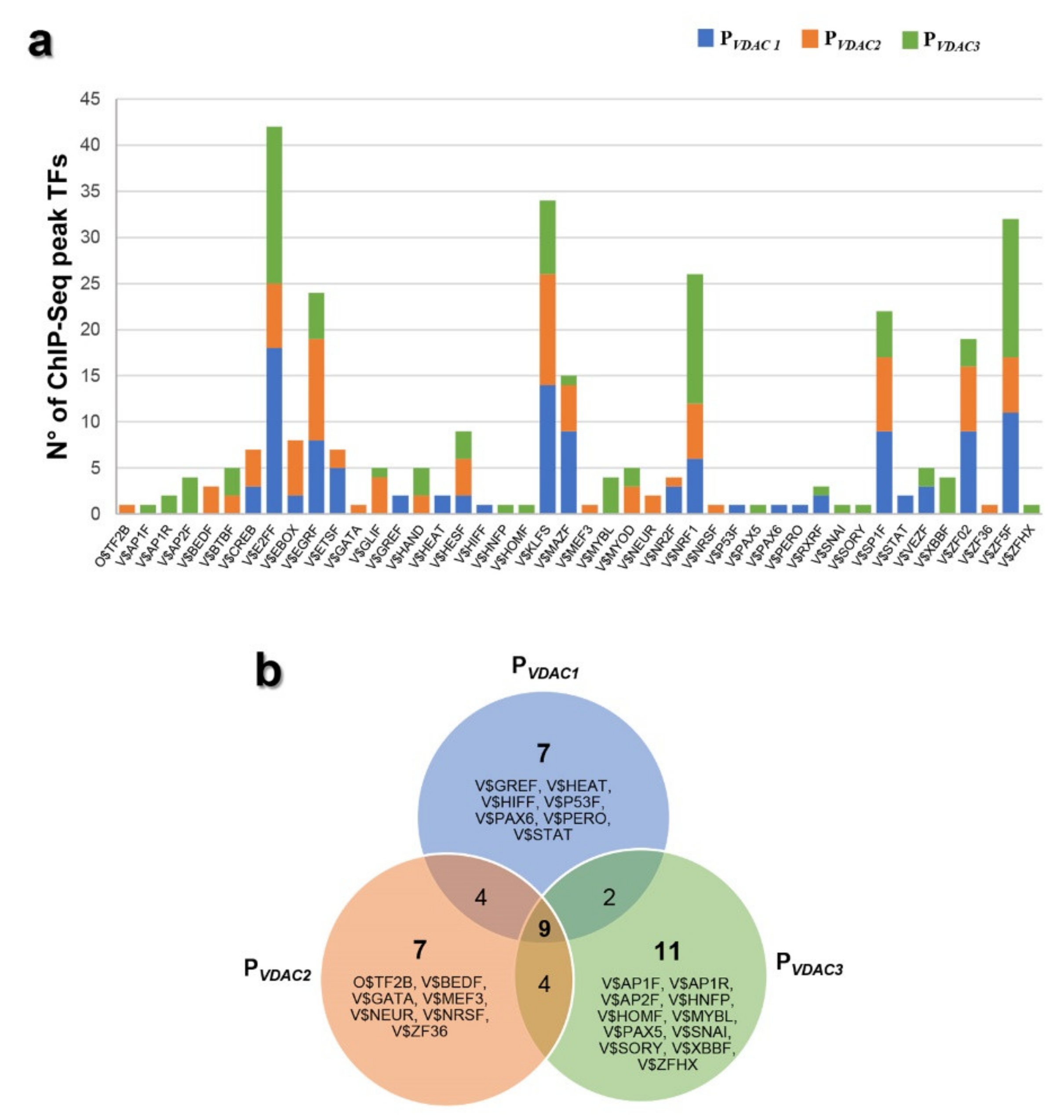
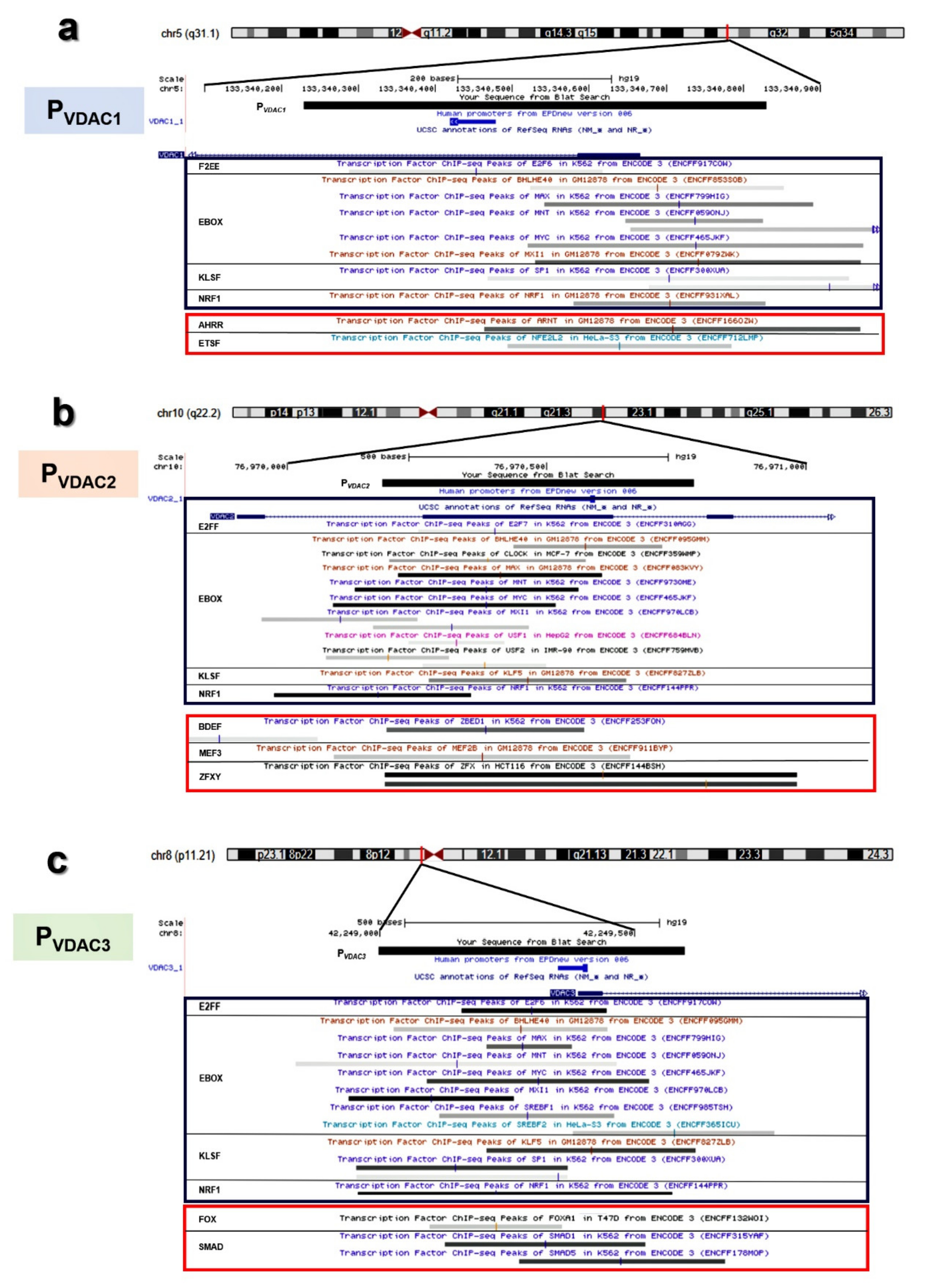
2.6. Characterization of VDACs’ Transcriptional Regulators
2.7. Analysis of VDACs Common Transcription Factor Binding Sites (TFBSs)
2.8. Analysis of VDACs Unique Transcription Factor Binding Sites (TFBSs)
3. Discussion
3.1. Structure of VDAC Genes, Transcripts, and Promoters
3.2. VDAC Expression in Expression Atlas Repository
3.3. VDAC Genes Core Promoters
3.4. Transcription Factors Binding Sites Common to Any VDAC Gene
3.5. Transcription Factors Binding Sites Specific to Each VDAC Gene
4. Materials and Methods
4.1. Bioinformatic Analysis of Promoter Region
4.2. Gene Expression Data Retrieval
4.3. Quantitative Real-Time PCR
4.4. Plasmid Constructs
4.5. Promoter Reporter Assay
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AHRR | AHR-arnt heterodimers and AHR-related factors |
| BCL6 | B-cell lymphoma 6 protein |
| BEDF | BED subclass of zinc-finger proteins |
| BRAC | Brachyury gene, mesoderm developmental factor |
| BRE | B recognition element |
| CAGE | Cap analysis gene expression |
| CDXF | Vertebrate caudal related homeodomain protein |
| CLOX | CLOX and CLOX homology (CDP) factors |
| DB | Database |
| DPE | Downstream promoter element |
| E2FF | E2F-myc activator/cell cycle |
| EBOX | E-box binding factors |
| ER | Endoplasmic Reticulum |
| ETSF | Human and murine ETS1 factors |
| FANTOM | Functional ANnotation of the Mammalian Genome |
| FPKM | Fragments Per Kilobase Million |
| FOX | Forkhead (FKH)/ Forkhead box (Fox) |
| GTEX | Genotype-Tissue Expression |
| HEAT | Heat shock factors |
| HMG | High-Mobility Group family |
| HOMF | Homeodomain transcription factors |
| Inr | Initiator Element |
| IRFF | Interferon regulatory factors |
| KLFS | Krueppel-like transcription factors |
| LBXF | Ladybird homeobox (lbx) gene family |
| MEF3 | MEF3 binding sites |
| MYBL | Cellular and viral myb-like transcriptional regulators |
| NEUR | NeuroD, Beta2, HLH domain |
| NRF1 | Nuclear respiratory factor 1 |
| PBXC | PBX-MEIS complexes |
| SMAD | Vertebrate SMAD family of transcription factors |
| SOHLH | Spermatogenesis and oogenesis basic helix-loop-helix |
| TF2B | RNA polymerase II transcription factor II B |
| TFBSs | Transcription Factor Binding Sites |
| TPM | Transcripts Per kilobase Million |
| TSS | Transcription Start Site |
| VDAC | Voltage-Dependent Anion selective Channel |
| XBBF | X-box binding factors |
| ZFXY | Zfx and Zfy-transcription factors |
References
- Shoshan-Barmatz, V.; de Pinto, V.; Zweckstetter, M.; Raviv, Z.; Keinan, N.; Arbel, N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol. Asp. Med. 2010, 31, 227–285. [Google Scholar] [CrossRef] [PubMed]
- Messina, A.; Reina, S.; Guarino, F.; de Pinto, V. VDAC isoforms in mammals. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
