Comparative Cytological and Transcriptome Analysis Revealed the Normal Pollen Development Process and Up-Regulation of Fertility-Related Genes in Newly Developed Tetraploid Rice
Abstract
1. Introduction
2. Results
2.1. Breeding Procedure of New Tetraploid Rice Huaduo1
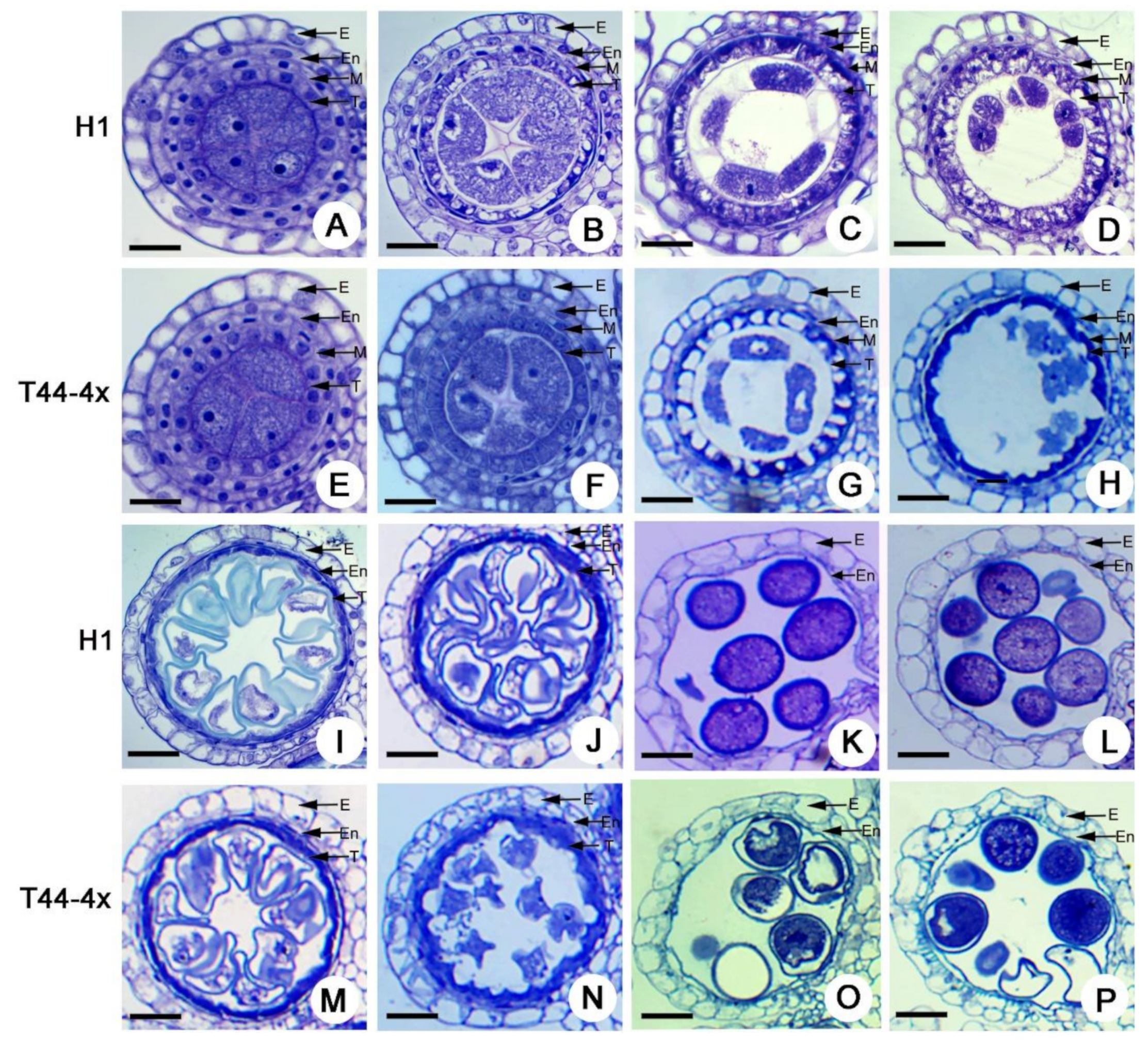
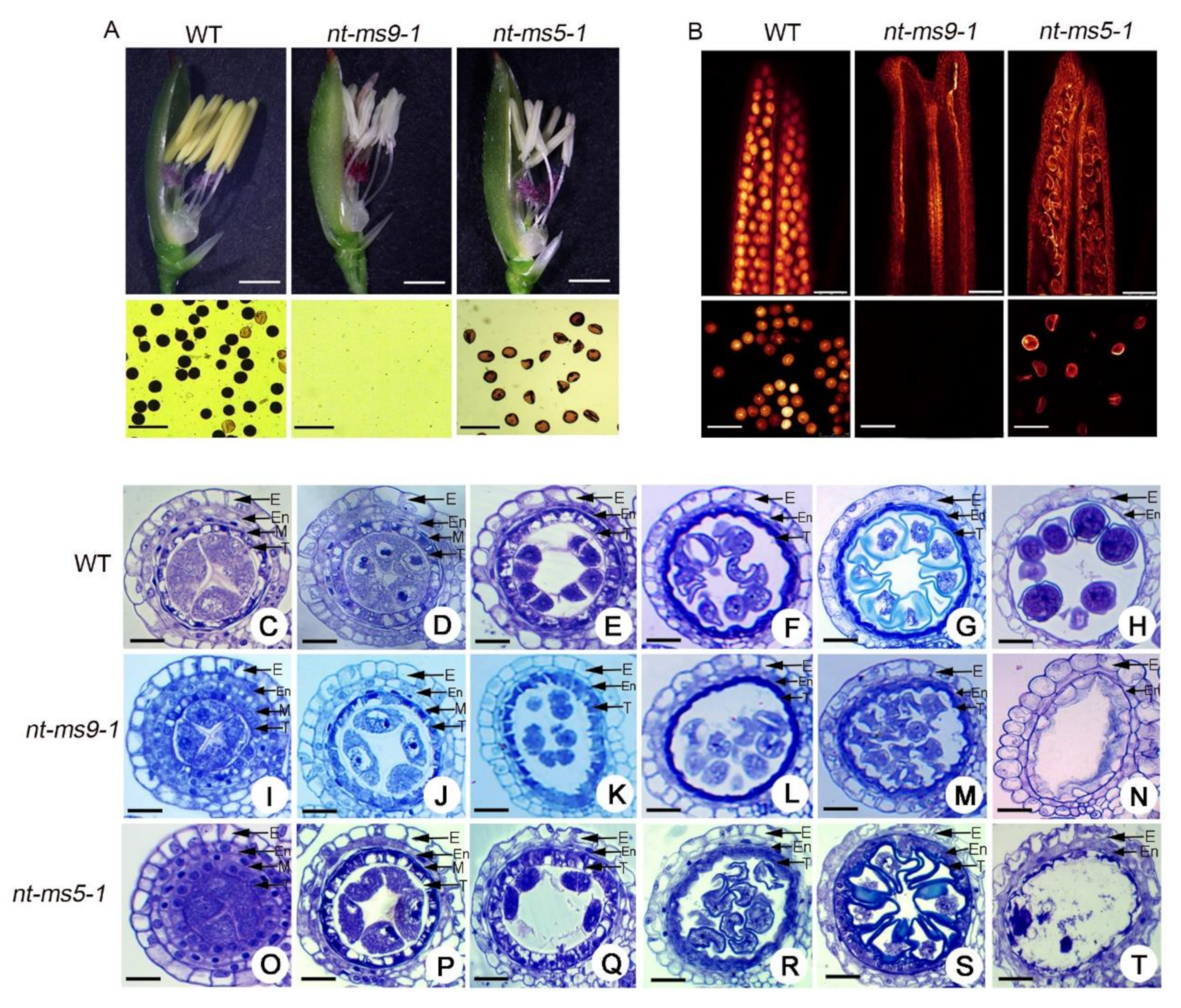
2.2. Comparison of Anther Development between Huaduo1 and Its Parents
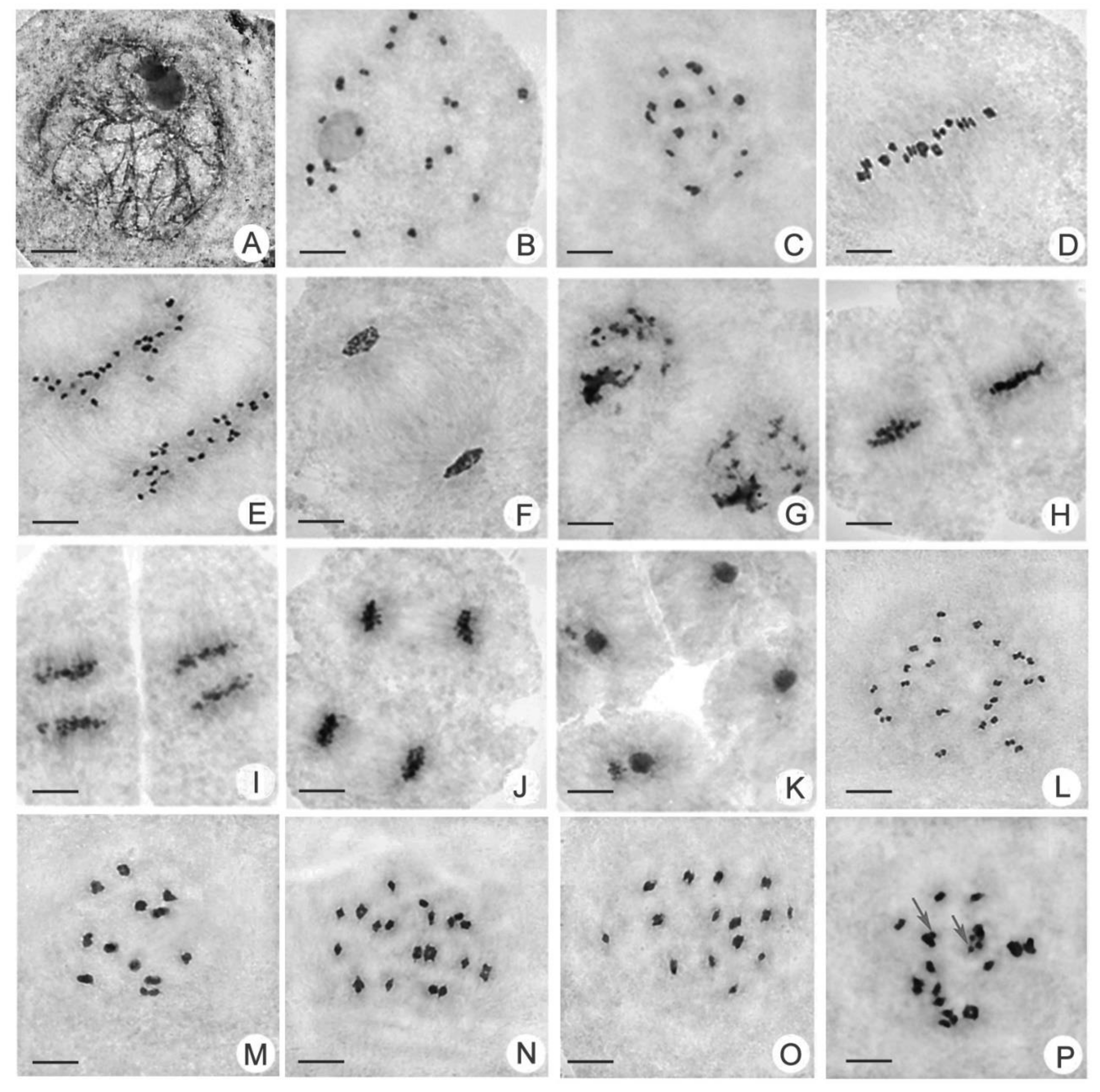
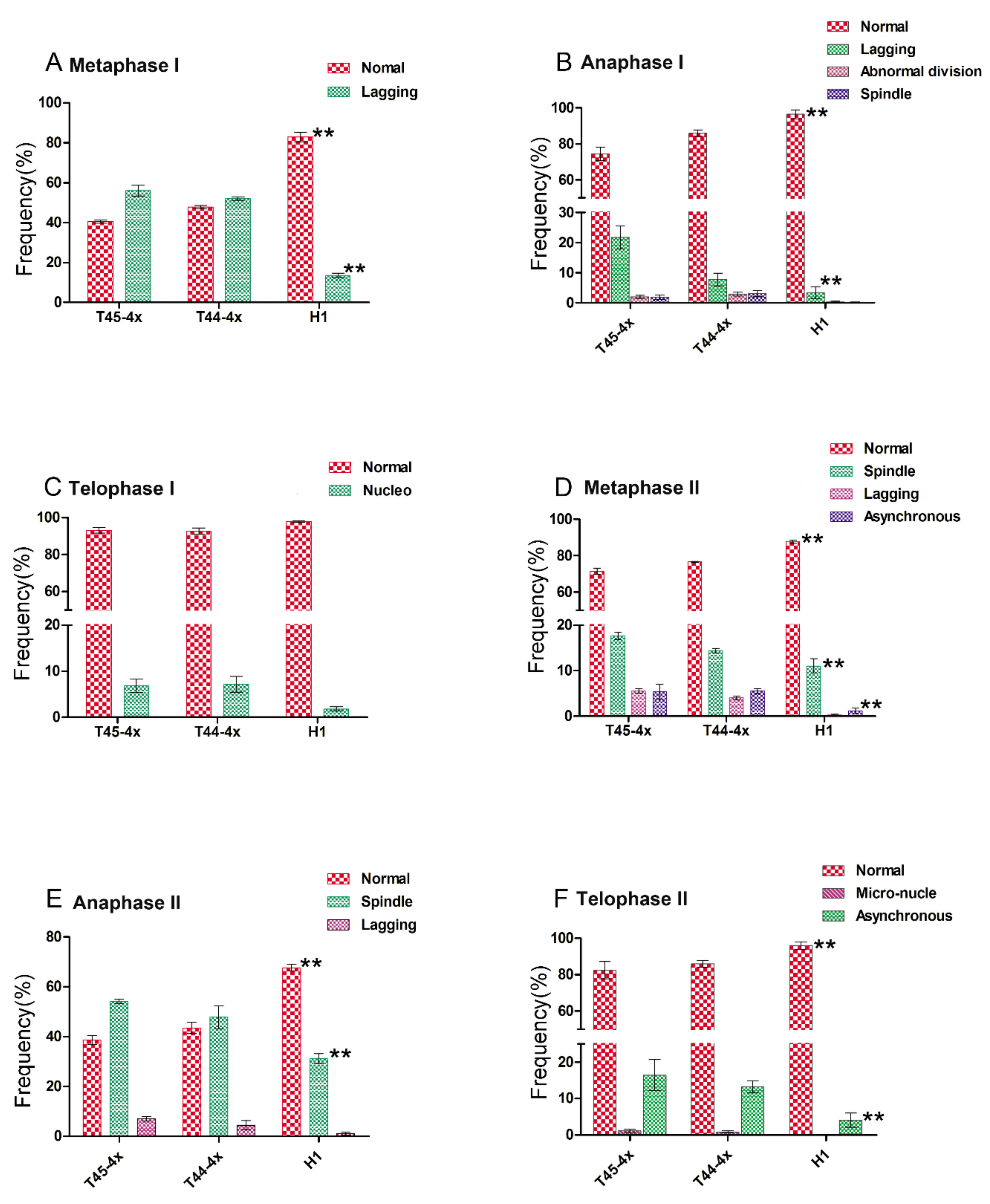
2.3. Comparison of Chromosomal Behavior between Huaduo1 and Its Parents
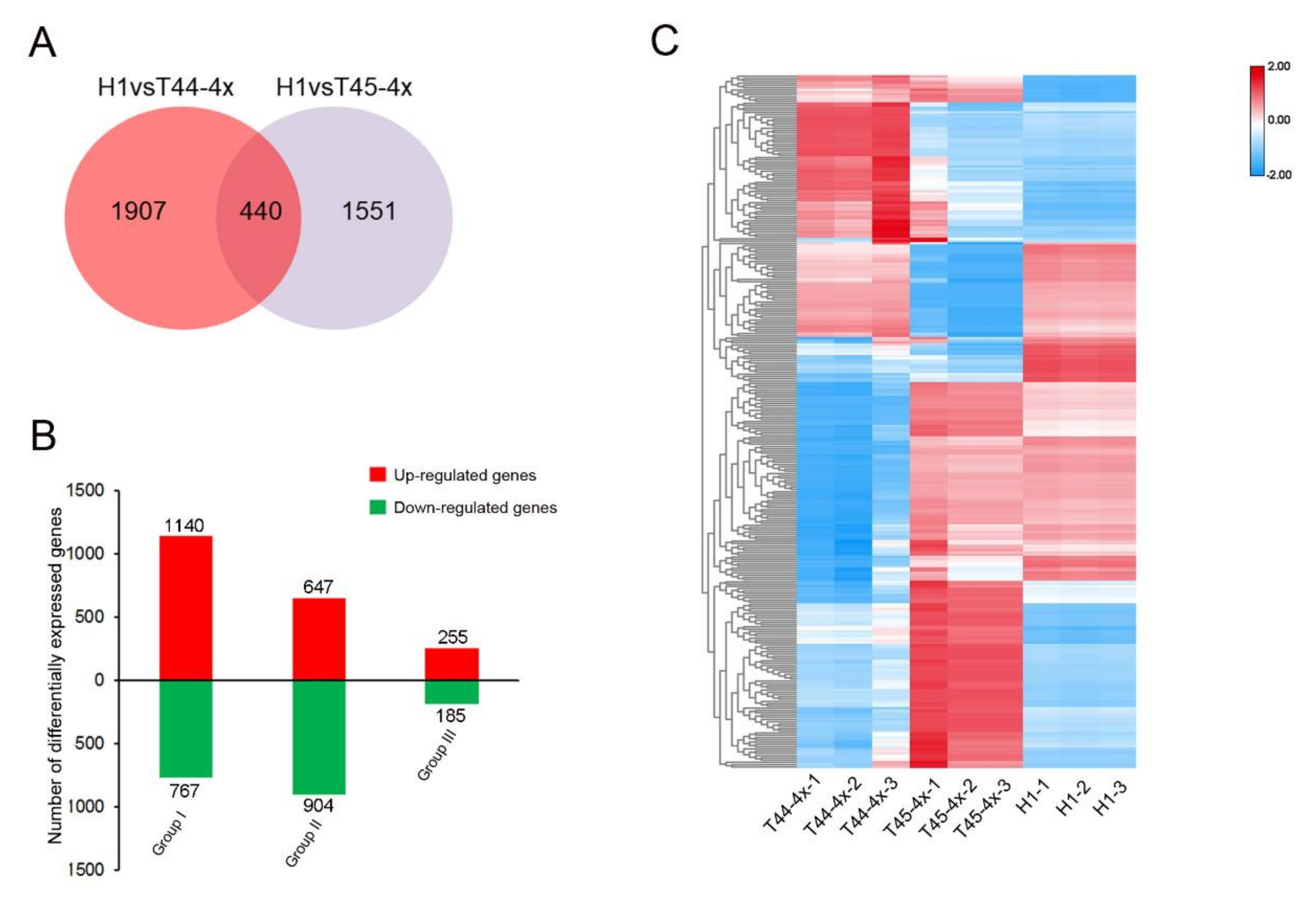
2.4. Comparative Transcriptome Analysis of Huaduo1 and Its Parents in Meiosis
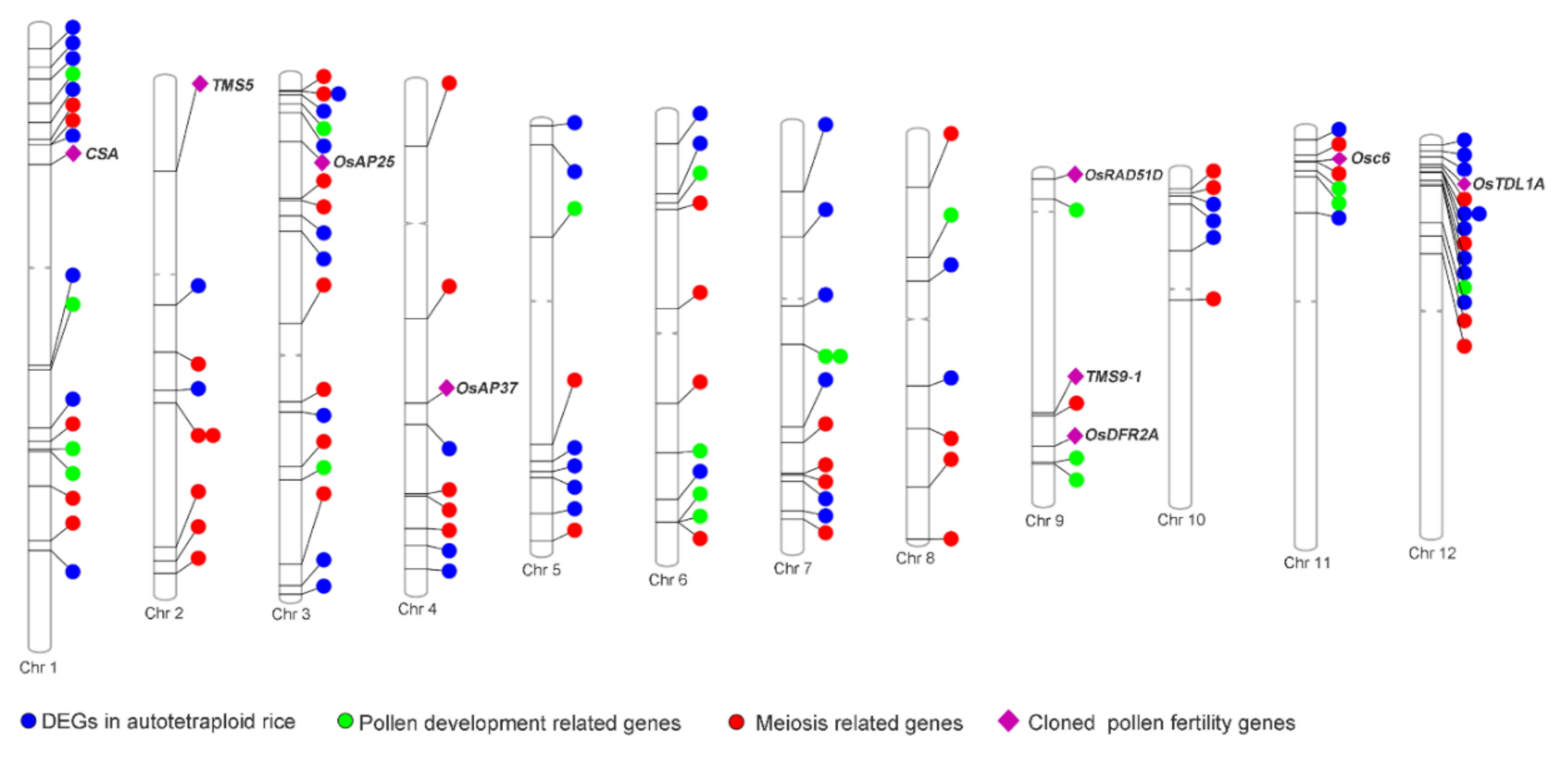
2.5. Up-Regulation of Pollen Fertility-Related Genes in Huaduo1 Compared with Its Two Parents
2.6. Knock-Out of Candidate Genes Causes Pollen Abortion in Huaduo1
3. Discussion
3.1. Significant Phenotypic Variation Exists in Huaduo1 Compared with Its Two Parents
3.2. Upregulation of Pollen Fertility Genes May a Play Critical Role in the High Pollen Fertility in Huaduo1
4. Materials and Methods
4.1. Plant Material
4.2. Analysis of Agronomic Traits and Heterosis
4.3. Pollen Fertility, Semi-Thin Section Analysis
4.4. Chromosome Behavior Observation
4.5. WE-CLSM Analysis
4.6. Sample Preparation, RNA Extraction, RNA-Sequencing, and qRT-PCR Verification
4.7. Generation and Mutation Sequencing of Mutant Plants in Huaduo1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PMC | Pollen mother cell |
| DEGs | Differentially expressed genes |
| PVP | Protection for new varieties of plants |
| GO | Gene ontology |
| TIGR | The Institute for Genomic Research Rice Genome Annotation |
| GEO | Gene Expression Omnibus |
| WE-CLSM | Whole-mount eosin B-staining confocal laser scanning microscopy |
| UbL40 | Ubiquitin fusion ribosomal protein L40 |
| PH | Plant height |
| PL | Panicle length |
