The Role of microRNAs in Epithelial Ovarian Cancer Metastasis
Abstract
1. Introduction
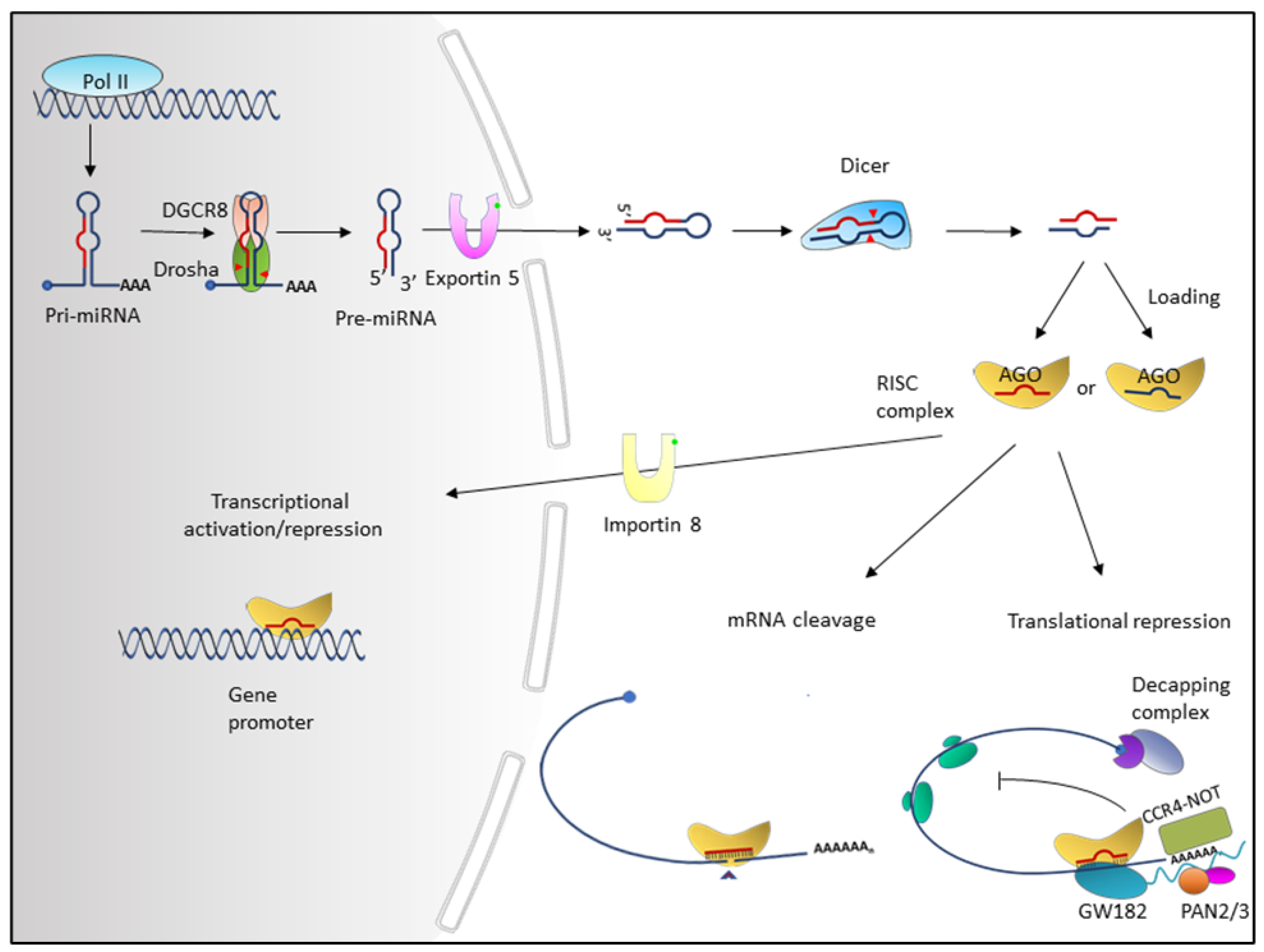
2. Overview of miRNAs
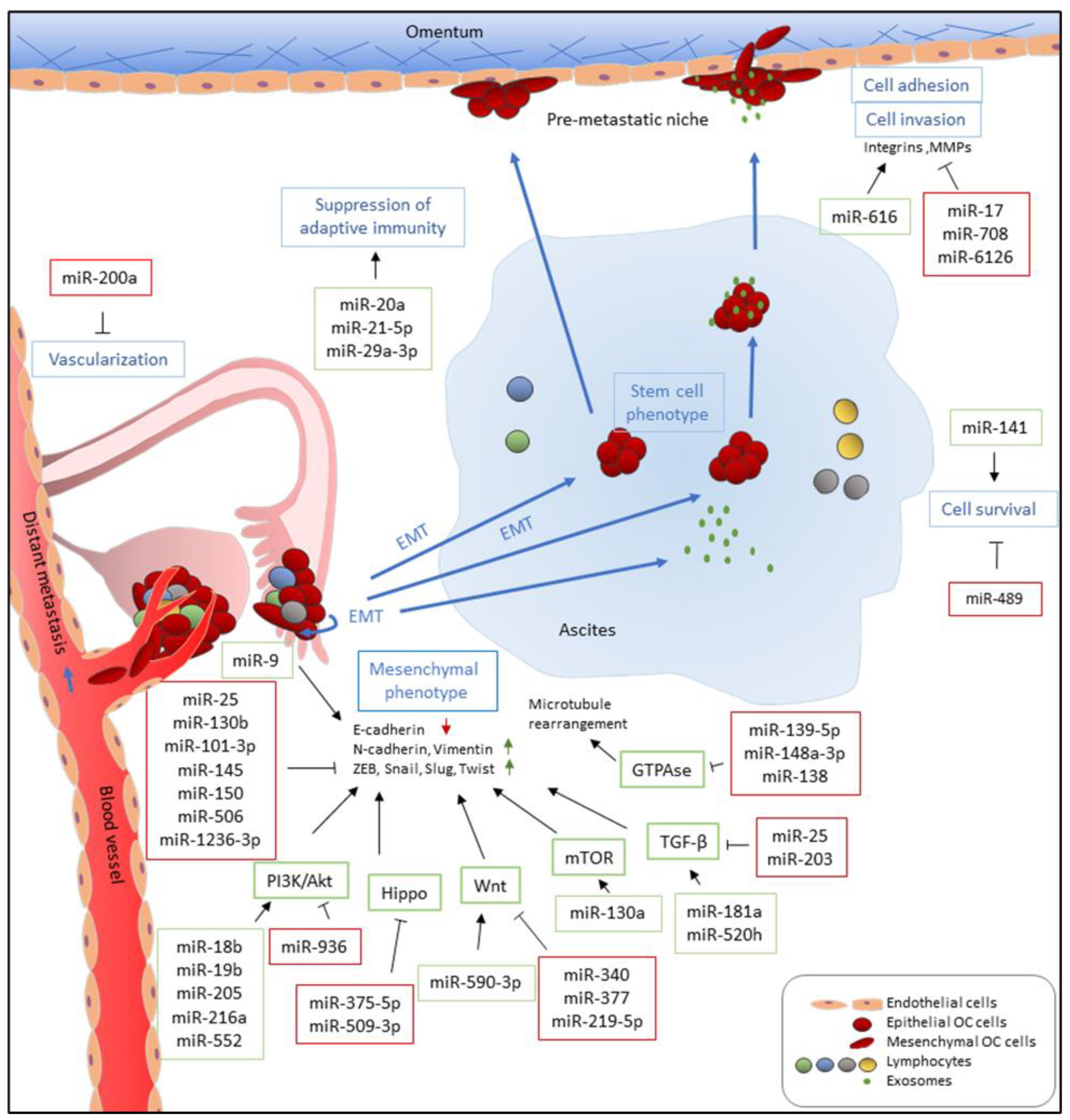
3. Ovarian Cancer Metastasis
4. Dysregulation of miRNA Expression in Ovarian Cancer
4.1. Aberrant Expression of Metastasis-Associated miRNAs in EOC
4.2. Dysregulations of miRNAs by Genetic and Epigenetic Alterations
4.3. Dysregulation of miRNAs by Alteration in miRNA Biogenesis
5. Roles of miRNAs in Ovarian Cancer Metastasis
5.1. MicroRNAs that Promote Metastasis
5.2. MicroRNAs that Suppress Metastasis
5.2.1. MicroRNAs Suppress Metastasis by Directly Targeting Transcription Factors that Regulate EMT Markers
5.2.2. MicroRNAs Suppress Metastasis by Targeting Growth Factors and Related Signaling Pathways
5.2.3. MicroRNAs Suppress Metastasis by Regulating Adhesion Molecules
5.2.4. MicroRNAs Suppress Metastasis by Directly Targeting HOX Genes
5.2.5. MicroRNAs Suppress Metastasis by Directly Targeting HIF
5.3. miRNAs that Have Been Reported to Promote and Inhibit Metastasis
6. Conclusions and Future Direction
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Boussios, S.; Moschetta, M.; Zarkavelis, G.; Papadaki, A.; Kefas, A.; Tatsi, K. Ovarian sex-cord stromal tumours and small cell tumours: Pathological, genetic and management aspects. Crit. Rev. Oncol. Hematol. 2017, 120, 43–51. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences, Engineering, and Medicine. Ovarian Cancers: Evolving Paradigms in Research and Care; The National Academies Press: Washington, DC, USA, 2016; p. 396. [Google Scholar] [CrossRef]
- Lupia, M.; Cavallaro, U. Ovarian cancer stem cells: Still an elusive entity? Mol. Cancer 2017, 16, 64. [Google Scholar] [CrossRef]
- Webb, P.M.; Jordan, S.J. Epidemiology of epithelial ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 3–14. [Google Scholar] [CrossRef]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Primers 2016, 2, 16061. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Ambros, V. microRNAs: Tiny regulators with great potential. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef]
- Staicu, C.E.; Predescu, D.V.; Rusu, C.M.; Radu, B.M.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. Role of microRNAs as Clinical Cancer Biomarkers for Ovarian Cancer: A Short Overview. Cells 2020, 9, 169. [Google Scholar] [CrossRef]
- Chen, S.N.; Chang, R.; Lin, L.T.; Chern, C.U.; Tsai, H.W.; Wen, Z.H.; Li, Y.H.; Li, C.J.; Tsui, K.H. MicroRNA in Ovarian Cancer: Biology, Pathogenesis, and Therapeutic Opportunities. Int. J. Environ. Res. Public Health 2019, 16, 1510. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.M.; Cardenas, C.; Tedja, R. The Role of Intra-Tumoral Heterogeneity and Its Clinical Relevance in Epithelial Ovarian Cancer Recurrence and Metastasis. Cancers 2019, 11, 1083. [Google Scholar] [CrossRef]
- Godnic, I.; Zorc, M.; Jevsinek Skok, D.; Calin, G.A.; Horvat, S.; Dovc, P.; Kovac, M.; Kunej, T. Genome-wide and species-wide in silico screening for intragenic MicroRNAs in human, mouse and chicken. PLoS ONE 2013, 8, e65165. [Google Scholar] [CrossRef] [PubMed]
- de Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Astrom, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef]
- Ramalingam, P.; Palanichamy, J.K.; Singh, A.; Das, P.; Bhagat, M.; Kassab, M.A.; Sinha, S.; Chattopadhyay, P. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA 2014, 20, 76–87. [Google Scholar] [CrossRef]
- Kirigin, F.F.; Lindstedt, K.; Sellars, M.; Ciofani, M.; Low, S.L.; Jones, L.; Bell, F.; Pauli, F.; Bonneau, R.; Myers, R.M.; et al. Dynamic microRNA gene transcription and processing during T cell development. J. Immunol. 2012, 188, 3257–3267. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881–E1889. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Triboulet, R.; Chang, H.M.; Lapierre, R.J.; Gregory, R.I. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 2009, 15, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Dahlberg, J.E. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Knight, S.W.; Bass, B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2001, 293, 2269–2271. [Google Scholar] [CrossRef]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, T.; Tomari, Y. Making RISC. Trends Biochem. Sci. 2010, 35, 368–376. [Google Scholar] [CrossRef]
- Su, H.; Trombly, M.I.; Chen, J.; Wang, X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009, 23, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Kadekar, S.; Nawale, G.N.; Karlsson, K.; Alander, C.; Oommen, O.P.; Varghese, O.P. Synthetic Design of Asymmetric miRNA with an Engineered 3′ Overhang to Improve Strand Selection. Mol. Ther. Nucleic Acids 2019, 16, 597–604. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004, 15, 185–197. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef]
- Pfaff, J.; Meister, G. Argonaute and GW182 proteins: An effective alliance in gene silencing. Biochem. Soc. Trans. 2013, 41, 855–860. [Google Scholar] [CrossRef]
- Christie, M.; Boland, A.; Huntzinger, E.; Weichenrieder, O.; Izaurralde, E. Structure of the PAN3 pseudokinase reveals the basis for interactions with the PAN2 deadenylase and the GW182 proteins. Mol. Cell 2013, 51, 360–373. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.E.; Truffault, V.; Boland, A.; Huntzinger, E.; Chang, C.T.; Haas, G.; Weichenrieder, O.; Coles, M.; Izaurralde, E. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5’ exonucleolytic degradation. Nat. Struct. Mol. Biol. 2012, 19, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; San Lucas, A.; Wang, Z.; Liu, Y. Identifying microRNA targets in different gene regions. BMC Bioinform. 2014, 15 (Suppl. 7), S4. [Google Scholar] [CrossRef] [PubMed]
- Dharap, A.; Pokrzywa, C.; Murali, S.; Pandi, G.; Vemuganti, R. MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PLoS ONE 2013, 8, e79467. [Google Scholar] [CrossRef]
- Forman, J.J.; Legesse-Miller, A.; Coller, H.A. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. USA 2008, 105, 14879–14884. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, W.; Liu, Y.; Liu, T.; Li, C.; Wang, L. Oncogenic role of microRNA-532-5p in human colorectal cancer via targeting of the 5’UTR of RUNX3. Oncol. Lett. 2018, 15, 7215–7220. [Google Scholar] [CrossRef]
- Truesdell, S.S.; Mortensen, R.D.; Seo, M.; Schroeder, J.C.; Lee, J.H.; LeTonqueze, O.; Vasudevan, S. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci. Rep. 2012, 2, 842. [Google Scholar] [CrossRef]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Tan, D.S.; Agarwal, R.; Kaye, S.B. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol. 2006, 7, 925–934. [Google Scholar] [CrossRef]
- Yeung, T.L.; Leung, C.S.; Yip, K.P.; Au Yeung, C.L.; Wong, S.T.; Mok, S.C. Cellular and molecular processes in ovarian cancer metastasis. A Review in the Theme: Cell and Molecular Processes in Cancer Metastasis. Am. J. Physiol. Cell Physiol. 2015, 309, C444–C456. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, E. Ovarian cancer development and metastasis. Am. J. Pathol. 2010, 177, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Micalizzi, D.S.; Farabaugh, S.M.; Ford, H.L. Epithelial-mesenchymal transition in cancer: Parallels between normal development and tumor progression. J. Mammary Gland Biol. Neoplasia 2010, 15, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef]
- Mani, S.A.; Yang, J.; Brooks, M.; Schwaninger, G.; Zhou, A.; Miura, N.; Kutok, J.L.; Hartwell, K.; Richardson, A.L.; Weinberg, R.A. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc. Natl. Acad. Sci. USA 2007, 104, 10069–10074. [Google Scholar] [CrossRef]
- Davidson, B.; Trope, C.G.; Reich, R. Epithelial-mesenchymal transition in ovarian carcinoma. Front. Oncol. 2012, 2, 33. [Google Scholar] [CrossRef]
- Palma Flores, C.; Garcia-Vazquez, R.; Gallardo Rincon, D.; Ruiz-Garcia, E.; Astudillo de la Vega, H.; Marchat, L.A.; Salinas Vera, Y.M.; Lopez-Camarillo, C. MicroRNAs driving invasion and metastasis in ovarian cancer: Opportunities for translational medicine (Review). Int. J. Oncol. 2017, 50, 1461–1476. [Google Scholar] [CrossRef]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef]
- Yang, J.; Weinberg, R.A. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef]
- Tsai, J.H.; Yang, J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013, 27, 2192–2206. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hu, L.; Zheng, H.; Bagnoli, M.; Guo, Y.; Rupaimoole, R.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Ji, P.; Chen, K.; et al. MiR-506 inhibits multiple targets in the epithelial-to-mesenchymal transition network and is associated with good prognosis in epithelial ovarian cancer. J. Pathol. 2015, 235, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.H.L.; Hough, R.; Bernaudo, S.; Peng, C. Wnt/beta-catenin signalling in ovarian cancer: Insights into its hyperactivation and function in tumorigenesis. J. Ovarian Res. 2019, 12, 122. [Google Scholar] [CrossRef]
- Rafehi, S.; Ramos Valdes, Y.; Bertrand, M.; McGee, J.; Prefontaine, M.; Sugimoto, A.; DiMattia, G.E.; Shepherd, T.G. TGFbeta signaling regulates epithelial-mesenchymal plasticity in ovarian cancer ascites-derived spheroids. Endocr. Relat. Cancer 2016, 23, 147–159. [Google Scholar] [CrossRef]
- Zhou, X.; Hu, Y.; Dai, L.; Wang, Y.; Zhou, J.; Wang, W.; Di, W.; Qiu, L. MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2 inactivation by targeting EGFR in epithelial ovarian cancer. PLoS ONE 2014, 9, e96718. [Google Scholar] [CrossRef]
- Moran-Jones, K. The Therapeutic Potential of Targeting the HGF/cMET Axis in Ovarian Cancer. Mol. Diagn. Ther. 2016, 20, 199–212. [Google Scholar] [CrossRef]
- Zhang, R.; Shi, H.; Ren, F.; Feng, W.; Cao, Y.; Li, G.; Liu, Z.; Ji, P.; Zhang, M. MicroRNA-338-3p suppresses ovarian cancer cells growth and metastasis: Implication of Wnt/catenin beta and MEK/ERK signaling pathways. J. Exp. Clin. Cancer Res. CR 2019, 38, 494. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, M.; Jin, K.; Wang, S.; Wei, H.; Fan, C.; Wu, Y.; Li, X.; Li, X.; Li, G.; et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer 2018, 17, 45. [Google Scholar] [CrossRef]
- Ke, Z.; Caiping, S.; Qing, Z.; Xiaojing, W. Sonic hedgehog-Gli1 signals promote epithelial-mesenchymal transition in ovarian cancer by mediating PI3K/AKT pathway. Med. Oncol. (Northwood Lond. Engl.) 2015, 32, 368. [Google Scholar] [CrossRef] [PubMed]
- Do, T.V.; Xiao, F.; Bickel, L.E.; Klein-Szanto, A.J.; Pathak, H.B.; Hua, X.; Howe, C.; O’Brien, S.W.; Maglaty, M.; Ecsedy, J.A.; et al. Aurora kinase A mediates epithelial ovarian cancer cell migration and adhesion. Oncogene 2014, 33, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Boussios, S.; Mikropoulos, C.; Samartzis, E.; Karihtala, P.; Moschetta, M.; Sheriff, M.; Karathanasi, A.; Sadauskaite, A.; Rassy, E.; Pavlidis, N. Wise Management of Ovarian Cancer: On the Cutting Edge. J. Pers. Med. 2020, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar] [CrossRef]
- Cance, W.G.; Golubovskaya, V.M. Focal adhesion kinase versus p53: Apoptosis or survival? Sci. Signal. 2008, 1, pe22. [Google Scholar] [CrossRef]
- Frisch, S.M.; Schaller, M.; Cieply, B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J. Cell Sci. 2013, 126, 21–29. [Google Scholar] [CrossRef]
- Shibue, T.; Brooks, M.W.; Inan, M.F.; Reinhardt, F.; Weinberg, R.A. The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer Discov. 2012, 2, 706–721. [Google Scholar] [CrossRef]
- Sood, A.K.; Coffin, J.E.; Schneider, G.B.; Fletcher, M.S.; DeYoung, B.R.; Gruman, L.M.; Gershenson, D.M.; Schaller, M.D.; Hendrix, M.J. Biological significance of focal adhesion kinase in ovarian cancer: Role in migration and invasion. Am. J. Pathol. 2004, 165, 1087–1095. [Google Scholar] [CrossRef]