- Colombini, M. VDAC: The channel at the interface between mitochondria and the cytosol. Mol. Cell. Biochem. 2004, 256, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Benz, R. Permeation of hydrophilic solutes through mitochondrial outer membranes: Review on mitochondrial porins. Biochim. Biophys. Acta Rev. Biomembr. 1994, 1197, 167–196. [Google Scholar] [CrossRef]
- Naghdi, S.; Hajnóczky, G. VDAC2-specific cellular functions and the underlying structure. Biochim. Biophys. Acta Bioenerg. 2016, 1863, 2503–2514. [Google Scholar] [CrossRef]
- Checchetto, V.; Reina, S.; Magri, A.; Szabò, I.; de Pinto, V. Recombinant Human Voltage Dependent Anion Selective Channel Isoform 3 (hVDAC3) Forms Pores with a Very Small Conductance. Cell. Physiol. Biochem. 2014, 34, 842–853. [Google Scholar] [CrossRef]
- Queralt-Martín, M.; Bergdoll, L.; Teijido, O.; Munshi, N.; Jacobs, D.; Kuszak, A.J.; Protchenko, O.; Reina, S.; Magri, A.; de Pinto, V.; et al. A lower affinity to cytosolic proteins reveals VDAC3 isoform-specific role in mitochondrial biology. J. Gen. Physiol. 2020, 152. [Google Scholar] [CrossRef]
- Sampson, M.J.; Lovell, R.S.; Davison, D.B.; Craigen, W.J. A Novel Mouse Mitochondrial Voltage-Dependent Anion Channel Gene Localizes to Chromosome 8. Genomics 1996, 36, 192–196. [Google Scholar] [CrossRef]
- Neumann, D.; Bückers, J.; Kastrup, L.; Hell, S.W.; Jakobs, S. Two-color STED microscopy reveals different degrees of colocalization between hexokinase-I and the three human VDAC isoforms. PMC Biophys. 2010, 3, 4. [Google Scholar] [CrossRef]
- Reina, S.; Palermo, V.; Guarnera, A.; Guarino, F.; Messina, A.; Mazzoni, C.; de Pinto, V. Swapping of the N-terminus of VDAC1 with VDAC3 restores full activity of the channel and confers anti-aging features to the cell. FEBS Lett. 2010, 584, 2837–2844. [Google Scholar] [CrossRef]
- Anflous, K.; Armstrong, D.D.; Craigen, W.J. Altered Mitochondrial Sensitivity for ADP and Maintenance of Creatine-stimulated Respiration in Oxidative Striated Muscles from VDAC1-deficient Mice. J. Biol. Chem. 2000, 276, 1954–1960. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.H.Y.; Sheiko, T.V.; Fisher, J.K.; Craigen, W.J.; Korsmeyer, S.J. VDAC2 Inhibits BAK Activation and Mitochondrial Apoptosis. Science 2003, 301, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Sampson, M.J.; Decker, W.K.; Beaudet, A.L.; Ruitenbeek, W.; Armstrong, D.; Hicks, M.J.; Craigen, W.J. Immotile Sperm and Infertility in Mice Lacking Mitochondrial Voltage-dependent Anion Channel Type 3. J. Biol. Chem. 2001, 276, 39206–39212. [Google Scholar] [CrossRef]
- Menzel, V.A.; Cassará, M.C.; Benz, R.; de Pinto, V.; Messina, A.; Cunsolo, V.; Saletti, R.; Hinsch, K.; Hinsch, E. Molecular and functional characterization of VDAC2 purified from mammal spermatozoa. Biosci. Rep. 2009, 29, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Manczak, M.; Sheiko, T.; Craigen, W.J.; Reddy, P.H. Reduced VDAC1 protects against Alzheimer’s disease, mitochondria, and synaptic deficiencies. J. Alzheimers Dis. 2013, 37, 679–690. [Google Scholar] [CrossRef]