| EPN | Effective panicle number |
| TGP | Total number of grains per plant |
| SS | Seed set ratio |
| HPH | High-parent heterosis |
| MPH | Mid-parent heterosis |
| FAA | Formalin–acetic acid–alcohol |
| PCR | Polymerase chain reaction |
References
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef] [PubMed]
- Masterson, J. Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science 1994, 264, 421–424. [Google Scholar] [CrossRef]
- Bingham, E.T.; Groose, R.W.; Woodfield, D.R.; Kidwell, K.K. Complementary gene interactions in alfalfa are greater in autotetraploids than diploids. Crop Sci. 1994, 34, 823–829. [Google Scholar] [CrossRef]
- Marhold, K.; Lihová, J. Polyploidy, hybridization and reticulate evolution: Lessons from the Brassicaceae. Plant Syst. Evol. 2006, 259, 143–174. [Google Scholar] [CrossRef]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17. [Google Scholar] [CrossRef]
- Shahid, M.Q.; Sun, J.; Wei, C.; Zhang, P.; Liu, X.D. Studies on the abnormality of embryo sac and pollen fertility in autotetraploid rice during different growing seasons. Pak. J. Bot. 2010, 42, 7–19. [Google Scholar]
- He, J.H.; Shahid, M.Q.; Li, Y.J.; Guo, H.B.; Cheng, X.A.; Liu, X.D.; Lu, Y.G. Allelic interaction of F1 pollen sterility loci and abnormal chromosome behaviour caused pollen sterility in intersubspecific autotetraploid rice hybrids. J. Exp. Bot. 2011, 62, 4433–4445. [Google Scholar] [CrossRef]
- Wu, J.W.; Hu, C.Y.; Shahid, M.Q.; Guo, H.B.; Lu, Y.G. Analysis on genetic diversification and heterosis in autotetraploid rice. Springerplus 2013, 2, 1–12. [Google Scholar] [CrossRef]
- Wu, J.W.; Shahid, M.Q.; Guo, H.B.; Yin, W.; Chen, Z.X.; Wang, L.; Liu, X.D.; Lu, Y.G. Comparative cytological and transcriptomic analysis of pollen development in autotetraploid and diploid rice. Plant Reprod. 2014, 27, 181–196. [Google Scholar] [CrossRef]