- Zhao, J.; Guan, J.L. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009, 28, 35–49. [Google Scholar] [CrossRef]
- Lin, K.T.; Yeh, Y.M.; Chuang, C.M.; Yang, S.Y.; Chang, J.W.; Sun, S.P.; Wang, Y.S.; Chao, K.C.; Wang, L.H. Glucocorticoids mediate induction of microRNA-708 to suppress ovarian cancer metastasis through targeting Rap1B. Nat. Commun. 2015, 6, 5917. [Google Scholar] [CrossRef]
- Mei, J.; Huang, Y.; Hao, L.; Liu, Y.; Yan, T.; Qiu, T.; Xu, R.; Xu, B.; Xiao, Z.; Jiang, X.; et al. DAAM1-mediated migration and invasion of ovarian cancer cells are suppressed by miR-208a-5p. Pathol. Res. Pract. 2019, 215, 152452. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zeng, M.; Zhao, Y.; Fang, X. Upregulation of Limk1 caused by microRNA-138 loss aggravates the metastasis of ovarian cancer by activation of Limk1/cofilin signaling. Oncol. Rep. 2014, 32, 2070–2076. [Google Scholar] [CrossRef] [PubMed]
- Prunier, C.; Prudent, R.; Kapur, R.; Sadoul, K.; Lafanechere, L. LIM kinases: Cofilin and beyond. Oncotarget 2017, 8, 41749–41763. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Mezencev, R.; Svajdler, M.; Benigno, B.B.; McDonald, J.F. Ectopic over-expression of miR-429 induces mesenchymal-to-epithelial transition (MET) and increased drug sensitivity in metastasizing ovarian cancer cells. Gynecol. Oncol. 2014, 134, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Dong, Z.; Chen, Y.; Yang, L.; Lai, D. Enrichment of ovarian cancer stem-like cells is associated with epithelial to mesenchymal transition through an miRNA-activated AKT pathway. Cell Prolif. 2013, 46, 436–446. [Google Scholar] [CrossRef]
- Persad, S.; Dedhar, S. The role of integrin-linked kinase (ILK) in cancer progression. Cancer Metastasis Rev. 2003, 22, 375–384. [Google Scholar] [CrossRef]
- Scheau, C.; Badarau, I.A.; Costache, R.; Caruntu, C.; Mihai, G.L.; Didilescu, A.C.; Constantin, C.; Neagu, M. The Role of Matrix Metalloproteinases in the Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma. Anal. Cell Pathol. (Amst.) 2019, 2019, 9423907. [Google Scholar] [CrossRef]
- Jin, H.; Yu, Y.; Zhang, T.; Zhou, X.; Zhou, J.; Jia, L.; Wu, Y.; Zhou, B.P.; Feng, Y. Snail is critical for tumor growth and metastasis of ovarian carcinoma. Int. J. Cancer 2010, 126, 2102–2111. [Google Scholar] [CrossRef]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta 2013, 1833, 3481–3498. [Google Scholar] [CrossRef]
- Braga, E.A.; Fridman, M.V.; Kushlinskii, N.E. Molecular Mechanisms of Ovarian Carcinoma Metastasis: Key Genes and Regulatory MicroRNAs. Biochemistry 2017, 82, 529–541. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol. Sci. 2012, 33, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Keith, B.; Johnson, R.S.; Simon, M.C. HIF1alpha and HIF2alpha: Sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 2011, 12, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Joshi, H.P.; Subramanian, I.V.; Schnettler, E.K.; Ghosh, G.; Rupaimoole, R.; Evans, C.; Saluja, M.; Jing, Y.; Cristina, I.; Roy, S.; et al. Dynamin 2 along with microRNA-199a reciprocally regulate hypoxia-inducible factors and ovarian cancer metastasis. Proc. Natl. Acad. Sci. USA 2014, 111, 5331–5336. [Google Scholar] [CrossRef] [PubMed]
- Schietke, R.; Warnecke, C.; Wacker, I.; Schodel, J.; Mole, D.R.; Campean, V.; Amann, K.; Goppelt-Struebe, M.; Behrens, J.; Eckardt, K.U.; et al. The lysyl oxidases LOX and LOXL2 are necessary and sufficient to repress E-cadherin in hypoxia: Insights into cellular transformation processes mediated by HIF-1. J. Biol. Chem. 2010, 285, 6658–6669. [Google Scholar] [CrossRef]
- Bindra, R.S.; Gibson, S.L.; Meng, A.; Westermark, U.; Jasin, M.; Pierce, A.J.; Bristow, R.G.; Classon, M.K.; Glazer, P.M. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005, 65, 11597–11604. [Google Scholar] [CrossRef]
- Boussios, S.; Karihtala, P.; Moschetta, M.; Karathanasi, A.; Sadauskaite, A.; Rassy, E.; Pavlidis, N. Combined Strategies with Poly (ADP-Ribose) Polymerase (PARP) Inhibitors for the Treatment of Ovarian Cancer: A Literature Review. Diagnostics 2019, 9, 87. [Google Scholar] [CrossRef]
- Balkwill, F. Cancer and the chemokine network. Nat. Rev. Cancer 2004, 4, 540–550. [Google Scholar] [CrossRef]
- Lv, Y.; Lei, Y.; Hu, Y.; Ding, W.; Zhang, C.; Fang, C. miR-448 negatively regulates ovarian cancer cell growth and metastasis by targeting CXCL12. Clin. Transl. Oncol. 2015, 17, 903–909. [Google Scholar] [CrossRef]
- Casey, R.C.; Skubitz, A.P. CD44 and beta1 integrins mediate ovarian carcinoma cell migration toward extracellular matrix proteins. Clin. Exp. Metastasis 2000, 18, 67–75. [Google Scholar] [CrossRef]
- Santoiemma, P.P.; Powell, D.J., Jr. Tumor infiltrating lymphocytes in ovarian cancer. Cancer Biol. Ther. 2015, 16, 807–820. [Google Scholar] [CrossRef]
- Zhou, J.; Li, X.; Wu, X.; Zhang, T.; Zhu, Q.; Wang, X.; Wang, H.; Wang, K.; Lin, Y.; Wang, X. Exosomes Released from Tumor-Associated Macrophages Transfer miRNAs That Induce a Treg/Th17 Cell Imbalance in Epithelial Ovarian Cancer. Cancer Immunol. Res. 2018, 6, 1578–1592. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, Q.; Lin, Y.; Wu, L.; Wu, X.; Wang, K.; He, Q.; Xu, C.; Wan, X.; Wang, X. Crosstalk between TEMs and endothelial cells modulates angiogenesis and metastasis via IGF1-IGF1R signalling in epithelial ovarian cancer. Br. J. Cancer 2017, 117, 1371–1382. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends. Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Jiang, B.; Zhu, S.J.; Xiao, S.S.; Xue, M. MiR-217 Inhibits M2-Like Macrophage Polarization by Suppressing Secretion of Interleukin-6 in Ovarian Cancer. Inflammation 2019, 42, 1517–1529. [Google Scholar] [CrossRef]
- Xie, J.; Liu, M.; Li, Y.; Nie, Y.; Mi, Q.; Zhao, S. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell Mol. Immunol. 2014, 11, 495–502. [Google Scholar] [CrossRef]
- Ghadially, H.; Brown, L.; Lloyd, C.; Lewis, L.; Lewis, A.; Dillon, J.; Sainson, R.; Jovanovic, J.; Tigue, N.J.; Bannister, D.; et al. MHC class I chain-related protein A and B (MICA and MICB) are predominantly expressed intracellularly in tumour and normal tissue. Br. J. Cancer 2017, 116, 1208–1217. [Google Scholar] [CrossRef]
- Nam, J.W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell 2014, 53, 1031–1043. [Google Scholar] [CrossRef]
- He, M.; Liu, Y.; Wang, X.; Zhang, M.Q.; Hannon, G.J.; Huang, Z.J. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 2012, 73, 35–48. [Google Scholar] [CrossRef]
- Parikh, A.; Lee, C.; Joseph, P.; Marchini, S.; Baccarini, A.; Kolev, V.; Romualdi, C.; Fruscio, R.; Shah, H.; Wang, F.; et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nat. Commun. 2014, 5, 2977. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, J.; Zhu, Y.; Wang, J. MicroRNA-616 promotes the progression of ovarian cancer by targeting TIMP2. Oncol. Rep. 2018, 39, 2960–2968. [Google Scholar] [CrossRef]
- Salem, M.; O’Brien, J.A.; Bernaudo, S.; Shawer, H.; Ye, G.; Brkic, J.; Amleh, A.; Vanderhyden, B.C.; Refky, B.; Yang, B.B.; et al. miR-590-3p Promotes Ovarian Cancer Growth and Metastasis via a Novel FOXA2-Versican Pathway. Cancer Res. 2018, 78, 4175–4190. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Wei, K.; Lin, Z.; Cui, Y.; Ding, J.; Chen, Y.; Xu, B. MicroRNA-125b Suppresses Ovarian Cancer Progression via Suppression of the Epithelial-Mesenchymal Transition Pathway by Targeting the SET Protein. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2016, 39, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Meng, Q.; Li, H.; Liang, K.; Li, B. LINC00339 promotes cell proliferation, migration, and invasion of ovarian cancer cells via miR-148a-3p/ROCK1 axes. Biomed. Pharmacother. 2019, 120, 109423. [Google Scholar] [CrossRef]
- Yan, H.; Li, H.; Li, P.; Li, X.; Lin, J.; Zhu, L.; Silva, M.A.; Wang, X.; Wang, P.; Zhang, Z. Long noncoding RNA MLK7-AS1 promotes ovarian cancer cells progression by modulating miR-375/YAP1 axis. J. Exp. Clin. Cancer Res. CR 2018, 37, 237. [Google Scholar] [CrossRef] [PubMed]
- Loginov, V.I.; Pronina, I.V.; Burdennyy, A.M.; Filippova, E.A.; Kazubskaya, T.P.; Kushlinsky, D.N.; Utkin, D.O.; Khodyrev, D.S.; Kushlinskii, N.E.; Dmitriev, A.A.; et al. Novel miRNA genes deregulated by aberrant methylation in ovarian carcinoma are involved in metastasis. Gene 2018, 662, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Q.; Zhao, Q.; Di, W. MiR-124 inhibits the migration and invasion of ovarian cancer cells by targeting SphK1. J. Ovarian Res. 2013, 6, 84. [Google Scholar] [CrossRef]
- Xiaohong, Z.; Lichun, F.; Na, X.; Kejian, Z.; Xiaolan, X.; Shaosheng, W. MiR-203 promotes the growth and migration of ovarian cancer cells by enhancing glycolytic pathway. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 14989–14997. [Google Scholar] [CrossRef]
- Zhao, G.; Guo, Y.; Chen, Z.; Wang, Y.; Yang, C.; Dudas, A.; Du, Z.; Liu, W.; Zou, Y.; Szabo, E.; et al. miR-203 Functions as a Tumor Suppressor by Inhibiting Epithelial to Mesenchymal Transition in Ovarian Cancer. J. Cancer Sci. Ther. 2015, 7, 34–43. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, G.; Wang, G.; Zhao, J.; Wang, B.; Yu, X.; Ding, Y. Profile of differentially expressed miRNAs in high-grade serous carcinoma and clear cell ovarian carcinoma, and the expression of miR-510 in ovarian carcinoma. Mol. Med. Rep. 2015, 12, 8021–8031. [Google Scholar] [CrossRef]
- Ye, G.; Bernaudo, S.; Peng, C. Etiology and overview. In Advances in Ovarian Cancer Management; Future Medicine Ltd.: London, UK, 2012. [Google Scholar] [CrossRef]
- Braicu, O.L.; Budisan, L.; Buiga, R.; Jurj, A.; Achimas-Cadariu, P.; Pop, L.A.; Braicu, C.; Irimie, A.; Berindan-Neagoe, I. miRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples. Onco Targets Ther. 2017, 10, 4225–4238. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. Exosomes promote pre-metastatic niche formation in ovarian cancer. Mol. Cancer 2019, 18, 124. [Google Scholar] [CrossRef]
- Yang, L.; Wei, Q.M.; Zhang, X.W.; Sheng, Q.; Yan, X.T. MiR-376a promotion of proliferation and metastases in ovarian cancer: Potential role as a biomarker. Life Sci. 2017, 173, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Zuberi, M.; Mir, R.; Das, J.; Ahmad, I.; Javid, J.; Yadav, P.; Masroor, M.; Ahmad, S.; Ray, P.C.; Saxena, A. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin. Transl. Oncol. 2015, 17, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Langhe, R.; Norris, L.; Saadeh, F.A.; Blackshields, G.; Varley, R.; Harrison, A.; Gleeson, N.; Spillane, C.; Martin, C.; O’Donnell, D.M.; et al. A novel serum microRNA panel to discriminate benign from malignant ovarian disease. Cancer Lett. 2015, 356, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Zuberi, M.; Khan, I.; Gandhi, G.; Ray, P.C.; Saxena, A. The conglomeration of diagnostic, prognostic and therapeutic potential of serum miR-199a and its association with clinicopathological features in epithelial ovarian cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 11259–11266. [Google Scholar] [CrossRef]
- Li, Y.; Liu, C.; Liao, Y.; Wang, W.; Hu, B.; Lu, X.; Cui, J. Characterizing the landscape of peritoneal exosomal microRNAs in patients with ovarian cancer by high-throughput sequencing. Oncol. Lett. 2019, 17, 539–547. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, W.; Shen, F.; Ma, X.; Liu, X.; Tian, F.; Zeng, W.; Xi, X.; Lin, Y. The activation of microRNA-520h-associated TGF-beta1/c-Myb/Smad7 axis promotes epithelial ovarian cancer progression. Cell Death Dis. 2018, 9, 884. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, L.; Huang, H.; Huang, Y.; Huang, L.; Wang, J.; Huang, S.; He, L.; Zhou, Y.; Jia, W.; et al. MiR-26b/KPNA2 axis inhibits epithelial ovarian carcinoma proliferation and metastasis through downregulating OCT4. Oncotarget 2015, 6, 23793–23806. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, W.; Ding, Y.; Li, X.; Song, J. Expression of miR-26b in ovarian carcinoma tissues and its correlation with clinicopathology. Oncol. Lett. 2019, 17, 4417–4422. [Google Scholar] [CrossRef]