- Nowak, G.; Megyesi, J.; Craigen, W.J. Deletion of VDAC1 Hinders Recovery of Mitochondrial and Renal Functions After Acute Kidney Injury. Biomolecules 2020, 10, 585. [Google Scholar] [CrossRef]
- Huizing, M.; Ruitenbeek, W.; Thinnes, F.P.; de Pinto, V.; Wendel, U.; Trijbels, F.J.M.; Smit, L.M.E.; Ter Laak, H.J.; Heuvel, L.P.V.D. Deficiency of the Voltage-Dependent Anion Channel: A Novel Cause of Mitochondriopathy. Pediatr. Res. 1996, 39, 760–765. [Google Scholar] [CrossRef]
- Huizing, M.; Ruitenbeek, W.; Thinnes, F.; de Pinto, V. Lack of voltage-dependent anion channel in human mitochondrial myopathies. Lancet 1994, 344, 762. [Google Scholar] [CrossRef]
- Baines, C.P.; Kaiser, R.A.; Sheiko, T.; Craigen, W.J.; Molkentin, J.D. Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat. Cell Biol. 2007, 9, 550–555. [Google Scholar] [CrossRef]
- Reina, S.; Pittalà, M.G.G.; Guarino, F.; Messina, A.; de Pinto, V.; Foti, S.; Saletti, R. Cysteine Oxidations in Mitochondrial Membrane Proteins: The Case of VDAC Isoforms in Mammals. Front. Cell Dev. Biol. 2020, 8, 397. [Google Scholar] [CrossRef]
- Saletti, R.; Reina, S.; Pittalà, M.G.; Magri, A.; Cunsolo, V.; Foti, S.; de Pinto, V. Post-translational modifications of VDAC1 and VDAC2 cysteines from rat liver mitochondria. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; de Pinto, V.; Barsanti, P.; Caggese, C. A genetic analysis of the porin gene encoding a voltage-dependent anion channel protein in Drosophila melanogaster. Mol. Genet. Genom. 2002, 267, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Magrì, A.; Di Rosa, M.C.; Orlandi, I.; Guarino, F.; Reina, S.; Guarnaccia, M.; Morello, G.; Spampinato, A.; Cavallaro, S.; Messina, A.; et al. Deletion of Voltage-Dependent Anion Channel 1 knocks mitochondria down triggering metabolic rewiring in yeast. Cell. Mol. Life Sci. 2019, 77, 3195–3213. [Google Scholar] [CrossRef] [PubMed]
- Specchia, V.; Guarino, F.; Messina, A.; Bozzetti, M.P.; de Pinto, V. Porin isoform 2 has a different localization in Drosophila melanogaster ovaries than porin 1. J. Bioenerg. Biomembr. 2008, 40, 219–226. [Google Scholar] [CrossRef]
- Hinsch, K.D.; Hinsch, E.; Konrad, L. VDAC2 (porin-2) expression pattern and localization in the bovine testis. Biochim. Biophys. Acta Gene Struct. Expr. 2001, 1518, 329–333. [Google Scholar] [CrossRef]
- Pan, L.; Qiu, D.; Li, J.; Li, J.; Xu, P.; Zhao, D.; Ma, J. Idiopathic male infertility in the Han population in China is affected by polymorphism in the VDAC2 gene. Oncotarget 2016, 7, 82594–82601. [Google Scholar] [CrossRef]
- Pan, L.; Liu, Q.; Li, J.; Wu, W.; Wang, X.; Zhao, D.; Ma, J. Association of the VDAC3 gene polymorphism with sperm count in Han-Chinese population with idiopathic male infertility. Oncotarget 2017, 8, 45242–45248. [Google Scholar] [CrossRef][Green Version]