- Chen, L.; Shahid, M.Q.; Wu, J.W.; Chen, Z.X.; Liu, X.D. Cytological and transcriptome analyses reveal abrupt gene expression for meiosis and saccharide metabolisms that associated with pollen abortion in autotetraploid rice. Mol. Genet. Genom. 2018, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- He, J.H.; Shahid, M.Q.; Chen, Z.X.; Chen, X.A.; Liu, X.D.; Lu, Y.G. Abnormal PMC microtubule distribution pattern and chromosome behavior resulted in low pollen fertility of an intersubspecific autotetraploid rice hybrid. Plant Syst. Evol. 2011, 291, 257–265. [Google Scholar] [CrossRef]
- Wu, J.W.; Shahid, M.Q.; Chen, L.; Chen, Z.X.; Wang, L.; Liu, X.D.; Lu, Y.G. Polyploidy enhances F1 pollen sterility loci interactions that increase meiosis abnormalities and pollen sterility in autotetraploid rice. Plant Physiol. 2015, 169, 2700–2717. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.; Chen, L.; Shahid, M.Q.; Chen, M.Y.; Dong, Q.L.; Li, J.R.; Xu, X.S.; Liu, X.D. Pervasive interactions of Sa and Sb loci cause high pollen sterility and abrupt changes in gene expression during meiosis that could be overcome by double neutral genes in autotetraploid rice. Rice 2017, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, H.; Jiao, Y.M.; Shahid, M.Q.; Wu, J.W.; Liu, X.D. Genome-wide analysis of DNA polymorphisms, the methylome and transcriptome revealed that multiple factors are associated with low pollen fertility in autotetraploid rice. PLoS ONE 2018, 13, e0201854. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.; Shahid, M.Q.; Chen, M.Y.; Li, X.; Li, J.R.; Xu, X.S.; Du, S.S.; Liu, X.D. Cytological and transcriptome analysis reveal that interaction at Sb pollen sterility locus cause down-regulation of important meiosis-related genes associated with high pollen sterility in autotetraploid rice hybrids. Plant Physiol. Biochem. 2019, 141, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shahid, M.Q.; Wu, J.W.; Wang, L.; Liu, X.D.; Lu, Y.G. Comparative small RNA analysis of pollen development in autotetraploid and diploid rice. Int. J. Mol. Sci. 2016, 17, 499. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.B.; Mendrikahy, J.N.; Xie, L.; Deng, J.F.; Lu, Z.J.; Wu, J.W.; Li, X.; Shahid, M.Q.; Liu, X.D. Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci. Rep. 2017, 7, 40139. [Google Scholar] [CrossRef]
- Bei, X.J.; Shahid, M.Q.; Wu, J.W.; Chen, Z.X.; Wang, L.; Liu, X.D. Re-sequencing and transcriptome analysis reveal rich DNA variations and differential expressions of fertility-related genes in neo-tetraploid rice. PLoS ONE 2019, 14, e0214953. [Google Scholar] [CrossRef]
- Ghaleb, M.A.A.; Li, C.; Shahid, M.Q.; Yu, H.; Liang, J.H.; Chen, R.X.; Wu, J.W.; Liu, X.D. Heterosis analysis and underlying molecular regulatory mechanism in a wide-compatible neo-tetraploid rice line with long panicles. BMC Plant Biol. 2020, 20, 83. [Google Scholar] [CrossRef]
- Zheng, Y.; Haage, K.; Streit, V.E.; Gierl, A.; Ruiz, R.A.T. A large number of tetraploid Arabidopsis thaliana lines, generated by a rapid strategy, reveal high stability of neo-tetraploids during consecutive generations. Theor. Appl. Genet. 2009, 118, 1107–1119. [Google Scholar]
- Yu, H.; Shahid, M.Q.; Li, Q.H.; Li, Y.D.; Li, C.; Lu, Z.J.; Wu, J.W.; Zhang, Z.M.; Liu, X.D. Production assessment and genome comparison revealed high yield potential and novel specific alleles associated with fertility and yield in neo-tetraploid rice. Rice 2020, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.N.; Campbell, M.; Childs, K.; Nissen, T.F.; Malek, R.L.; Lee, Y.D.; Zheng, L.; et al. The TIGR rice genome annotation resource: Improvements and new features. Nucleic Acids Res. 2007, 35, 883–887. [Google Scholar] [CrossRef]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro Version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2012, 41, 1206–1213. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Horiuchi, Y.; Ueda, Y.; Mizuta, Y.; Kubo, T.; Yano, K.; Yamaki, S.; Tsuda, K.; Nagata, T.; Niihama, M.; et al. Rice expression atlas in reproductive development. Plant Cell Physiol. 2010, 51, 2060–2081. [Google Scholar] [CrossRef]