- Vang, S.; Wu, H.T.; Fischer, A.; Miller, D.H.; MacLaughlan, S.; Douglass, E.; Comisar, L.; Steinhoff, M.; Collins, C.; Smith, P.J.; et al. Identification of ovarian cancer metastatic miRNAs. PLoS ONE 2013, 8, e58226. [Google Scholar] [CrossRef] [PubMed]
- Wahab, N.A.; Othman, Z.; Nasri, N.W.M.; Mokhtar, M.H.; Ibrahim, S.F.; Hamid, A.A.; Raja Ali, R.A.; Mokhtar, N.M. Inhibition of miR-141 and miR-200a Increase DLC-1 and ZEB2 Expression, Enhance Migration and Invasion in Metastatic Serous Ovarian Cancer. Int. J. Environ. Res. Public Health 2020, 17, 2766. [Google Scholar] [CrossRef] [PubMed]
- McMillen, B.D.; Aponte, M.M.; Liu, Z.; Helenowski, I.B.; Scholtens, D.M.; Buttin, B.M.; Wei, J.J. Expression analysis of MIR182 and its associated target genes in advanced ovarian carcinoma. Mod. Pathol. 2012, 25, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, J.; Segura, M.F.; Shao, C.; Lee, P.; Gong, Y.; Hernando, E.; Wei, J.J. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J. Pathol. 2012, 228, 204–215. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Banerjee, A.; Cui, T.; Han, C.; Cai, S.; Liu, L.; Wu, D.; Cui, R.; Li, Z.; Zhang, X.; et al. Inhibition of miR-328-3p Impairs Cancer Stem Cell Function and Prevents Metastasis in Ovarian Cancer. Cancer Res. 2019, 79, 2314–2326. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Tang, W.; Lin, Z.; Xu, L.; Dong, R.; Li, Y.; Li, J.; Zhang, Z.; Li, X.; et al. miR-130a upregulates mTOR pathway by targeting TSC1 and is transactivated by NF-kappaB in high-grade serous ovarian carcinoma. Cell Death Differ. 2017, 24, 2089–2100. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, G.; Lv, S.; Wen, X.; Liu, P. miRNA-301b-3p accelerates migration and invasion of high-grade ovarian serous tumor via targeting CPEB3/EGFR axis. J. Cell Biochem. 2019, 120, 12618–12627. [Google Scholar] [CrossRef]
- Lou, Y.; Yang, X.; Wang, F.; Cui, Z.; Huang, Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int. J. Mol. Med. 2010, 26, 819–827. [Google Scholar] [CrossRef]
- Su, N.; Qiu, H.; Chen, Y.; Yang, T.; Yan, Q.; Wan, X. miR-205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol. Rep. 2013, 29, 2297–2302. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, Y.; Wang, Y.; Zhang, Y.; Luo, Q.; Man, X.; Wei, F.; Yu, X. Downregulation of Foxo3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med. Oncol. (Northwood Lond. Engl.) 2016, 33, 126. [Google Scholar] [CrossRef]
- Chaluvally-Raghavan, P.; Jeong, K.J.; Pradeep, S.; Silva, A.M.; Yu, S.; Liu, W.; Moss, T.; Rodriguez-Aguayo, C.; Zhang, D.; Ram, P.; et al. Direct Upregulation of STAT3 by MicroRNA-551b-3p Deregulates Growth and Metastasis of Ovarian Cancer. Cell Rep. 2016, 15, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, Y.; Wang, D.; Fu, X.; Li, M.; Wang, A. Upregulation of microRNA-18b induces phosphatase and tensin homolog to accelerate the migration and invasion abilities of ovarian cancer. Oncol. Lett. 2017, 14, 5631–5637. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.T.; Yao, H.R.; Li, Y.Y.; Song, Y.Y.; Su, M.Y. MicroRNA-19b promotes the migration and invasion of ovarian cancer cells by inhibiting the PTEN/AKT signaling pathway. Oncol. Lett. 2018, 16, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, R.J.; Bai, X.X.; Liu, W. MicroRNA-23a depletion promotes apoptosis of ovarian cancer stem cell and inhibits cell migration by targeting DLG2. Cancer Biol. Ther. 2019, 20, 897–911. [Google Scholar] [CrossRef]
- Xia, Y.; Gao, Y. MicroRNA-181b promotes ovarian cancer cell growth and invasion by targeting LATS2. Biochem. Biophys. Res. Commun. 2014, 447, 446–451. [Google Scholar] [CrossRef]
- Wang, A.; Jin, C.; Li, H.; Qin, Q.; Li, L. LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway. Int. J. Biol. Macromol. 2018, 120, 1705–1713. [Google Scholar] [CrossRef]
- Liang, T.; Li, L.; Cheng, Y.; Ren, C.; Zhang, G. MicroRNA-194 promotes the growth, migration, and invasion of ovarian carcinoma cells by targeting protein tyrosine phosphatase nonreceptor type 12. Onco. Targets Ther. 2016, 9, 4307–4315. [Google Scholar] [CrossRef]
- Fan, Y.; Fan, J.; Huang, L.; Ye, M.; Huang, Z.; Wang, Y.; Li, Q.; Huang, J. Increased expression of microRNA-196a predicts poor prognosis in human ovarian carcinoma. Int. J. Clin Exp. Pathol. 2015, 8, 4132–4137. [Google Scholar]
- Wei, J.; Zhang, L.; Li, J.; Zhu, S.; Tai, M.; Mason, C.W.; Chapman, J.A.; Reynolds, E.A.; Weiner, C.P.; Zhou, H.H. MicroRNA-205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J. Ovarian Res. 2017, 10, 33. [Google Scholar] [CrossRef]
- Chu, P.; Liang, A.; Jiang, A.; Zong, L. miR-205 regulates the proliferation and invasion of ovarian cancer cells via suppressing PTEN/SMAD4 expression. Oncol. Lett. 2018, 15, 7571–7578. [Google Scholar] [CrossRef]
- Li, J.; Hu, K.; Gong, G.; Zhu, D.; Wang, Y.; Liu, H.; Wu, X. Upregulation of MiR-205 transcriptionally suppresses SMAD4 and PTEN and contributes to human ovarian cancer progression. Sci. Rep. 2017, 7, 41330. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Pan, Y.; Han, X.; Liu, J.; Li, R. MicroRNA-216a promotes the metastasis and epithelial-mesenchymal transition of ovarian cancer by suppressing the PTEN/AKT pathway. Onco. Targets Ther. 2017, 10, 2701–2709. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Han, T.; Li, B.; Ma, Q.; Yang, P.; Li, H. miR-552 promotes ovarian cancer progression by regulating PTEN pathway. J. Ovarian Res. 2019, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Ivan, C.; Yang, D.; Gharpure, K.M.; Wu, S.Y.; Pecot, C.V.; Previs, R.A.; Nagaraja, A.S.; Armaiz-Pena, G.N.; McGuire, M.; et al. Hypoxia-upregulated microRNA-630 targets Dicer, leading to increased tumor progression. Oncogene 2016, 35, 4312–4320. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Li-ya, Q.; Feng, Z.; Yin, W.; Ji-hong, L. MiR-939 promotes the proliferation of human ovarian cancer cells by repressing APC2 expression. Biomed. Pharmacother. 2015, 71, 64–69. [Google Scholar] [CrossRef]
- Tang, M.; Jiang, L.; Lin, Y.; Wu, X.; Wang, K.; He, Q.; Wang, X.; Li, W. Platelet microparticle-mediated transfer of miR-939 to epithelial ovarian cancer cells promotes epithelial to mesenchymal transition. Oncotarget 2017, 8, 97464–97475. [Google Scholar] [CrossRef]
- Salem, M.; Shan, Y.; Bernaudo, S.; Peng, C. miR-590-3p Targets Cyclin G2 and FOXO3 to Promote Ovarian Cancer Cell Proliferation, Invasion, and Spheroid Formation. Int. J. Mol. Sci. 2019, 20, 1810. [Google Scholar] [CrossRef]
- Zhou, J.; Gong, G.; Tan, H.; Dai, F.; Zhu, X.; Chen, Y.; Wang, J.; Liu, Y.; Chen, P.; Wu, X.; et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol. Rep. 2015, 33, 2915–2923. [Google Scholar] [CrossRef]
- Dong, R.; Liu, X.; Zhang, Q.; Jiang, Z.; Li, Y.; Wei, Y.; Li, Y.; Yang, Q.; Liu, J.; Wei, J.J.; et al. miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget 2014, 5, 10816–10829. [Google Scholar] [CrossRef]
- Lam, S.S.; Ip, C.K.; Mak, A.S.; Wong, A.S. A novel p70 S6 kinase-microRNA biogenesis axis mediates multicellular spheroid formation in ovarian cancer progression. Oncotarget 2016, 7, 38064–38077. [Google Scholar] [CrossRef]
- Kim, T.H.; Song, J.Y.; Park, H.; Jeong, J.Y.; Kwon, A.Y.; Heo, J.H.; Kang, H.; Kim, G.; An, H.J. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett. 2015, 356, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Robertson, G.; Pedersen, L.; Lim, E.; Hernandez-Herrera, A.; Rowat, A.C.; Patil, S.L.; Chan, C.K.; Wen, Y.; Zhang, X.; et al. miR-509-3p is clinically significant and strongly attenuates cellular migration and multi-cellular spheroids in ovarian cancer. Oncotarget 2016, 7, 25930–25948. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, S.; Liu, X.; Zhang, W.; Li, Y.; Dong, R.; Zhang, Q.; Yang, Q.; Yuan, C.; Shen, K.; et al. miR-1236-3p represses the cell migration and invasion abilities by targeting ZEB1 in high-grade serous ovarian carcinoma. Oncol. Rep. 2014, 31, 1905–1910. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhu, J.; Zheng, Y.; Zhang, H.; Sun, H.; Gao, S. miRNA-574-3p inhibits metastasis and chemoresistance of epithelial ovarian cancer (EOC) by negatively regulating epidermal growth factor receptor (EGFR). Am. J. Transl. Res. 2019, 11, 4151–4165. [Google Scholar] [PubMed]
- Liang, H.; Zhao, X.; Wang, C.; Sun, J.; Chen, Y.; Wang, G.; Fang, L.; Yang, R.; Yu, M.; Gu, Y.; et al. Systematic analyses reveal long non-coding RNA (PTAF)-mediated promotion of EMT and invasion-metastasis in serous ovarian cancer. Mol. Cancer 2018, 17, 96. [Google Scholar] [CrossRef]
- Liang, H.; Yu, T.; Han, Y.; Jiang, H.; Wang, C.; You, T.; Zhao, X.; Shan, H.; Yang, R.; Yang, L.; et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol. Cancer 2018, 17, 119. [Google Scholar] [CrossRef]
- Liang, H.; Yu, M.; Yang, R.; Zhang, L.; Zhang, L.; Zhu, D.; Luo, H.; Hong, Y.; Yu, T.; Sun, J.; et al. A PTAL-miR-101-FN1 Axis Promotes EMT and Invasion-Metastasis in Serous Ovarian Cancer. Mol. Ther. Oncolytics 2020, 16, 53–62. [Google Scholar] [CrossRef]
- Gong, C.; Yang, Z.; Wu, F.; Han, L.; Liu, Y.; Gong, W. miR-17 inhibits ovarian cancer cell peritoneal metastasis by targeting ITGA5 and ITGB1. Oncol. Rep. 2016, 36, 2177–2183. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Xiu, Y.L.; Sun, K.X.; Zhao, Y. Inhibition of Ovarian Epithelial Carcinoma Tumorigenesis and Progression by microRNA 106b Mediated through the RhoC Pathway. PLoS ONE 2015, 10, e0125714. [Google Scholar] [CrossRef]
- Yan, J.; Jiang, J.Y.; Meng, X.N.; Xiu, Y.L.; Zong, Z.H. MiR-23b targets cyclin G1 and suppresses ovarian cancer tumorigenesis and progression. J. Exp. Clin. Cancer Res. CR 2016, 35, 31. [Google Scholar] [CrossRef]
- Xu, H.; Mao, H.L.; Zhao, X.R.; Li, Y.; Liu, P.S. MiR-29c-3p, a target miRNA of LINC01296, accelerates tumor malignancy: Therapeutic potential of a LINC01296/miR-29c-3p axis in ovarian cancer. J. Ovarian Res. 2020, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.R.; Wang, L.M.; Shen, J.J. Overexpression of miR-32 inhibits the proliferation and metastasis of ovarian cancer cells by targeting BTLA. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4671–4678. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Watari, H.; Hanley, S.J.; Konno, Y.; Ihira, K.; Yamada, T.; Kudo, M.; Yue, J.; Sakuragi, N. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J. Exp. Clin. Cancer Res. CR 2016, 35, 132. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.X.; Luo, M.; Qiu, L.W.; He, Y.L.; Wang, X.F. Prognostic implications of microRNA-100 and its functional roles in human epithelial ovarian cancer. Oncol. Rep. 2012, 27, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Li, S.; Zhou, Q.; Wang, D.; Zou, D.; Shu, J.; Huang, Y. MiR-124 inhibits invasion and induces apoptosis of ovarian cancer cells by targeting programmed cell death 6. Oncol. Lett. 2017, 14, 7311–7317. [Google Scholar] [CrossRef][Green Version]
- Cao, Y.; Shi, H.; Ren, F.; Jia, Y.; Zhang, R. Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in epithelial ovarian cancer. Exp. Cell Res. 2017, 359, 185–194. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.; Cheng, Q.; Zhou, C.; Ding, Z. Role of microRNA-133a in epithelial ovarian cancer pathogenesis and progression. Oncol. Lett. 2014, 7, 1043–1048. [Google Scholar] [CrossRef]
- Zhang, Y.; Dun, Y.; Zhou, S.; Huang, X.H. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/beta-catenin signaling pathway. Biomed. Pharmacother. 2017, 96, 1216–1221. [Google Scholar] [CrossRef]
- Yang, L.; Hou, J.; Cui, X.H.; Suo, L.N.; Lv, Y.W. MiR-133b regulates the expression of CTGF in epithelial-mesenchymal transition of ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5602–5609. [Google Scholar] [CrossRef]