- Sampson, M.J.; Lovell, R.S.; Craigen, W.J. The murine voltage-dependent anion channel gene family. Conserved structure and function. J. Biol. Chem. 1997, 272, 18966–18973. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, Y.; Sheng, Y.; Fu, X.; Cheng, H.; Zhou, R. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy 2015, 11, 1081–1098. [Google Scholar] [CrossRef]
- Xu, A.; Hua, Y.; Zhang, J.; Chen, W.; Zhao, K.; Xi, W.; Wang, H.; Fang, J.; Su, S.; Tang, M.; et al. Abnormal Hypermethylation of the VDAC2 Promoter is a Potential Cause of Idiopathic Asthenospermia in Men. Sci. Rep. 2016, 6, 37836. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Q.; Liao, M.; Diao, L.; Wu, T.; Liao, W.; Wang, Z.; Li, B.; Zhang, S.; Wang, S.; et al. Overexpression of lncRNA EPB41L4A-AS1 Induces Metabolic Reprogramming in Trophoblast Cells and Placenta Tissue of Miscarriage. Mol. Ther. Nucleic Acids 2019, 18, 518–532. [Google Scholar] [CrossRef] [PubMed]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.-P.; George, N.; Fexova, S.; Fonseca, N.A.; Füllgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas update: From tissues to single cells. Nucleic Acids Res. 2019, 48, D77–D83. [Google Scholar] [CrossRef] [PubMed]
- Ardlie, K.G.; Deluca, D.S.; Segre, A.V.; Sullivan, T.J.; Young, T.R.; Gelfand, E.T.; Trowbridge, C.A.; Maller, J.B.; Tukiainen, T.; Lek, M.; et al. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar]
- Noguchi, S.; Arakawa, T.; Fukuda, S.; Furuno, M.; Hasegawa, A.; Hori, F.; Ishikawa-Kato, S.; Kaida, K.; Kaiho, A.; Kanamori-Katayama, M.; et al. FANTOM5 CAGE profiles of human and mouse samples. Sci. Data 2017, 4, 170112. [Google Scholar] [CrossRef]
- De Pinto, V.; Guarino, F.; Guarnera, A.; Messina, A.; Reina, S.; Tomasello, F.M.; Palermo, V.; Mazzoni, C. Characterization of human VDAC isoforms: A peculiar function for VDAC3? Biochim. Biophys. Acta Bioenerg. 2010, 6–7, 1268–1275. [Google Scholar] [CrossRef]
- Deng, W. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005, 19, 2418–2423. [Google Scholar] [CrossRef]
- Nepal, C.; Hadzhiev, Y.; Balwierz, P.J.; Tarifeño-Saldivia, E.; Cardenas, R.; Wragg, J.W.; Suzuki, A.M.; Carninci, P.; Peers, B.; Lenhard, B.; et al. Dual-initiation promoters with intertwined canonical and TCT/TOP transcription start sites diversify transcript processing. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Grandori, C.; Cowley, S.M.; James, L.P.; Eisenman, R.N. The Myc/Max/Mad Network and the Transcriptional Control of Cell Behavior. Annu. Rev. Cell Dev. Biol. 2000, 16, 653–699. [Google Scholar] [CrossRef]
- Thiel, G.; Cibelli, G. Regulation of life and death by the zinc finger transcription factor Egr-1. J. Cell. Physiol. 2002, 193, 287–292. [Google Scholar] [CrossRef]
- Liu, Z.H.; Dai, X.M.; Du, B. Hes1: A key role in stemness, metastasis and multidrug resistance. Cancer Biol. Ther. 2015, 16, 353–359. [Google Scholar] [CrossRef]