- Deveshwar, P.; Bovill, W.D.; Sharma, R.; Able, J.A.; Kapoor, S. Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biol. 2011, 11, 78. [Google Scholar] [CrossRef]
- Yant, L.; Hollister, J.D.; Wright, K.M.; Arnold, B.J.; Higgins, J.D.; Franklin, F.C.H.; Bomblies, K. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 2013, 23, 2151–2156. [Google Scholar] [CrossRef]
- Wright, K.M.; Arnold, B.; Xue, K.; Urinova, M.; Connell, J.O.; Bomblies, K. Selection on meiosis gnes in diploid and tetraploid Arabidopsis arenosa. Mol. Biol. Evol. 2015, 32, 944–955. [Google Scholar] [CrossRef]
- Byun, M.Y.; Kim, W.T. Suppression of OsRAD51D results in defects in reproductive development in rice (Oryza sativa L.). Plant J. 2014, 79, 256–269. [Google Scholar] [CrossRef]
- Hong, L.; Tang, D.; Shen, Y.; Hu, Q.; Wang, K.; Li, M.; Lu, T.; Cheng, Z. MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytol. 2012, 196, 402–413. [Google Scholar] [CrossRef]
- Zhao, X.; Palma, J.D.; Oane, R.; Gamuyao, R.; Bennett, J. OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J. 2010, 54, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Liang, W.; Yang, X.; Jin, W.; Wilson, Z.A.; Hu, J.; Zhang, D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 2013, 4, 1445. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, S.; Kifuji, Y.; Komiya, R.; Kitashiba, H. Identification of a single nucleotide deletion causing frameshift mutation in OsDFR2A in a genic male sterile mutant of rice and its possible application to F1 hybrid breeding. Mol. Breed. 2013, 31, 805–814. [Google Scholar] [CrossRef]