- Tang, W.; Jiang, Y.; Mu, X.; Xu, L.; Cheng, W.; Wang, X. MiR-135a functions as a tumor suppressor in epithelial ovarian cancer and regulates HOXA10 expression. Cell Signal. 2014, 26, 1420–1426. [Google Scholar] [CrossRef]
- Duan, S.; Dong, X.; Hai, J.; Jiang, J.; Wang, W.; Yang, J.; Zhang, W.; Chen, C. MicroRNA-135a-3p is downregulated and serves as a tumour suppressor in ovarian cancer by targeting CCR2. Biomed. Pharmacother. 2018, 107, 712–720. [Google Scholar] [CrossRef]
- Yeh, Y.M.; Chuang, C.M.; Chao, K.C.; Wang, L.H. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int. J. Cancer 2013, 133, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Zhu, Y.; Jin, M. MicroRNA-138 inhibits SOX12 expression and the proliferation, invasion and migration of ovarian cancer cells. Exp. Ther. Med. 2018, 16, 1629–1638. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jin, S.; Wang, R. MicroRNA139 suppressed tumor cell proliferation, migration and invasion by directly targeting HDGF in epithelial ovarian cancer. Mol. Med. Rep. 2017, 16, 3379–3386. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Li, Q.R.; Xu, Y.H.; Zhou, H.B. MicroRNA-139-3p Inhibits the Growth and Metastasis of Ovarian Cancer by Inhibiting ELAVL1. Onco Targets Ther. 2019, 12, 8935–8945. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Xu, C.; Zhang, X. MicroRNA-139-5p Inhibits Cell Proliferation and Invasion by Targeting RHO-Associated Coiled-Coil-Containing Protein Kinase 2 in Ovarian Cancer. Oncol. Res. 2018, 26, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Wen, J.; Qi, T.; Wang, Y. Long non-coding RNA TTN-AS1 promotes tumorigenesis of ovarian cancer through modulating the miR-139-5p/ROCK2 axis. Biomed. Pharmacother. 2020, 125, 109882. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, X.; Li, W.; Chen, Y. MicroRNA-145-5p regulates the proliferation of epithelial ovarian cancer cells via targeting SMAD4. J. Ovarian Res. 2020, 13, 54. [Google Scholar] [CrossRef]
- Li, L.W.; Xiao, H.Q.; Ma, R.; Yang, M.; Li, W.; Lou, G. miR-152 is involved in the proliferation and metastasis of ovarian cancer through repression of ERBB3. Int. J. Mol. Med. 2018, 41, 1529–1535. [Google Scholar] [CrossRef]
- Jin, M.; Yang, Z.; Ye, W.; Xu, H.; Hua, X. MicroRNA-150 predicts a favorable prognosis in patients with epithelial ovarian cancer, and inhibits cell invasion and metastasis by suppressing transcriptional repressor ZEB1. PLoS ONE 2014, 9, e103965. [Google Scholar] [CrossRef]
- Zhu, X.; Shen, H.; Yin, X.; Long, L.; Xie, C.; Liu, Y.; Hui, L.; Lin, X.; Fang, Y.; Cao, Y.; et al. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene 2016, 35, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.K.; Chiang, C.Y.; Tiwari, P.; Tomar, S.; Watters, K.M.; Peter, M.E.; Lengyel, E. Microenvironment-induced downregulation of miR-193b drives ovarian cancer metastasis. Oncogene 2015, 34, 5923–5932. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, Y.; Qiu, W.; Zhao, D.; Zhang, Y. Tissue miR-193b as a Novel Biomarker for Patients with Ovarian Cancer. Med. Sci. Monit. 2015, 21, 3929–3934. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Y.; Pan, S.S. MiR-202-5p suppressed cell proliferation, migration and invasion in ovarian cancer via regulating HOXB2. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2256–2263. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Xie, Y.; Zhuang, X.; Yuan, Z. MiR-206 inhibits epithelial ovarian cancer cells growth and invasion via blocking c-Met/AKT/mTOR signaling pathway. Biomed. Pharmacother. 2018, 104, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Guo, R.; Yuan, Z.; Shi, H.; Zhang, D. LncRNA HOTAIR Regulates CCND1 and CCND2 Expression by Sponging miR-206 in Ovarian Cancer. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 49, 1289–1303. [Google Scholar] [CrossRef]
- Lin, Y.; Jin, Y.; Xu, T.; Zhou, S.; Cui, M. MicroRNA-215 targets NOB1 and inhibits growth and invasion of epithelial ovarian cancer. Am. J. Transl. Res. 2017, 9, 466–477. [Google Scholar]
- Li, J.; Li, D.; Zhang, W. Tumor suppressor role of miR-217 in human epithelial ovarian cancer by targeting IGF1R. Oncol. Rep. 2016, 35, 1671–1679. [Google Scholar] [CrossRef]
- Li, N.; Wang, L.; Tan, G.; Guo, Z.; Liu, L.; Yang, M.; He, J. MicroRNA-218 inhibits proliferation and invasion in ovarian cancer by targeting Runx2. Oncotarget 2017, 8, 91530–91541. [Google Scholar] [CrossRef]
- Wei, C.; Zhang, X.; He, S.; Liu, B.; Han, H.; Sun, X. MicroRNA-219-5p inhibits the proliferation, migration, and invasion of epithelial ovarian cancer cells by targeting the Twist/Wnt/beta-catenin signaling pathway. Gene 2017, 637, 25–32. [Google Scholar] [CrossRef]
- Xing, F.; Song, Z.; He, Y. MiR-219-5p inhibits growth and metastasis of ovarian cancer cells by targeting HMGA2. Biol. Res. 2018, 51, 50. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Cai, J.; Huang, D.; Han, Q.; Chen, Y.; Yang, Q.; Yang, C.; Kuang, Y.; Li, D.; Wang, Z. miR-335 represents an independent prognostic marker in epithelial ovarian cancer. Am. J. Clin. Pathol. 2014, 141, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Shi, H.; Ren, F.; Liu, Z.; Ji, P.; Zhang, W.; Wang, W. Down-regulation of miR-338-3p and Up-regulation of MACC1 Indicated Poor Prognosis of Epithelial Ovarian Cancer Patients. J. Cancer 2019, 10, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Liu, X.; Ma, H.; Zhang, W.; Li, H. miR3383p suppresses tumor growth of ovarian epithelial carcinoma by targeting Runx2. Int. J. Oncol. 2015, 46, 2277–2285. [Google Scholar] [CrossRef]
- Huang, Z.; Li, Q.; Luo, K.; Zhang, Q.; Geng, J.; Zhou, X.; Xu, Y.; Qian, M.; Zhang, J.A.; Ji, L.; et al. miR-340-FHL2 axis inhibits cell growth and metastasis in ovarian cancer. Cell Death Dis. 2019, 10, 372. [Google Scholar] [CrossRef]
- Li, P.; Sun, Y.; Liu, Q. MicroRNA-340 Induces Apoptosis and Inhibits Metastasis of Ovarian Cancer Cells by Inactivation of NF-x03BA;B1. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2016, 38, 1915–1927. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, T.; Zhou, S.; Cui, M. MicroRNA-363 inhibits ovarian cancer progression by inhibiting NOB1. Oncotarget 2017, 8, 101649–101658. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Wang, Y.; Zhang, X. MicroRNA-365 inhibits ovarian cancer progression by targeting Wnt5a. Am. J. Cancer Res. 2017, 7, 1096–1106. [Google Scholar]
- Zhang, Y.; Zhao, F.J.; Chen, L.L.; Wang, L.Q.; Nephew, K.P.; Wu, Y.L.; Zhang, S. MiR-373 targeting of the Rab22a oncogene suppresses tumor invasion and metastasis in ovarian cancer. Oncotarget 2014, 5, 12291–12303. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, J.; Wu, Y.; Qiu, J.; Sun, Y.; Tong, X. LncRNA HOTAIR controls the expression of Rab22a by sponging miR-373 in ovarian cancer. Mol. Med. Rep. 2016, 14, 2465–2472. [Google Scholar] [CrossRef]
- Yu, R.; Cai, L.; Chi, Y.; Ding, X.; Wu, X. miR377 targets CUL4A and regulates metastatic capability in ovarian cancer. Int. J. Mol. Med. 2018, 41, 3147–3156. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Wang, X.; Yu, C.; Jin, Z.; Wei, L.; Cao, J.; Wang, Q.; Zhang, M.; Zhang, L.; Zhang, L.; et al. microRNA-494 is a potential prognostic marker and inhibits cellular proliferation, migration and invasion by targeting SIRT1 in epithelial ovarian cancer. Oncol. Lett. 2017, 14, 3177–3184. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yang, Y.J.; An, Q. LINC00963 Promotes Ovarian Cancer Proliferation, Migration and EMT via the miR-378g /CHI3L1 Axis. Cancer Manag. Res. 2020, 12, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Ni, Z.; Yicheng, S.; Pan, H.; Huang, Y.; Xiong, Y.; Liu, T. Anisomycin inhibits angiogenesis in ovarian cancer by attenuating the molecular sponge effect of the lncRNAMeg3/miR421/PDGFRA axis. Int. J. Oncol. 2019, 55, 1296–1312. [Google Scholar] [CrossRef] [PubMed]
- Muys, B.R.; Sousa, J.F.; Placa, J.R.; de Araujo, L.F.; Sarshad, A.A.; Anastasakis, D.G.; Wang, X.; Li, X.L.; de Molfetta, G.A.; Ramao, A.; et al. miR-450a Acts as a Tumor Suppressor in Ovarian Cancer by Regulating Energy Metabolism. Cancer Res. 2019, 79, 3294–3305. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, H.; Su, L.; Lu, Q.; Liu, Z. MicroRNA455 inhibits cell proliferation and invasion of epithelial ovarian cancer by directly targeting Notch1. Mol. Med. Rep. 2017, 16, 9777–9785. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.W.; Li, L.; Jiang, P.; Wang, Y.F. MicroRNA-489 targets XIAP to inhibit the biological progression of ovarian cancer via regulating PI3K/Akt signaling pathway and epithelial-to-mesenchymal transition. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4113–4122. [Google Scholar] [CrossRef]
- Wang, W.; Ren, F.; Wu, Q.; Jiang, D.; Li, H.; Peng, Z.; Wang, J.; Shi, H. MicroRNA-497 inhibition of ovarian cancer cell migration and invasion through targeting of SMAD specific E3 ubiquitin protein ligase 1. Biochem. Biophys. Res. Commun. 2014, 449, 432–437. [Google Scholar] [CrossRef]
- Park, G.B.; Kim, D. MicroRNA-503-5p Inhibits the CD97-Mediated JAK2/STAT3 Pathway in Metastatic or Paclitaxel-Resistant Ovarian Cancer Cells. Neoplasia 2019, 21, 206–215. [Google Scholar] [CrossRef]
- Yang, D.; Sun, Y.; Hu, L.; Zheng, H.; Ji, P.; Pecot, C.V.; Zhao, Y.; Reynolds, S.; Cheng, H.; Rupaimoole, R.; et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell 2013, 23, 186–199. [Google Scholar] [CrossRef]
- Wei, H.; Tang, Q.L.; Zhang, K.; Sun, J.J.; Ding, R.F. miR-532-5p is a prognostic marker and suppresses cells proliferation and invasion by targeting TWIST1 in epithelial ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5842–5850. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cui, M.; Zhang, A.; Tong, L.; Wang, K.; Li, K.; Wang, X.; Sun, Z.; Zhang, H. MiR-548c impairs migration and invasion of endometrial and ovarian cancer cells via downregulation of Twist. J. Exp. Clin. Cancer Res. CR 2016, 35, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ma, H.L. MiRNA-584 suppresses the progression of ovarian cancer by negatively regulating LPIN1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, W.; Xu, Y.; Tao, E.; Mo, M.; Xu, W.; Cai, X.; Chen, X.; Yuan, J.; Wu, X. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany NY) 2020, 12, 4558–4572. [Google Scholar] [CrossRef]
- Yu, H.; Xu, Y.; Zhang, D.; Liu, G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem Biophys. Res. Commun. 2018, 503, 2095–2100. [Google Scholar] [CrossRef]
- Duan, M.; Fang, M.; Wang, C.; Wang, H.; Li, M. LncRNA EMX2OS Induces Proliferation, Invasion and Sphere Formation of Ovarian Cancer Cells via Regulating the miR-654-3p/AKT3/PD-L1 Axis. Cancer Manag. Res. 2020, 12, 2141–2154. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, Y.; Wan, Y.; Zhou, S.; Thapa, S.; Cheng, W. MicroRNA665 suppresses the growth and migration of ovarian cancer cells by targeting HOXA10. Mol. Med. Rep. 2018, 18, 2661–2668. [Google Scholar] [CrossRef]
- Xia, B.; Lin, M.; Dong, W.; Chen, H.; Li, B.; Zhang, X.; Hou, Y.; Lou, G. Upregulation of miR-874-3p and miR-874-5p inhibits epithelial ovarian cancer malignancy via SIK2. J. Biochem. Mol. Toxicol. 2018, 32, e22168. [Google Scholar] [CrossRef]
- Li, C.; Yu, S.; Wu, S.; Ni, Y.; Pan, Z. MicroRNA-936 targets FGF2 to inhibit epithelial ovarian cancer aggressiveness by deactivating the PI3K/Akt pathway. Onco Targets Ther. 2019, 12, 5311–5322. [Google Scholar] [CrossRef]
- Dasari, S.; Pandhiri, T.; Grassi, T.; Visscher, D.W.; Multinu, F.; Agarwal, K.; Mariani, A.; Shridhar, V.; Mitra, A.K. Signals from the Metastatic Niche Regulate Early and Advanced Ovarian Cancer Metastasis through miR-4454 Downregulation. Mol. Cancer Res. MCR 2020, 18, 1202–1217. [Google Scholar] [CrossRef]