- Triner, D.; Castillo, C.; Hakim, J.B.; Xue, X.; Greenson, J.K.; Núñez, G.; Chen, G.Y.; Colacino, J.A.; Shah, Y.M. Myc-Associated Zinc Finger Protein Regulates the Proinflammatory Response in Colitis and Colon Cancer via STAT3 Signaling. Mol. Cell. Biol. 2018, 38. [Google Scholar] [CrossRef] [PubMed]
- Woo, A.J.; Kim, J.; Xu, J.; Huang, H.; Cantor, A.B. Role of ZBP-89 in human globin gene regulation and erythroid differentiation. Blood 2011, 118, 3684–3693. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qu, H.; Qu, D.; Chen, F.; Zhang, Z.; Liu, B.; Liu, H. ZBTB7 Overexpression Contributes to Malignancy in Breast Cancer. Cancer Investig. 2010, 28, 672–678. [Google Scholar] [CrossRef]
- Niederreither, K.; Dollé, P. Retinoic acid in development: Towards an integrated view. Nat. Rev. Genet. 2008, 9, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Matson, J.P.; Cook, J.G. Cell cycle proliferation decisions: The impact of single cell analyses. FEBS J. 2016, 284, 362–375. [Google Scholar] [CrossRef]
- Allevato, M.; Bolotin, E.; Grossman, M.; Mane-Padros, D.; Sladek, F.M.; Martinez, E. Sequence-specific DNA binding by MYC/MAX to low-affinity non-E-box motifs. PLoS ONE 2017, 12, e0180147. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.T.; Chen, C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell. Mol. Life Sci. 2009, 66, 2691–2706. [Google Scholar] [CrossRef]
- Scarpulla, R.C.; Vega, R.B.; Kelly, D.P. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol. Metab. 2012, 23, 459–466. [Google Scholar] [CrossRef]
- Nilsson, L.I.H.; Pettersen, I.K.N.; Nikolaisen, J.; Micklem, D.; Dale, H.A.; Rosland, G.V.; Lorens, J.; Tronstad, K.J. A new live-cell reporter strategy to simultaneously monitor mitochondrial biogenesis and morphology. Sci. Rep. 2015, 5, 17217. [Google Scholar] [CrossRef]
- Costoya, J.A. Functional analysis of the role of POK transcriptional repressors. Briefings Funct. Genom. Proteom. 2007, 6, 8–18. [Google Scholar] [CrossRef]
- Labrecque, M.P.; Prefontaine, G.G.; Beischlag, T.V. The aryl hydrocarbon receptor nuclear translocator (ARNT) family of proteins: Transcriptional modifiers with multi-functional protein interfaces. Curr. Mol. Med. 2013, 13, 1047–1065. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef] [PubMed]
- Anckar, J.; Sistonen, L. Regulation of HSF1 Function in the Heat Stress Response: Implications in Aging and Disease. Annu. Rev. Biochem. 2011, 80, 1089–1115. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.; Pandha, H.S. PBX3 in Cancer. Cancers 2020, 12, 431. [Google Scholar] [CrossRef]
- Saghizadeh, M.; Akhmedov, N.B.; Yamashita, C.K.; Gribanova, Y.; Theendakara, V.; Mendoza, E.; Nelson, S.F.; Ljubimov, A.V.; Farber, D.B. ZBED4, a BED-type zinc-finger protein in the cones of the human retina. Investig. Opthalmolgy Vis. Sci. 2009, 50, 3580–3588. [Google Scholar] [CrossRef]