- Zhang, D.S.; Liang, W.Q.; Yin, C.S.; Zong, J.; Gu, F.W.; Zhang, D.B. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 2010, 154, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Liu, Q.; Zhang, L.; Mao, B.; Yan, D.; Jin, Q.; He, Z. Fine mapping and candidate gene analysis of the novel thermo-sensitive genic male sterility tms9-1 gene in rice. Theor. Appl. Genet. 2014, 127, 1173. [Google Scholar] [CrossRef]
- Li, H.; Yuan, Z.; Vizcay-Barrena, G.; Yang, C.; Liang, W.; Zong, J.; Wilson, Z.A.; Zhang, D.B. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 2011, 156, 615–630. [Google Scholar] [CrossRef]
- Yang, Z.F.; Liu, L.; Sun, L.P.; Yu, P.; Zhang, P.P.; Abbas, A.; Xiang, X.J.; Wu, W.X.; Zhang, Y.X.; Cao, L.Y.; et al. OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 2019, 99, 175–191. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, W.Q.; Yang, X.J.; Luo, X.; Jiang, N.; Ma, H.; Zhang, D.B. Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 2010, 22, 672–689. [Google Scholar] [CrossRef]
- Zhou, H.; Zhou, M.; Yang, Y.; Li, J.; Zhu, L.; Jiang, D.; Dong, J.; Liu, Q.; Gu, L.; Zhou, L.; et al. RNase Z (S1) processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat. Commun. 2014, 5, 4884. [Google Scholar] [CrossRef]
- Tu, S.; Luan, L.; Liu, Y.; Long, W.; Kong, F.; He, T.; Xu, Q.; Yan, W.; Yu, M. Production and Heterosis analysis of rice autotetraploid hybrids. Crop Sci. 2007, 47, 2356–2363. [Google Scholar] [CrossRef]
- Shahid, M.Q.; Liu, G.; Li, J.; Naeem, M.; Liu, X.D. Heterosis and gene action study of agronomic traits in diploid and autotetraploid rice. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2011, 61, 23–32. [Google Scholar] [CrossRef]
- Chen, L.; Yuan, Y.; Wu, J.W.; Chen, Z.X.; Liu, X.D. Carbohydrate metabolism and fertility related genes high expression levels promote heterosis in autotetraploid rice harboring double neutral genes. Rice 2019, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Hollister, J.D.; Arnold, B.J.; Svedin, E.; Xue, K.S.; Dilkes, B.P.; Bomblies, K. Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa. PLoS Genet 2012, 8, e1003093. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.Q.; Li, Y.J.; Saleem, M.F.; Naeem, M.; Wei, C.M.; Liu, X.D. Yield and yield components in autotetraploid and diploid rice genotypes (indica and japonica) sown in early and late seasons. Aust. J. Crop Sci. 2013, 7, 632–641. [Google Scholar]
- Zeng, Y.X.; Hu, C.Y.; Lu, Y.G.; Li, J.Q.; Liu, X.D. Diversity of abnormal embryo sacs in indica/japonica hybrids in rice demonstrated by confocal microscopy of ovaries. Plant Breed. 2010, 126, 574–580. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xie, X.R.; Ma, X.L.; Zhu, Q.L.; Zeng, D.C.; Li, G.S.; Liu, Y.G. CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 2017, 10, 1246–1249. [Google Scholar] [CrossRef]
- Ma, K.; Han, J.L.; Yao, Y.; Yang, Z.F.; Chen, J.Y.; Liu, Y.G.; Zhu, Q.L.; Chen, L.T. An effective strategy to establish a male sterility mutant mini-library by CRISPR/Cas9-mediated knockout of anther-specific genes in rice. J. Genet. Genom. 2019, 20, 273–275. [Google Scholar] [CrossRef]


| Seasons | Material Name | PH (cm) | EPN | PL (cm) | FG | TGP | SS (%) | GWT (g) |
|---|---|---|---|---|---|---|---|---|
| Late season | T45-4x | 94.40 ± 1.19 ** | 3.40 ± 0.27 ** | 25.97 ± 0.60 | 63.27 ± 4.78 | 120.90 ± 9.17 * | 53.27 ± 2.34 ** | 30.74 ± 1.39 * |
| T44-4x | 78.10 ± 1.58 ** | 5.65 ± 0.44 | 23.53 ± 0.33 | 15.58 ± 1.41 ** | 55.89 ± 1.94 * | 26.45 ± 1.88 ** | 31.39 ± 0.46 | |
| H1 | 104.40 ± 0.93 | 6.60 ± 0.40 | 25.46 ± 0.52 | 66.34 ± 4.63 | 87.02 ± 6.34 | 76.46 ± 1.43 | 35.44 ± 0.34 | |
| Early season | T45-4x | 117.70 ± 0.64 ** | 4.45 ± 0.28 | 31.74 ± 0.46 ** | 40.26 ± 1.33 ** | 132.38 ± 4.05 | 30.80 ± 0.97 ** | 30.82 ± 1.04 ** |
| T44-4x | 96.05 ± 0.46 ** | 5.85 ± 0.27 * | 28.81 ± 0.34 | 30.17 ± 0.65 ** | 77.90 ± 0.55 ** | 39.16 ± 7.16 ** | 33.74 ± 0.49 | |
| H1 | 124.45 ± 0.84 | 4.95 ± 0.22 | 28.09 ± 0.24 | 92.75 ± 3.57 | 120.41 ± 4.23 | 76.55 ± 0.84 | 33.86 ± 0.29 |