- Liu, L.; Ning, Y.; Yi, J.; Yuan, J.; Fang, W.; Lin, Z.; Zeng, Z. miR-6089/MYH9/beta-catenin/c-Jun negative feedback loop inhibits ovarian cancer carcinogenesis and progression. Biomed. Pharmacother. 2020, 125, 109865. [Google Scholar] [CrossRef]
- Kanlikilicer, P.; Rashed, M.H.; Bayraktar, R.; Mitra, R.; Ivan, C.; Aslan, B.; Zhang, X.; Filant, J.; Silva, A.M.; Rodriguez-Aguayo, C.; et al. Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells. Cancer Res. 2016, 76, 7194–7207. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, G.; Zhang, K. MiR-125a regulates ovarian cancer proliferation and invasion by repressing GALNT14 expression. Biomed. Pharmacother. 2016, 80, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Cowden Dahl, K.D.; Dahl, R.; Kruichak, J.N.; Hudson, L.G. The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia 2009, 11, 1208–1215. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, X.; Ma, Y.; Zhao, X.; Li, B.; Wang, H. Ascites promotes cell migration through the repression of miR-125b in ovarian cancer. Oncotarget 2017, 8, 51008–51015. [Google Scholar] [CrossRef] [PubMed]
- Zuberi, M.; Khan, I.; Mir, R.; Gandhi, G.; Ray, P.C.; Saxena, A. Utility of Serum miR-125b as a Diagnostic and Prognostic Indicator and Its Alliance with a Panel of Tumor Suppressor Genes in Epithelial Ovarian Cancer. PLoS ONE 2016, 11, e0153902. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.Q.; Liang, H.T.; Qin, D.C.; Jin, H.F.; Zhao, Y.; She, M.C. MiR-212 exerts suppressive effect on SKOV3 ovarian cancer cells through targeting HBEGF. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 12427–12434. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Dong, Y.; Dang, R.; Hu, Z.; Yang, Y.; Hu, Y.; Cheng, J. MiR-122 inhibits epithelial mesenchymal transition by regulating P4HA1 in ovarian cancer cells. Cell Biol. Int. 2018, 42, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Wang, C.; Gao, Y.; Wang, J. Decreased expression of microRNA-148a predicts poor prognosis in ovarian cancer and associates with tumor growth and metastasis. Biomed. Pharmacother. 2016, 83, 58–63. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, H.; Xia, M.; Sun, C.; Li, N.; Guo, E.; Guo, L.; Shan, W.; Lu, H.; Wu, Y.; et al. Overexpressed miR-9 promotes tumor metastasis via targeting E-cadherin in serous ovarian cancer. Front. Med. 2017, 11, 214–222. [Google Scholar] [CrossRef]
- Laios, A.; O’Toole, S.; Flavin, R.; Martin, C.; Kelly, L.; Ring, M.; Finn, S.P.; Barrett, C.; Loda, M.; Gleeson, N.; et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol. Cancer 2008, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Yao, L.; Tao, X.; Yu, Y.; Chen, M.; Zhang, R.; Xu, C. miR-9 functions as a tumor suppressor in ovarian serous carcinoma by targeting TLN1. Int. J. Mol. Med. 2013, 32, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Mak, C.S.; Yung, M.M.; Hui, L.M.; Leung, L.L.; Liang, R.; Chen, K.; Liu, S.S.; Qin, Y.; Leung, T.H.; Lee, K.F.; et al. MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting KLF12/Sp1/survivin axis. Mol. Cancer 2017, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Mateescu, B.; Batista, L.; Cardon, M.; Gruosso, T.; de Feraudy, Y.; Mariani, O.; Nicolas, A.; Meyniel, J.P.; Cottu, P.; Sastre-Garau, X.; et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 2011, 17, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Lei, L.; Shao, L.; Shi, J.; Jia, J.; Tong, X. MicroRNA141 inhibits epithelialmesenchymal transition, and ovarian cancer cell migration and invasion. Mol. Med. Rep. 2017, 16, 6743–6749. [Google Scholar] [CrossRef]
- Suo, H.B.; Zhang, K.C.; Zhao, J. MiR-200a promotes cell invasion and migration of ovarian carcinoma by targeting PTEN. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4080–4089. [Google Scholar] [CrossRef]
- Pecot, C.V.; Rupaimoole, R.; Yang, D.; Akbani, R.; Ivan, C.; Lu, C.; Wu, S.; Han, H.D.; Shah, M.Y.; Rodriguez-Aguayo, C.; et al. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013, 4, 2427. [Google Scholar] [CrossRef]
- Sun, Q.; Zou, X.; Zhang, T.; Shen, J.; Yin, Y.; Xiang, J. The role of miR-200a in vasculogenic mimicry and its clinical significance in ovarian cancer. Gynecol. Oncol. 2014, 132, 730–738. [Google Scholar] [CrossRef]
- Nam, E.J.; Yoon, H.; Kim, S.W.; Kim, H.; Kim, Y.T.; Kim, J.H.; Kim, J.W.; Kim, S. MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 2690–2695. [Google Scholar] [CrossRef]
- Lu, Y.M.; Shang, C.; Ou, Y.L.; Yin, D.; Li, Y.N.; Li, X.; Wang, N.; Zhang, S.L. miR-200c modulates ovarian cancer cell metastasis potential by targeting zinc finger E-box-binding homeobox 2 (ZEB2) expression. Med. Oncol. (Northwood Lond. Engl.) 2014, 31, 134. [Google Scholar] [CrossRef]
- Cochrane, D.R.; Spoelstra, N.S.; Howe, E.N.; Nordeen, S.K.; Richer, J.K. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol. Cancer Ther. 2009, 8, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, X.; Zhao, G.; Yan, H.; Dong, P.; Watari, H.; Sims, M.; Li, W.; Pfeffer, L.M.; Guo, Y.; et al. miR-203 inhibits ovarian tumor metastasis by targeting BIRC5 and attenuating the TGFbeta pathway. J. Exp. Clin. Cancer Res. CR 2018, 37, 235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, J.; Yang, N.; Greshock, J.; Megraw, M.S.; Giannakakis, A.; Liang, S.; Naylor, T.L.; Barchetti, A.; Ward, M.R.; et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 2006, 103, 9136–9141. [Google Scholar] [CrossRef] [PubMed]
- Hirata, Y.; Murai, N.; Yanaihara, N.; Saito, M.; Saito, M.; Urashima, M.; Murakami, Y.; Matsufuji, S.; Okamoto, A. MicroRNA-21 is a candidate driver gene for 17q23-25 amplification in ovarian clear cell carcinoma. BMC Cancer 2014, 14, 799. [Google Scholar] [CrossRef]
- Filippova, E.A.; Loginov, V.I.; Burdennyi, A.M.; Braga, E.A.; Pronina, I.V.; Kazubskaya, T.P.; Kushlinskii, D.N.; Utkin, D.O.; Fridman, M.V.; Khodyrev, D.S.; et al. Hypermethylated Genes of MicroRNA in Ovarian Carcinoma: Metastasis Prediction Marker Systems. Bull. Exp. Biol. Med. 2019, 167, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Braga, E.A.; Loginov, V.I.; Filippova, E.A.; Burdennyi, A.M.; Pronina, I.V.; Kazubskaya, T.P.; Khodyrev, D.S.; Utkin, D.O.; Kushlinskii, D.N.; Adamyan, L.V.; et al. Diagnostic Value of a Group of MicroRNA Genes Hypermethylated in Ovarian Carcinoma. Bull. Exp. Biol. Med. 2018, 166, 253–256. [Google Scholar] [CrossRef]
- Hermeking, H. MicroRNAs in the p53 network: Micromanagement of tumour suppression. Nat. Rev. Cancer 2012, 12, 613–626. [Google Scholar] [CrossRef]
- Welponer, H.; Tsibulak, I.; Wieser, V.; Degasper, C.; Shivalingaiah, G.; Wenzel, S.; Sprung, S.; Marth, C.; Hackl, H.; Fiegl, H.; et al. The miR-34 family and its clinical significance in ovarian cancer. J. Cancer 2020, 11, 1446–1456. [Google Scholar] [CrossRef]
- Tian, M.; Tian, D.; Qiao, X.; Li, J.; Zhang, L. Modulation of Myb-induced NF-kB -STAT3 signaling and resulting cisplatin resistance in ovarian cancer by dietary factors. J. Cell. Physiol. 2019, 234, 21126–21134. [Google Scholar] [CrossRef]
- Luo, S.; Wang, J.; Ma, Y.; Yao, Z.; Pan, H. PPARgamma inhibits ovarian cancer cells proliferation through upregulation of miR-125b. Biochem. Biophys. Res. Commun. 2015, 462, 85–90. [Google Scholar] [CrossRef]
- Vignati, S.; Albertini, V.; Rinaldi, A.; Kwee, I.; Riva, C.; Oldrini, R.; Capella, C.; Bertoni, F.; Carbone, G.M.; Catapano, C.V. Cellular and molecular consequences of peroxisome proliferator-activated receptor-gamma activation in ovarian cancer cells. Neoplasia 2006, 8, 851–861. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, S.; Lee, J.J.; Heo, D.S. PPARgamma ligands induce growth inhibition and apoptosis through p63 and p73 in human ovarian cancer cells. Biochem. Biophys. Res. Commun. 2011, 406, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, R.; Muller, C.Y.; Stack, M.S.; Hudson, L.G. Targeting the EGF receptor for ovarian cancer therapy. J. Oncol. 2010, 2010, 414676. [Google Scholar] [CrossRef] [PubMed]
- Merritt, W.M.; Lin, Y.G.; Han, L.Y.; Kamat, A.A.; Spannuth, W.A.; Schmandt, R.; Urbauer, D.; Pennacchio, L.A.; Cheng, J.F.; Nick, A.M.; et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008, 359, 2641–2650. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Triboulet, R.; Mohseni, M.; Schlegelmilch, K.; Shrestha, K.; Camargo, F.D.; Gregory, R.I. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 2014, 156, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Yamagata, K.; Sugimoto, K.; Iwamoto, T.; Kato, S.; Miyazono, K. Modulation of microRNA processing by p53. Nature 2009, 460, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Liu, Y.; Wan, G.; Choi, H.J.; Zhao, L.; Ivan, C.; He, X.; Sood, A.K.; Zhang, X.; Lu, X. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep. 2014, 8, 1447–1460. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Kuang, Z.; Sheng, C.; Zhang, Y.; Xu, H.; Cheng, Q. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig. Dis. Sci. 2010, 55, 2288–2293. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.C.; Avissar-Whiting, M.; Ouellet, L.G.; Butler, R.A.; Nelson, H.H.; McClean, M.D.; Marsit, C.J.; Kelsey, K.T. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin. Cancer Res. Off. 2010, 16, 3713–3720. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, A.E.; Zheng, T.; Yi, C.; Leaderer, D.; Weidhaas, J.; Slack, F.; Zhang, Y.; Paranjape, T.; Zhu, Y. microRNA miR-196a-2 and breast cancer: A genetic and epigenetic association study and functional analysis. Cancer Res. 2009, 69, 5970–5977. [Google Scholar] [CrossRef]
- Song, Z.S.; Wu, Y.; Zhao, H.G.; Liu, C.X.; Cai, H.Y.; Guo, B.Z.; Xie, Y.A.; Shi, H.R. Association between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancer. Oncol. Lett. 2016, 11, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, J.; Soltesz, B.; Penyige, A.; Nagy, B.; Poka, R. Identification of miR-146a and miR-196a-2 single nucleotide polymorphisms at patients with high-grade serous ovarian cancer. J. Biotechnol. 2019, 297, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Martin, T.A.; Jordan, N.J.; Ruge, F.; Ye, L.; Jiang, W.G. Metastasis suppressor 1 expression in human ovarian cancer: The impact on cellular migration and metastasis. Int. J. Oncol. 2015, 47, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, M.M. PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control. Genes Cancer 2010, 1, 1170–1177. [Google Scholar] [CrossRef]
- Niu, K.; Shen, W.; Zhang, Y.; Zhao, Y.; Lu, Y. MiR-205 promotes motility of ovarian cancer cells via targeting ZEB1. Gene 2015, 574, 330–336. [Google Scholar] [CrossRef]
- Li, J.; Li, L.; Li, Z.; Gong, G.; Chen, P.; Liu, H.; Wang, J.; Liu, Y.; Wu, X. The role of miR-205 in the VEGF-mediated promotion of human ovarian cancer cell invasion. Gynecol. Oncol. 2015, 137, 125–133. [Google Scholar] [CrossRef]
- Ghoneum, A.; Said, N. PI3K-AKT-mTOR and NFkappaB Pathways in Ovarian Cancer: Implications for Targeted Therapeutics. Cancers 2019, 11, 949. [Google Scholar] [CrossRef]
- Xu, G.; Bernaudo, S.; Fu, G.; Lee, D.Y.; Yang, B.B.; Peng, C. Cyclin G2 is degraded through the ubiquitin-proteasome pathway and mediates the antiproliferative effect of activin receptor-like kinase 7. Mol. Biol. Cell 2008, 19, 4968–4979. [Google Scholar] [CrossRef]
- Bernaudo, S.; Salem, M.; Qi, X.; Zhou, W.; Zhang, C.; Yang, W.; Rosman, D.; Deng, Z.; Ye, G.; Yang, B.B.; et al. Cyclin G2 inhibits epithelial-to-mesenchymal transition by disrupting Wnt/beta-catenin signaling. Oncogene 2016, 35, 4816–4827. [Google Scholar] [CrossRef]