- Oh, S.; Hwang, E.S. The Role of Protein Modifications of T-Bet in Cytokine Production and Differentiation of T Helper Cells. J. Immunol. Res. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Li, N.; Zhao, C.T.; Wang, Y.; Yuan, X.B. The Transcription Factor Cux1 Regulates Dendritic Morphology of Cortical Pyramidal Neurons. PLoS ONE 2010, 5, e10596. [Google Scholar] [CrossRef]
- Hidaka, K.; Yamamoto, I.; Arai, Y.; Mukai, T. The MEF-3 motif is required for MEF-2-mediated skeletal muscle-specific induction of the rat aldolase A gene. Mol. Cell. Biol. 1993, 13, 6469–6478. [Google Scholar] [CrossRef]
- Kageyama, R.; Shimojo, H.; Ohtsuka, T. Dynamic control of neural stem cells by bHLH factors. Neurosci. Res. 2019, 138, 12–18. [Google Scholar] [CrossRef]
- Buratowski, S.; Zhou, H. Functional domains of transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 1993, 90, 5633–5637. [Google Scholar] [CrossRef]
- Fang, X.; Huang, Z.; Zhou, W.; Wu, Q.; Sloan, A.E.; Ouyang, G.; McLendon, R.E.; Yu, J.S.; Rich, J.N.; Bao, S. The Zinc Finger Transcription Factor ZFX is Required for Maintaining the Tumorigenic Potential of Glioblastoma Stem Cells. Stem Cells 2014, 32, 2033–2047. [Google Scholar] [CrossRef] [PubMed]
- Basso, K.; Dalla-Favera, R. BCL6: Master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 2010, 105, 193–210. [Google Scholar] [PubMed]
- Bai, Y.Q.; Miyake, S.; Iwai, T.; Yuasa, Y. CDX2, a homeobox transcription factor, upregulates transcription of the p21/WAF1/CIP1 gene. Oncogene 2003, 22, 7942–7949. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.R.; Kaestner, K.H. The Foxa family of transcription factors in development and metabolism. Cell. Mol. Life Sci. 2006, 63, 2317–2328. [Google Scholar] [CrossRef]
- Suzuki, H.; Ahn, H.W.; Chu, T.; Bowden, W.; Gassei, K.; Orwig, K.E.; Rajkovic, A. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev. Biol. 2012, 361, 301–312. [Google Scholar] [CrossRef]
- He, S.; Yang, S.; Niu, M.; Zhong, Y.; Dan, G.; Zhang, Y.; Ma, H.; Xiong, W.; Zhou, M.; Zhou, Y.; et al. HMG-box transcription factor 1: A positive regulator of the G1/S transition through the Cyclin-CDK-CDKI molecular network in nasopharyngeal carcinoma. Cell Death Dis. 2018, 9, 100. [Google Scholar] [CrossRef]
- Wang, W.; Lufkin, T. Hmx homeobox gene function in inner ear and nervous system cell-type specification and development. Exp. Cell Res. 2005, 306, 373–379. [Google Scholar] [CrossRef]
- Mahnke, J.; Schumacher, V.; Ahrens, S.; Käding, N.; Feldhoff, L.M.; Huber, M.; Rupp, J.; Raczkowski, F.; Mittrücker, H.W. Interferon Regulatory Factor 4 controls TH1 cell effector function and metabolism. Sci. Rep. 2016, 6, 35521. [Google Scholar] [CrossRef]
- Jennings, W.; Hou, M.; Peterson, D.; Missiuna, P.; Thabane, L.; Tarnopolsky, M.; Samaan, M.C. Paraspinal muscle ladybird homeobox 1 (LBX1) in adolescent idiopathic scoliosis: A cross-sectional study. Spine J. 2019, 19, 1911–1916. [Google Scholar] [CrossRef]