| Material Name | Pollen Fertility (%±SE) | Typical Aborted Pollen (%±SE) | Stained Aborted Pollens (%±SE) | Small Pollen (%±SE) |
|---|---|---|---|---|
| T45-4x | 54.10 ± 3.81 | 8.18 ± 0.72 | 37.26 ± 3.92 | 0.46 ± 0.12 |
| T44-4x | 57.66 ± 2.78 | 15.08 ± 1.56 | 23.07 ± 1.98 | 4.19 ± 0.51 ** |
| H1 | 95.62 ± 0.34 ** | 3.16 ± 0.34 ** | 0.60 ± 0.13 ** | 0.62 ± 0.12 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Chen, Y.; Lin, H.; Chen, Y.; Yu, H.; Lu, Z.; Li, X.; Zhou, H.; Chen, Z.; Liu, X. Comparative Cytological and Transcriptome Analysis Revealed the Normal Pollen Development Process and Up-Regulation of Fertility-Related Genes in Newly Developed Tetraploid Rice. Int. J. Mol. Sci. 2020, 21, 7046. https://doi.org/10.3390/ijms21197046
Wu J, Chen Y, Lin H, Chen Y, Yu H, Lu Z, Li X, Zhou H, Chen Z, Liu X. Comparative Cytological and Transcriptome Analysis Revealed the Normal Pollen Development Process and Up-Regulation of Fertility-Related Genes in Newly Developed Tetraploid Rice. International Journal of Molecular Sciences. 2020; 21(19):7046. https://doi.org/10.3390/ijms21197046
Chicago/Turabian StyleWu, Jinwen, Yuanmou Chen, Hong Lin, Yang Chen, Hang Yu, Zijun Lu, Xiang Li, Hai Zhou, Zhixiong Chen, and Xiangdong Liu. 2020. "Comparative Cytological and Transcriptome Analysis Revealed the Normal Pollen Development Process and Up-Regulation of Fertility-Related Genes in Newly Developed Tetraploid Rice" International Journal of Molecular Sciences 21, no. 19: 7046. https://doi.org/10.3390/ijms21197046
APA StyleWu, J., Chen, Y., Lin, H., Chen, Y., Yu, H., Lu, Z., Li, X., Zhou, H., Chen, Z., & Liu, X. (2020). Comparative Cytological and Transcriptome Analysis Revealed the Normal Pollen Development Process and Up-Regulation of Fertility-Related Genes in Newly Developed Tetraploid Rice. International Journal of Molecular Sciences, 21(19), 7046. https://doi.org/10.3390/ijms21197046