- Fu, G.; Peng, C. Nodal enhances the activity of FoxO3a and its synergistic interaction with Smads to regulate cyclin G2 transcription in ovarian cancer cells. Oncogene 2011, 30, 3953–3966. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Q. Relationship between matrix metalloproteinases and the occurrence and development of ovarian cancer. Braz. J. Med. Biol. Res. 2017, 50, e6104. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Jin, P.; Xu, S.; Zhang, T.; Yang, X.; Li, X.; Wei, X.; Sun, C.; Chen, G.; Ma, D.; et al. Dicer reprograms stromal fibroblasts to a pro-inflammatory and tumor-promoting phenotype in ovarian cancer. Cancer Lett. 2018, 415, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Stone, R.L.; Nick, A.M.; McNeish, I.A.; Balkwill, F.; Han, H.D.; Bottsford-Miller, J.; Rupairmoole, R.; Armaiz-Pena, G.N.; Pecot, C.V.; Coward, J.; et al. Paraneoplastic thrombocytosis in ovarian cancer. N. Engl. J. Med. 2012, 366, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Chen, X.; Chen, L.; Wan, F.; Chen, K.; Sun, Y.; Zhu, X. STAT3 signaling in ovarian cancer: A potential therapeutic target. J. Cancer 2020, 11, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Hill, L.; Browne, G.; Tulchinsky, E. ZEB/miR-200 feedback loop: At the crossroads of signal transduction in cancer. Int. J. Cancer 2013, 132, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Eichler, W.; Hamann, J.; Aust, G. Expression characteristics of the human CD97 antigen. Tissue Antigens 1997, 50, 429–438. [Google Scholar] [CrossRef]
- Yona, S.; Lin, H.H.; Siu, W.O.; Gordon, S.; Stacey, M. Adhesion-GPCRs: Emerging roles for novel receptors. Trends Biochem. Sci. 2008, 33, 491–500. [Google Scholar] [CrossRef]
- Li, J.; Zhou, B.P. Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 2011, 11, 49. [Google Scholar] [CrossRef]
- Pan, Y.; Tong, J.H.M.; Lung, R.W.M.; Kang, W.; Kwan, J.S.H.; Chak, W.P.; Tin, K.Y.; Chung, L.Y.; Wu, F.; Ng, S.S.M.; et al. RASAL2 promotes tumor progression through LATS2/YAP1 axis of hippo signaling pathway in colorectal cancer. Mol. Cancer 2018, 17, 102. [Google Scholar] [CrossRef]
- Li, S.; Yu, Z.; Chen, S.S.; Li, F.; Lei, C.Y.; Chen, X.X.; Bao, J.M.; Luo, Y.; Lin, G.Z.; Pang, S.Y.; et al. The YAP1 oncogene contributes to bladder cancer cell proliferation and migration by regulating the H19 long noncoding RNA. Urol. Oncol. 2015, 33, 427.e1–427.e10. [Google Scholar] [CrossRef]
- Guo, C.; Wang, X.; Liang, L. LATS2-mediated YAP1 phosphorylation is involved in HCC tumorigenesis. Int. J. Clin. Exp. Pathol. 2015, 8, 1690–1697. [Google Scholar] [PubMed]
- Lu, Q.; Longo, F.M.; Zhou, H.; Massa, S.M.; Chen, Y.H. Signaling through Rho GTPase pathway as viable drug target. Curr. Med. Chem. 2009, 16, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Bagley, A.F.; Na, Y.J.; Birrer, M.J.; Bhatia, S.N.; Belcher, A.M. Deep, noninvasive imaging and surgical guidance of submillimeter tumors using targeted M13-stabilized single-walled carbon nanotubes. Proc. Natl. Acad. Sci. USA 2014, 111, 13948–13953. [Google Scholar] [CrossRef]
- Lessan, K.; Aguiar, D.J.; Oegema, T.; Siebenson, L.; Skubitz, A.P. CD44 and beta1 integrin mediate ovarian carcinoma cell adhesion to peritoneal mesothelial cells. Am. J. Pathol. 1999, 154, 1525–1537. [Google Scholar] [CrossRef]
- Morgan, M.R.; Byron, A.; Humphries, M.J.; Bass, M.D. Giving off mixed signals--distinct functions of alpha5beta1 and alphavbeta3 integrins in regulating cell behaviour. IUBMB Life 2009, 61, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Iwanicki, M.P.; Davidowitz, R.A.; Ng, M.R.; Besser, A.; Muranen, T.; Merritt, M.; Danuser, G.; Ince, T.A.; Brugge, J.S. Ovarian cancer spheroids use myosin-generated force to clear the mesothelium. Cancer Discov. 2011, 1, 144–157. [Google Scholar] [CrossRef]
- Abate-Shen, C. Deregulated homeobox gene expression in cancer: Cause or consequence? Nat. Rev. Cancer 2002, 2, 777–785. [Google Scholar] [CrossRef]
- Li, B.; Jin, H.; Yu, Y.; Gu, C.; Zhou, X.; Zhao, N.; Feng, Y. HOXA10 is overexpressed in human ovarian clear cell adenocarcinoma and correlates with poor survival. Int. J. Gynecol. Cancer 2009, 19, 1347–1352. [Google Scholar] [CrossRef]
- Seeber, L.M.; Horree, N.; Vooijs, M.A.; Heintz, A.P.; van der Wall, E.; Verheijen, R.H.; van Diest, P.J. The role of hypoxia inducible factor-1alpha in gynecological cancer. Crit. Rev. Oncol. Hematol. 2011, 78, 173–184. [Google Scholar] [CrossRef]
- Markert, E.K.; Levine, A.J.; Vazquez, A. Proliferation and tissue remodeling in cancer: The hallmarks revisited. Cell Death Dis. 2012, 3, e397. [Google Scholar] [CrossRef]
- Hu, X.; Macdonald, D.M.; Huettner, P.C.; Feng, Z.; El Naqa, I.M.; Schwarz, J.K.; Mutch, D.G.; Grigsby, P.W.; Powell, S.N.; Wang, X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol. Oncol. 2009, 114, 457–464. [Google Scholar] [CrossRef]
- van Jaarsveld, M.T.; Helleman, J.; Boersma, A.W.; van Kuijk, P.F.; van Ijcken, W.F.; Despierre, E.; Vergote, I.; Mathijssen, R.H.; Berns, E.M.; Verweij, J.; et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene 2013, 32, 4284–4293. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Chen, F.; Zhang, T.T.; Liu, N.F. Suppression of SIK1 by miR-141 in human ovarian cancer cell lines and tissues. Int. J. Mol. Med. 2016, 37, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Kang, Y. Multilayer control of the EMT master regulators. Oncogene 2014, 33, 1755–1763. [Google Scholar] [CrossRef] [PubMed]
- Dykxhoorn, D.M. MicroRNAs and metastasis: Little RNAs go a long way. Cancer Res. 2010, 70, 6401–6406. [Google Scholar] [CrossRef]
- Olson, P.; Lu, J.; Zhang, H.; Shai, A.; Chun, M.G.; Wang, Y.; Libutti, S.K.; Nakakura, E.K.; Golub, T.R.; Hanahan, D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009, 23, 2152–2165. [Google Scholar] [CrossRef]
- Imai, T.; Horiuchi, A.; Shiozawa, T.; Osada, R.; Kikuchi, N.; Ohira, S.; Oka, K.; Konishi, I. Elevated expression of E-cadherin and alpha-, beta-, and gamma-catenins in metastatic lesions compared with primary epithelial ovarian carcinomas. Hum. Pathol. 2004, 35, 1469–1476. [Google Scholar] [CrossRef]
- Nowek, K.; Wiemer, E.A.C.; Jongen-Lavrencic, M. The versatile nature of miR-9/9(*) in human cancer. Oncotarget 2018, 9, 20838–20854. [Google Scholar] [CrossRef]
- Gang, W.; Tanjun, W.; Yong, H.; Jiajun, Q.; Yi, Z.; Hao, H. Inhibition of miR-9 decreases osteosarcoma cell proliferation. Bosn. J. Basic Med. Sci. 2020, 20, 218–225. [Google Scholar] [CrossRef]
- Hersi, H.M.; Raulf, N.; Gaken, J.; Folarin, N.; Tavassoli, M. MicroRNA-9 inhibits growth and invasion of head and neck cancer cells and is a predictive biomarker of response to plerixafor, an inhibitor of its target CXCR4. Mol. Oncol. 2018, 12, 2023–2041. [Google Scholar] [CrossRef]
- Dochez, V.; Caillon, H.; Vaucel, E.; Dimet, J.; Winer, N.; Ducarme, G. Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review. J. Ovarian Res. 2019, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.G.; Chang, T.H.; Liu, Y.N.; Shih, J.Y. MicroRNA in Lung Cancer Metastasis. Cancers 2019, 11, 265. [Google Scholar] [CrossRef] [PubMed]
| miRNA/ miRNA Family | Altered Expression in EOC and Clinical Significance | Target Gene | In Vitro/In Vivo Effects | Citations |
|---|---|---|---|---|
| Pro-Metastatic miRNAs | ||||
| miRNAs upregulated in tumor tissues | ||||
| miR-19a | Upregulated in metastatic HGSC tissues compared to normal ovarian tissues | ND | ND | [134] |
| miR-182 | Upregulated in HGSC tissues compared to fallopian tube tissues | MTSS1 | Promotes cell invasion in vitro, and tumor growth and metastasis in vivo | [135,136] |
| miR-328-3p | Upregulated in cancer stem-like cells isolated from HGSC tissues compared to bulk cancer cells | DDB2 | Increases ALDH+ population and promotes spheroid formation and CSC properties in vitro and tumor growth and metastasis in vivo | [137] |
| miR-130a | Upregulated in HGSC tissues compared to normal fallopian tube tissues | TSC1 | Promotes cell proliferation, invasion, and EMT and tumor growth and metastasis in vivo | [138] |
| miR-301b-3p | Upregulated in HGSC tissues compared to paired adjacent normal tissues; positively correlated with tumor stage, lymph node metastasis, and poor survival | CPEB3 | Promotes cell migration and invasion in vitro | [139] |
| miR-520h | Upregulated in EOC tissues compared to benign ovarian tumors and highest in HGSC compared to MC, EC, and CCC subtypes; correlated with tumor stage, increased ascites, lymph node metastasis, and poor survival | SMAD7 | Promotes cell proliferation, invasion, and EMT in vitro and tumor growth and metastasis in vivo | [130] |
| miR-21 | Upregulated in serous EOC, EC, and MC tissues compared to ovarian cysts and normal ovarian tissues; positively correlated with tumor stage and lymph node metastasis | ND | Promotes cell proliferation, migration, and invasion in vitro | [140] |
| miR-205 | Upregulated in EC subtype compared to normal endometrial tissues | ESRRG | Promotes cell proliferation, migration, and invasion in vitro | [141] |
| miR-146a | Upregulated in omental metastatic serous EOC tumors compared to primary EOC tumors | ND | Promotes spheroid formation and cisplatin resistance in vitro | [133] |
| miR-551b | Upregulated in recurrent serous EOC tissues compared to primary EOC tumors; associated with advanced stage | FOXO3 TRIM31 | Promotes cell proliferation, invasion, and colony formation in vitro and tumor burden in vivo | [142] |
| miR-551b-3p | Upregulated in HGSC tissues compared to normal ovarian tissues; associated with poor outcome | STAT3 promoter | Promotes cell proliferation, spheroid formation, and survival in vitro and tumor burden in vivo | [143] |
| miR-18b | Upregulated in EOC tissues compared to normal ovarian tissues; positively correlated with tumor grade and lymph node metastasis | PTEN | Promotes cell migration and invasion in vitro | [144] |
| miR-19b | Upregulated in EOC tissues compared to matched non-tumor tissues; positively correlated with tumor stage and lymph node metastasis | PTEN | Promotes cell migration and invasion in vitro | [145] |
| miR-23a | Upregulated in EOC tissues compared to adjacent normal tissues | DLG2 | Promotes cell proliferation, migration, and invasion in vitro and tumor growth in vivo | [146] |
| miR-181a | Upregulated in recurrent EOC tissues compared to primary EOC tissues; associated with poor survival | SMAD7 | Promotes cell migration, invasion, survival, and EMT in vitro and tumor growth and metastasis in vivo | [110] |
| miR-181b | Upregulated in EOC tissues compared to normal ovarian tissues | LATS2 | Promotes cell proliferation and invasion in vitro | [147] |
| miR-182-5p | Upregulated in EOC tissues compared to non-tumor ovarian tissues | FOXF2 | Promotes cell proliferation, and invasion in vitro and tumor growth in vivo | [148] |
| miR-194 | Upregulated in EOC tissues compared to normal ovarian epithelium tissues | PTPN12 | Promotes cell proliferation, migration and invasion in vitro | [149] |
| miR-196a | Upregulated in EOC compared to paired normal ovarian tissues; positively correlated with tumor stage, and lymph node metastasis | ND | ND | [150] |
| miR-205 | Upregulated in EOC tissues compared to normal ovarian tissues; correlated with tumor stage and poor survival | TCF21 PTEN SMAD4 | Promotes cell proliferation, migration, and invasion in vitro and tumor growth and metastasis in vivo | [151,152,153] |
| miR-216a | Upregulated in EOC tissues compared to normal ovarian tissues; correlated with tumor stage, lymph node metastasis, and poor survival | PTEN | Promotes cell migration and invasion and EMT in vitro | [154] |
| miR-552 | Upregulated in EOC tissues compared to paired non-tumor tissues; associated with metastasis, recurrence, and poor survival | PTEN | Promotes cell proliferation, migration, and invasion in vitro | [155] |
| miR-616 | Upregulated in EOC tissues compared to adjacent non-tumor tissues; associated with metastasis, tumor stage and grade, and poor survival | TIMP2 | Promotes cell migration, invasion, and EMT in vitro and metastasis in vivo | [111] |
| miR-630 | Upregulated in EOC tissues with high levels of hypoxia compared to low levels of hypoxia; associated with poor survival | DICER1 | Promotes cell migration, invasion, and EMT in vitro and tumor growth and metastasis in vivo | [156] |