- Musa, J.; Aynaud, M.M.; Mirabeau, O.; Delattre, O.; Grünewald, T.G. MYBL2 (B-Myb): A central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017, 8, e2895. [Google Scholar] [CrossRef]
- Zhao, M.; Mishra, L.; Deng, C.X. The role of TGF-beta/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018, 14, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.D.; Jin, R.U.; Lo, H.; Jung, M.; Wang, H.; Battle, M.A.; Wollheim, C.B.; Urano, F.; Mills, J.C. Transcriptional Regulation of X-Box-binding Protein One (XBP1) by Hepatocyte Nuclear Factor 4α (HNF4A) is Vital to Beta-cell Function. J. Biol. Chem. 2016, 291, 6146–6157. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Qiang, Y.; Zhu, L.; Jiang, Y.; Wang, Y.; Shao, X.; Yin, L.; Chen, J.; Chen, Z. MicroRNA-7 downregulates the oncogene VDAC1 to influence hepatocellular carcinoma proliferation and metastasis. Tumor Biol. 2016, 37, 10235–10246. [Google Scholar] [CrossRef] [PubMed]
- Guarino, F.; Zinghirino, F.; Mela, L.; Pappalardo, X.G.; Ichas, F.; De, V.; Messina, A. NRF-1 and HIF-1α contribute to modulation of human VDAC1 gene promoter during starvation and hypoxia in HeLa cells. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148289. [Google Scholar] [CrossRef]
- Cam, H.; Balciunaite, E.; Blais, A.; Spektor, A.; Scarpulla, R.C.; Young, R.; Kluger, Y.; Dynlacht, B.D. A Common Set of Gene Regulatory Networks Links Metabolism and Growth Inhibition. Mol. Cell 2004, 16, 399–411. [Google Scholar] [CrossRef]
- Hughes, T.R. A Handbook of Transcription Factors; Subcellular Biochemistry 52; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar]
- Camara, A.K.S.; Zhou, Y.; Wen, P.C.; Tajkhorshid, E.; Kwok, W.M. Mitochondrial VDAC1: A Key Gatekeeper as Potential Therapeutic Target. Front. Physiol. 2017, 8, 460. [Google Scholar] [CrossRef]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2019, 48, D87–D92. [Google Scholar] [CrossRef]
- Gheorghe, M.; Sandve, G.K.; Khan, A.; Chèneby, J.; Ballester, B.; Mathelier, A. A map of direct TF-DNA interactions in the human genome. Nucleic Acids Res. 2019, 47, 7715. [Google Scholar] [CrossRef]


| Rate of Frequency in Database | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Matrix Family | Description | PVDAC1 | PVDAC2 | PVDAC3 | ||||||
| Genomatix | Jaspar | Unibind | Genomatix | Jaspar | Unibind | Genomatix | Jaspar | Unibind | ||
| V$E2FF | E2F-myc cell cycle regulator | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | - |
| V$EBOX | E-box binding factors | ✓ | ✓ | ✓ | ✓ | - | - | ✓ | ✓ | ✓ |
| V$KLFS | Krueppel like transcription factors | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| V$NRF1 | Nuclear respiratory factor 1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Rate of Frequency in Database | ||||
|---|---|---|---|---|
| Matrix Family | Description | PVDAC1 | ||
| Genomatix | Jaspar | UniBind | ||
| V$AHRR | AHR-arnt heterodimers and AHR-related factors | - | - | ✓ |
| V$ETSF | Human and murine ETS1 factors | ✓ | - | - |