| miR-939 | Upregulated in EOC tissues compared to matched adjacent normal tissues | APC2 | Promotes cell proliferation, colony formation, cell migration, invasion, and EMT in vitro | [157,158] |
| miRNAs upregulated in secreted exosomes and circulating body fluids | ||||
| miR-376a | Upregulated in EOC tissues compared to paired adjacent normal tissues and in blood samples of EOC patients compared to healthy controls; associated with advanced stages | KLF15 CASP8 | Promotes cell proliferation, migration, and invasion in vitro and tumor growth in vivo | [125] |
| miR-590-3p | Upregulated in EOC tissues compared to normal ovarian tissues, and in plasma of EOC patients compared to those with benign gynecologic disorders; correlated with tumor grade | FOXA2 FOXO3 CCNG2 | Promotes colony and spheroid formation, cell migration, and invasion in vitro and tumor burden in vivo | [112,159] |
| miR-29-3p | Upregulated in exosomes secreted by M2 macrophages compared to those derived from THP-1 cells; associated with poor survival | STAT3 | Promotes Tregs/Th17 imbalance in vitro and tumor growth and metastasis in vivo | [102] |
| miR-30a-5p | Upregulated in urine samples of serous EOC patients compared to healthy controls and higher in stage I/II compared to stage III/V; associated with lymphatic metastasis | ND | Promotes cell proliferation and migration in vitro | [160] |
| miR-149-3p | Upregulated in peritoneal exosomes of EOC patients compared to healthy controls; associated with poor survival | ND | ND | [129] |
| miR-222-5p | Upregulated in peritoneal exosomes of EOC patients compared to healthy controls; associated with poor survival | ND | ND | [129] |
| Anti-Metastatic miRNAs | ||||
| miRNAs down-regulated in tumor tissues | ||||
| miR-145 | Downregulated in HGSC compared to normal FT tissues | MTDH TWIST SOX9 HMGA2 | Inhibits cell proliferation, invasion, migration, EMT, and spheroid formation in vitro and tumor growth and metastasis in vivo | [161,162,163] |
| miR-509-3p | Positively associated with survival in HGSC | YAP1 | Inhibits cell invasion, migration, and spheroid formation in vitro | [164] |
| miR-1236-3p | Downregulated in HGSC tissues compared to normal FT tissues | ZEB1 | Inhibits cell invasion, migration, and EMT in vitro | [165] |
| miR-574-3p | Decreased in EOC tissues compared to normal ovarian tissues, significantly lower in serous EOC tissues compared to non-serous EOC tissues; negatively associated with tumor stage | EGFR | Inhibits cell invasion and migration in vitro | [166] |
| miR-25 | Downregulated in integrated mesenchymal EOC subtype compared to epithelial EOC subtype based on TCGA database | SNAI2 | Inhibits cell invasion, migration, and EMT in vitro and tumor growth and metastasis in the orthotopic xenograft mouse model | [167] |
| miR-101 | Decreased in integrated mesenchymal OC subtype compared to integrated epithelial OC subtype from TCGA database | ZEB1 FN1 | Inhibits cell invasion, migration, and EMT in vitro and tumor growth and intraperitoneal metastasis in vivo | [168,169] |
| miR-7 | Downregulated in metastatic EOC tissues from omentum or peritoneum compared to primary EOC tissues; associated with metastasis | EGFR | Inhibits cell invasion, migration, and EMT in vitro | [68] |
| miR-17-5p | ND | ITGA5 ITGB1 | Suppresses cell adhesion and invasion in vitro and peritoneal metastasis in vivo | [170] |
| miR-106b | Decreased in EOC tissues compared to normal ovarian tissues and benign tumors; negatively associated with tumor stage and grade | RHOC | Inhibits cell proliferation, invasion, and migration in vitro and tumor growth in vivo | [171] |
| miR-23b | Decreased in EOC tissues compared to normal ovarian tissues and benign tumors | CCNG1 | Inhibits cell proliferation, invasion, and migration in vitro and tumor growth in vivo | [172] |
| miR-26b | Downregulated in EOC tissues compared to normal ovarian surface epithelial tissues; inversely correlated with stage and grade, and higher risk with distant metastasis, recurrence, and poor survival | KPNA2 | Inhibits cell proliferation, migration, spheroid formation, and EMT in vitro and tumor growth and lung metastasis in vivo | [131,132] |
| miR-29c-3p | Downregulated in EOC tissues compared to normal ovarian tissues | ND | Inhibits cell proliferation, invasion, migration, and EMT in vitro | [173] |
| miR-32 | Downregulated in EOC tissues compared to adjacent normal tissues and in recurrent EOC tissues compared to primary tumors | BTLA | Inhibits cell proliferation, migration, and invasion in vitro | [174] |
| miR-34a | Downregulated in EOC tissues compared to paired adjacent normal ovarian tissues; negatively associated with late stage | SNAI1 | Inhibits cell invasion, EMT, spheroid formation, and apoptosis in vitro | [175] |
| miR-100 | Downregulated in EOC tissues compared to matched adjacent normal ovarian tissues; negatively associated with advanced stage, lymph node metastasis, and poor survival | PLK1 | Inhibits cell proliferation in vitro | [176] |
| miR-124 | Downregulated in EOC tissues compared to normal ovarian tissues, and lower in metastatic EOC tissues compared to primary EOC tissues | SPHK1 PDCD6 | Inhibits cell proliferation, colony formation, cell invasion, and migration in vitro | [117,177] |
| miR-130b | Downregulated in EOC tissues compared to adjacent non-tumor tissues | ZEB1 STAT3 | Inhibits cell invasion, migration, and EMT in vitro | [178] |
| miR-133a | Downregulated in EOC tissues compared to normal ovarian tissues; negatively associated with late stage and lymph node metastasis | ND | Inhibits cell proliferation, invasion, and migration and induces apoptosis in vitro | [179] |
| miR-133a-3p | Downregulated in EOC tissues compared to adjacent non-tumor tissues | ND | Inhibits cell proliferation, invasion, and EMT in vitro | [180] |
| miR-133b | Downregulated in EOC tissues compared to normal ovarian epithelial tissues and benign ovarian cyst tissues; negatively associated with tumor grade and lymph node metastasis | CTGF | Inhibits cell invasion, migration, and EMT in vitro | [181] |
| miR-135a | Downregulated in EOC tissues compared to ovarian cystadenomas; negatively associated with stage, lymph node metastasis, and poor survival | HOXA10 | Inhibits cell proliferation and adhesion and promotes apoptosis in vitro | [182] |
| miR-135a-3p | Downregulated in EOC tumors compared to paired adjacent non-tumor tissues; negatively correlated with advanced stage and poor OS | CCR2 | Inhibits cell proliferation, migration, invasion, and EMT in vitro and tumor growth and lung metastasis in vivo | [183] |
| miR-137 | Downregulated in EOC tissues compared to paired adjacent tissues | SNAI1 | Inhibits cell invasion, spheroid formation, and EMT in vitro | [175] |
| miR-138 | Downregulated in EOC tissues compared to contralateral normal ovarian tissues; negatively associated with lymph node metastasis | SOX4 HIF1A SOX12 LIMK1 | Inhibits cell proliferation and invasion in vitro and tumor metastasis in the orthotopic xenograft mouse model | [83,184,185] |
| miR-139 | Downregulated in EOC tissues compared to paired adjacent normal tissues | HDGF | Inhibits cell proliferation, migration, and invasion in vitro | [186] |
| miR-139-3p | Downregulated in EOC compared to adjacent normal ovarian; negatively associated with advanced stage, lymph node metastasis, and poor survival | ELAVL1 | Inhibits cell proliferation, colony formation, invasion, and migration in vitro and tumor growth and lung metastasis in vivo | [187] |
| miR-139-5p | Downregulated in EOC tissues compared to precancerous tissues; negatively associated with stage, lymph node metastasis and poor survival | ROCK2 | Inhibits cell proliferation, colony formation, migration, and invasion in vitro and tumor growth in vivo | [188,189] |
| miR-145-5p | Downregulated in EOC tissues compared to paired adjacent normal ovarian tissues | SMAD4 | Inhibits cell proliferation and migration and promotes apoptosis in vitro | [190] |
| miR-148a-3p | Downregulated in EOC tissues compared to adjacent non-tumor tissues | ROCK1 | Inhibits cell proliferation, invasion, and migration in vitro, as well as tumor growth in vivo | [114] |
| miR-152 | Downregulated in EOC tissues compared to paired adjacent normal ovarian tissues | ADAM17 WNT1 ERBB3 | Inhibits cell proliferation, invasion, migration, and EMT in vitro | [178,191] |
| miR-150 | Downregulated in EOC tissues compared to normal ovarian tissues; negatively correlated with advanced tumor stage and grade and poor survival | ZEB1 | Inhibits cell proliferation, invasion, migration, EMT, and spheroid formation in vitro | [133,192] |
| miR-186 | Downregulated in cisplatin-resistant EOC cells compared to cisplatin-sensitive EOC cells; decreased expression is associated poor OS | TWIST1 | Inhibits cell proliferation, invasion, migration, and EMT in vitro and tumor growth and EMT in vivo | [193] |
| miR-193b | Downregulated in EOC compared to matched adjacent normal ovarian tissues and in omental metastasis compared to paired adjacent normal omentum; negatively correlated with stage, grade, ascites, lymph node metastasis, tumor size, and poor survival | uPA | Inhibits cell adhesion, proliferation, colony formation, invasion and migration in vitro, and inhibits tumor growth and metastasis in the orthotopic xenograft mouse model | [194,195] |
| miR-199a-5p | Downregulated in EOC cells under hypoxia compared to in normoxic condition | HIF1A HIF2A | Inhibits cell migration in vitro and inhibits tumor growth and peritoneal seeding in vivo | [94] |
| miR-202-5p | Downregulated in EOC tissues compared to paired adjacent normal ovarian tissues; positively associated with patient survival | HOXB2 | Inhibits cell proliferation, invasion, migration, and EMT in vitro | [196] |
| miR-206 | Downregulated in EOC tissues compared to noncancerous glioma tissues; negatively associated with lymph node and distant metastasis | c-MET CCND1 CCND2 | Suppresses cell proliferation, migration, and invasion in vitro | [197,198] |
| miR-208a-5p | Downregulated in metastatic EOC tissues compared to nonmetastatic EOC tissues | DAAM1 | Inhibits cell invasion, migration, and microfilaments formation in vitro | [82] |
| miR-215 | Downregulated in EOC tissues compared to adjacent normal; negatively associated with stage and lymph node metastasis | NOB1 | Inhibits cell proliferation, colony formation, migration, and invasion in vitro and tumor growth in vivo. | [199] |
| miR-217 | Downregulated in EOC tissues compared to paired adjacent normal ovarian tissues; negatively associated with stage, histological grade, and lymph node metastasis | IGF1R IL6 | Inhibits cell proliferation, colony formation, invasion, and migration, and reduces M0 macrophages differentiation in vitro and tumor growth in vivo | [105,200] |
| miR-218 | Downregulated in EOC tissues compared to adjacent normal; negatively associated with stage and lymph node metastasis | RUNX2 | Inhibits cell proliferation, colony formation, invasion, and migration in vitro and tumor growth in vivo | [201] |
| miR-219-5p | Decreased in EOC tissues compared to adjacent normal tissues | TWIST1 HMGA2 | Inhibits cell proliferation, invasion, and migration in vitro and tumor growth in vivo | [202,203] |
| miR-335 | Downregulated in EOC tissues compared to normal ovarian tissues, in omental metastatic tissues compared to primary tumors; negatively associated with poor survival and recurrence | ND | ND | [204] |
| miR-338-3p | Downregulated in EOC tissues compared to normal fallopian samples based on TCGA database; negatively associated with stage, grade, and metastasis | MACC1 RUNX2 | Inhibits cell proliferation, colony formation, invasion, migration, and EMT in vitro and tumor growth and metastasis in vivo | [70,205,206] |
| miR-340 | Downregulated in EOC tissues compared to normal adjacent ovarian | FHL2 NFKB1 | Inhibits cell proliferation, invasion, and migration in vitro and tumor growth and intraperitoneal metastasis in vivo | [207,208] |
| miR-363 | Downregulated in EOC compared to paired adjacent normal ovarian tissues; negatively associated with advanced stage, lymph node metastasis, and poor prognosis | NOB1 | Inhibits cell proliferation, colony formation, invasion, and migration in vitro and tumor growth in vivo | [209] |
| miR-365 | Downregulated in EOC tissues compared to adjacent normal ovarian tissues; negatively associated with stage, grade, and lymph node metastasis | WNT5A | Inhibits cell proliferation, colony formation, invasion, migration, and EMT in vitro, and tumor growth in vivo | [210] |
| miR-373 | Downregulated in EOC tumors compared to benign epithelial ovarian tumors | RAB22A | Inhibits cell invasion, migration and EMT in vitro and peritoneal metastasis in vivo | [211,212] |
| miR-375 | Downregulated EOC tissues compared to normal ovarian tissues | YAP1 | Inhibits cell proliferation, invasion, migration, and EMT in vitro and tumor growth, metastasis, and EMT in vivo | [115] |
| miR-377 | Downregulated in EOC tissues compared to adjacent normal ovarian tissues; positively correlated with survival | CUL4A | Suppresses cell proliferation, invasion, migration, and EMT in vitro | [213] |
| miR-494 | Downregulated in EOC tissues compared to normal adjacent tissues; negatively associated with stage, tumor size, and lymph node metastasis | SIRT1 | Inhibits cell proliferation, invasion, and migration in vitro | [214] |
| miR-378g | Downregulated in EOC tissues compared to normal ovarian tissues | CHI3L1 | Inhibits cell proliferation, migration, and TGF-β1-induced EMT in vitro | [215] |
| miR-421 | ND | PDGFRA | Inhibits cell proliferation, invasion, and tubule formation in vitro, and tumor growth and angiogenesis in vivo | [216] |
| miR-448 | Downregulated in EOC tissues compared to adjacent normal ovarian tissues | CXCL12 | Inhibits cell proliferation, migration and invasion in vitro and tumor growth in vivo | [99] |
| miR-450a | Downregulated in EOC tissues compared to normal ovarian tissues | TIMMDC1 MT-ND2 ACO2 ATP5B | Inhibits cell clonogenicity, invasion, migration, and EMT and promotes anoikis in vitro and intraperitoneal tumor growth in vivo | [217] |
| miR-455 | Downregulated in EOC tissues compared to normal adjacent tissues; negatively associated with stage, tumor size, and lymph node metastasis | NOTCH1 | Inhibits cell proliferation and invasion in vitro | [218] |
| miR-489 | Downregulated in EOC tissues compared to normal ovarian tissues; negatively associated with poor survival, stage, grade, lymph node, and distant metastasis | XIAP | Inhibits cell proliferation, invasion, migration and EMT in vitro | [219] |
| miR-497 | Downregulated in EOC tissues compared to normal ovarian tissues; negatively associated with tumor stage, lymph node metastasis, and poor survival | SMURF1 | Inhibition of cell migration and invasion in vitro | [220] |
| miR-503-5p | Downregulated in response to NF-kB pathway activation in SKOV3 cells | CD97 | Inhibits colony formation, migration and invasion in vitro | [221] |
| miR-506 | Decreased expression is associated with stage and poor survival in EOC | SNAI2 VIM CDH2 | Inhibits cell invasion, migration, and EMT in vitro, and EMT, tumor growth, and metastasis in the orthotopic xenograft mouse model | [63,222] |
| miR-532-5p | Downregulated in EOC compared to normal ovarian; negatively associated with stage, grade, and distant metastasis | TWIST1 | Inhibits cell proliferation, colony formation, and invasion in vitro | [223] |
| miR-548c | Decreased in EOC tissues compared to normal ovarian tissues; negatively associated with advanced stage and poor survival | TWIST1 | Inhibits cell proliferation, migration, invasion, stemness, and EMT in vitro | [224] |
| miR-584 | Downregulated in EOC tissues compared to paracancerous tissues; negatively associated with distant and lymphatic metastasis and poor survival | LPIN1 | Inhibits cell proliferation, colony formation, and migration in vitro | [225] |
| miR-596 | ND | LETM1 | Inhibits cell proliferation, invasion, and migration in vitro and tumor growth in vivo | [226] |
| miR-612 | Downregulated in EOC tissues compared to matched non-tumor tissues | HOXA13 | Inhibits cell proliferation, invasion, and migration, and promotes apoptosis in vitro | [227] |
| miR-654-3p | Downregulated in EOC cells compared to IOSE80 cells | AKT3 | Inhibits cell invasion, migration and sphere formation in vitro and tumor growth in vivo | [228] |
| miR-665 | Decreased in EOC tissues compared to normal ovarian tissues | HOXA10 | Inhibits cell proliferation and migration in vitro | [229] |
| miR-708 | Downregulated in EOC tissues compared to normal ovarian tissues; negatively associated with stage | RAP1B | Inhibits cell invasion, migration, cell adhesion, and EMT in vitro and tumor growth and metastasis in vivo | [81] |
| miR-874-3p/5p | Downregulated in EOC tissues compared to normal ovarian epithelial tissues | SIK2 | Inhibits cell proliferation, colony formation, invasion, and migration in vitro | [230] |
| miR-936 | Downregulated in EOC tissues compared to adjacent normal tissues; negatively associated with tumor size, stage, and lymphatic metastasis | FGF2 | Inhibits cell proliferation, invasion, and migration in vitro and tumor growth in vivo. | [231] |
| miR-4454 | Downregulated in metastatic EOC tissues compared to primary EOC tissues; positively associated with patient survival | SPARC BAG5 | Inhibits cell proliferation, colony formation, migration, and invasion in vitro, and peritoneal metastasis in vivo. | [232] |
| miR-6089 | Downregulated in EOC tissues compared to paratumor tissues | MYH9 | Suppresses cell proliferation, migration, invasion, and EMT in vitro, and tumor growth and metastasis in vivo | [233] |
| miR-6126 | Downregulated in EOC tissues compared to normal ovarian tissues; inversely correlated with tumor progression and positively associated with survival | ITGB1 | Inhibits cell invasion, migration, and tube formation in vitro and angiogenesis and tumor growth in the orthotopic xenograft mouse model | [234] |
| miRNAs down-regulated in secreted exosomes and circulating body fluids | ||||
| miR-125a | Downregulated in serum of EOC patients compared to healthy controls | GALNT14 ARID3B | Inhibits cell proliferation, invasion, and EMT in vitro | [235,236] |
| miR-125b | Downregulated in tissues and serum of EOC patients compared to adjacent normal ovarian tissues and serum of healthy control respectively; negatively associated with stage and lymph node and distant metastasis | SET GAB2 | Inhibits cell invasion, migration, and EMT in vitro and metastasis in vivo | [113,237,238] |
| miR-212 | Downregulated in EOC tissues and serum compared to paired adjacent normal ovarian tissues and healthy controls respectively; negatively correlated with tumor stage and metastasis | HBEGF | Inhibits cell proliferation, invasion, and migration in vitro | [239] |
| miR-122 | Downregulated in serum of serous EOC patients compared to benign controls | P4HA1 | Inhibits cell invasion, migration, and EMT in vitro, and intraperitoneal metastasis in vivo | [127,240] |
| miR-148a | Downregulated in plasma of EOC patients compared to healthy controls; negatively associated with tumor grade, stage, lymph node metastasis, and poor survival | ND | Inhibits cell proliferation, invasion, and migration in vitro | [241] |
| miR-199a | Downregulated in serum of EOC patients compared to normal controls; negatively associated with tumor grade, lymph node and distant metastasis, and poor survival | ND | ND | [128] |
| miRNAs with both pro- and anti-metastatic effects | ||||
| miR-9 | Upregulated in metastatic EOC tissues compared to paired primary EOC tissues | CDH1 | Promotes cell migration, invasion, and EMT in vitro | [242] |
| Downregulated in recurrent serous EOC tissues compared to primary EOC tissues | TLN1 | Inhibits cell proliferation, migration, and invasion in vitro | [243,244] | |
| miR-141 | Upregulated in EOC tissues compared to normal ovarian tissues, and serum of EOC patients compared to healthy controls; positively associated with tumor stage, lymph node metastasis, and poor survival | KLF12 SIK1 MAPK14 | Promotes proliferation, anoikis resistance, and survival in vitro, and tumor growth and metastasis in vivo | [126,245,246] |
| Downregulated in integrated mesenchymal subtype of EOC compared to normal ovarian epithelial tissues | ND | Inhibits cell migration, invasion, and EMT in vitro | [222,247] | |
| miR-200a | Upregulated in EOC tumors compared to normal ovarian tissues and in serum of EOC patients compared to healthy controls; positively associated with aggressiveness, late stage, and lymph node metastasis | PTEN MAPK14 | Promotes cell migration and invasion in vitro | [126,246,248] |
| Downregulated in vasculogenic mimicry positive EOC tissues compared to vascular mimicry negative ovarian tissues and in integrated mesenchymal subtype compared to ovarian normal epithelial tissues; negatively associated with stage, ascites, and metastasis and positively correlated with patient survival | IL8 CXCL1 EPHA2 | Inhibits tube formation, vasculogenic mimicry, and cell invasion in vitro, and angiogenesis and metastasis in vivo | [222,249,250] | |
| miR-200b | Upregulated serum of EOC patients compared to healthy controls; positively associated with tumor stage, lymph node metastasis, and poor survival | ND | ND | [126] |
| Downregulated in vasculogenic mimicry positive EOC tissues compared to vascular mimicry negative ovarian tissues; negatively associated with stage, ascites, and metastasis and positively correlated with patient survival | IL8 CXCL1 | Inhibits tube formation, vasculogenic mimicry, and cell invasion in vitro, and angiogenesis and metastasis in vivo | [249] | |
| miR-200c | Upregulated in SOC tissues and serum of EOC patients compared to heathy controls; associated with tumor stage, lymph node metastasis, and poor survival | ND | ND | [126,251] |
| Upregulated in EOC tumors compared to normal ovarian tissues; inversely associated with tumor stage and lymph node metastasis | ZEB2 TUBB3 | Inhibits cell migration and invasion in vitro | [252,253] | |
| miR-203 | Upregulated in EOC tissues compared with adjacent normal tissues | PDHB | Promotes cell proliferation, migration, and glycolysis in vitro, and tumor metastasis in vivo | [118] |
| Downregulated in SOC tissues compared to adjacent normal ovarian tissue; positively associated with patient survival | SNAI2 BIRC5 | Inhibits cell proliferation, invasion, migration, and EMT in vitro, and tumor growth in vivo | [119,254] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, V.H.L.; Yue, C.; Du, K.Y.; Salem, M.; O'Brien, J.; Peng, C. The Role of microRNAs in Epithelial Ovarian Cancer Metastasis. Int. J. Mol. Sci. 2020, 21, 7093. https://doi.org/10.3390/ijms21197093
Nguyen VHL, Yue C, Du KY, Salem M, O'Brien J, Peng C. The Role of microRNAs in Epithelial Ovarian Cancer Metastasis. International Journal of Molecular Sciences. 2020; 21(19):7093. https://doi.org/10.3390/ijms21197093
Chicago/Turabian StyleNguyen, Vu Hong Loan, Chenyang Yue, Kevin Y. Du, Mohamed Salem, Jacob O'Brien, and Chun Peng. 2020. "The Role of microRNAs in Epithelial Ovarian Cancer Metastasis" International Journal of Molecular Sciences 21, no. 19: 7093. https://doi.org/10.3390/ijms21197093
APA StyleNguyen, V. H. L., Yue, C., Du, K. Y., Salem, M., O'Brien, J., & Peng, C. (2020). The Role of microRNAs in Epithelial Ovarian Cancer Metastasis. International Journal of Molecular Sciences, 21(19), 7093. https://doi.org/10.3390/ijms21197093