| V$HEAT | Heat shock factors | ✓ | - | - |
| V$PBXC | PBX - MEIS complexes | - | - | ✓ |
| Rate of Frequency in Database | ||||
|---|---|---|---|---|
| Matrix Family | Description | PVDAC2 | ||
| Genomatix | Jaspar | UniBind | ||
| V$BEDF | BED subclass of zinc-finger proteins | ✓ | - | - |
| V$BRAC | Brachyury gene, mesoderm developmental factor | - | - | ✓ |
| V$CLOX | CLOX and CLOX homology (CDP) factors | - | - | ✓ |
| V$MEF3 | MEF3 binding sites | ✓ | - | - |
| V$NEUR | NeuroD, Beta2, HLH domain | ✓ | - | - |
| O$TF2B | RNA polymerase II transcription factor II B | ✓ | - | - |
| V$ZFXY | Zfx and Zfy—transcription factors | - | - | ✓ |
| Rate of Frequency in Database | ||||
|---|---|---|---|---|
| Matrix Family | Description | PVDAC3 | ||
| Genomatix | Jaspar | UniBind | ||
| V$BCL6 | POZ domain zinc finger expressed in B-Cells | - | - | ✓ |
| V$CDXF | Vertebrate caudal related homeodomain protein | - | - | ✓ |
| V$FOX | Forkhead (FKH)/Forkhead box (Fox) | - | - | ✓ |
| V$SOHLH | Spermatogenesis and oogenesis basic helix-loop-helix | - | ✓ | - |
| V$HMG | HMG family | ✓ | - | - |
| V$HOMF | Homeodomain transcription factors | ✓ | ✓ | - |
| V$IRFF | Interferon regulatory factors | - | - | ✓ |
| V$LBXF | Ladybird homeobox (lbx) gene family | - | ✓ | - |
| V$MYBL | Cellular and viral myb-like transcriptional regulators | - | ✓ | - |
| V$SMAD | Vertebrate SMAD family of transcription factors | - | ✓ | - |
| V$XBBF | X-box binding factors | ✓ | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zinghirino, F.; Pappalardo, X.G.; Messina, A.; Guarino, F.; De Pinto, V. Is the Secret of VDAC Isoforms in Their Gene Regulation? Characterization of Human VDAC Genes Expression Profile, Promoter Activity, and Transcriptional Regulators. Int. J. Mol. Sci. 2020, 21, 7388. https://doi.org/10.3390/ijms21197388
Zinghirino F, Pappalardo XG, Messina A, Guarino F, De Pinto V. Is the Secret of VDAC Isoforms in Their Gene Regulation? Characterization of Human VDAC Genes Expression Profile, Promoter Activity, and Transcriptional Regulators. International Journal of Molecular Sciences. 2020; 21(19):7388. https://doi.org/10.3390/ijms21197388
Chicago/Turabian StyleZinghirino, Federica, Xena Giada Pappalardo, Angela Messina, Francesca Guarino, and Vito De Pinto. 2020. "Is the Secret of VDAC Isoforms in Their Gene Regulation? Characterization of Human VDAC Genes Expression Profile, Promoter Activity, and Transcriptional Regulators" International Journal of Molecular Sciences 21, no. 19: 7388. https://doi.org/10.3390/ijms21197388
APA StyleZinghirino, F., Pappalardo, X. G., Messina, A., Guarino, F., & De Pinto, V. (2020). Is the Secret of VDAC Isoforms in Their Gene Regulation? Characterization of Human VDAC Genes Expression Profile, Promoter Activity, and Transcriptional Regulators. International Journal of Molecular Sciences, 21(19), 7388. https://doi.org/10.3390/ijms21197388








