Expression of a Large Single-Chain 13F6 Antibody with Binding Activity against Ebola Virus-Like Particles in a Plant System
Abstract
1. Introduction
2. Results
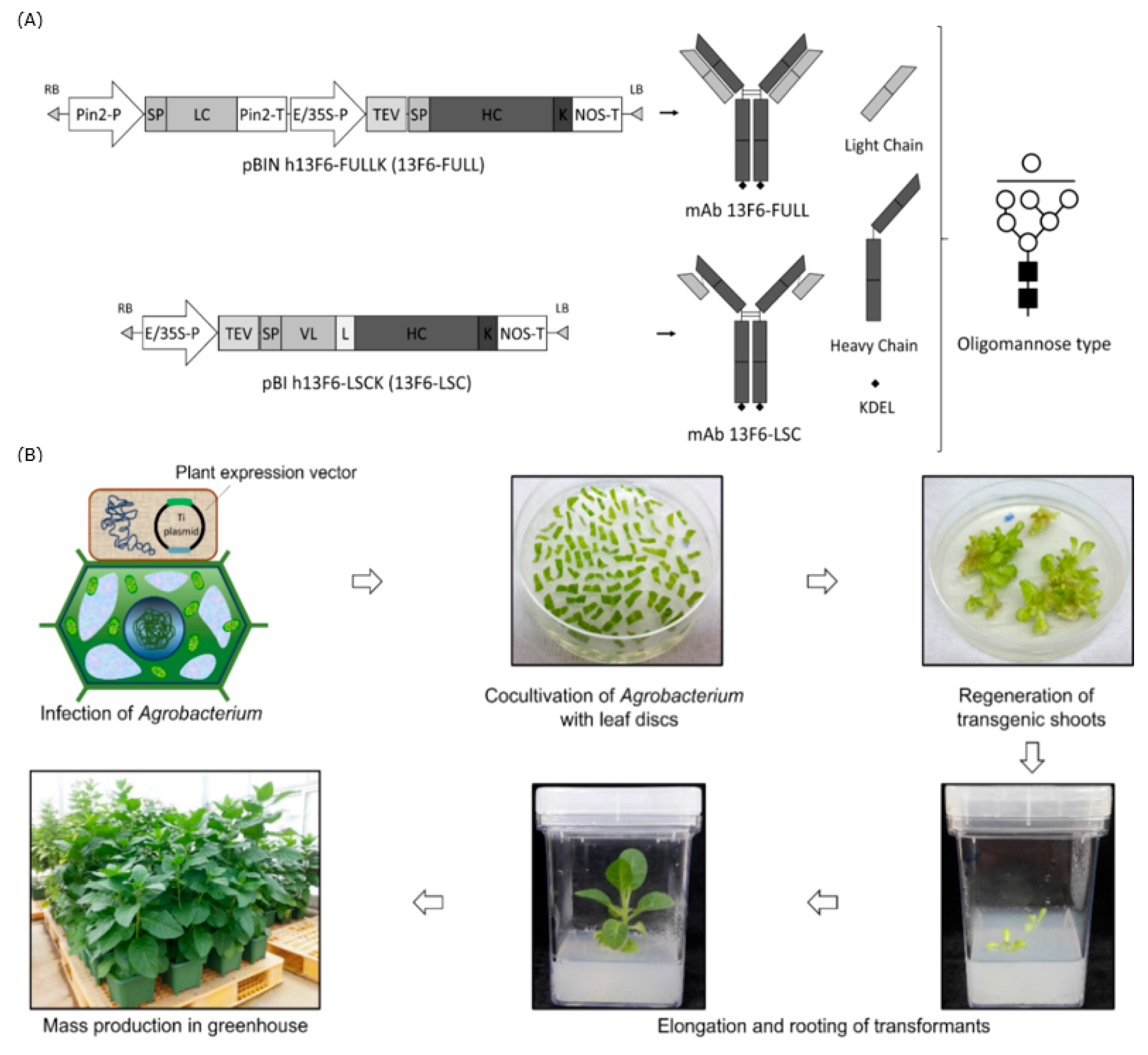
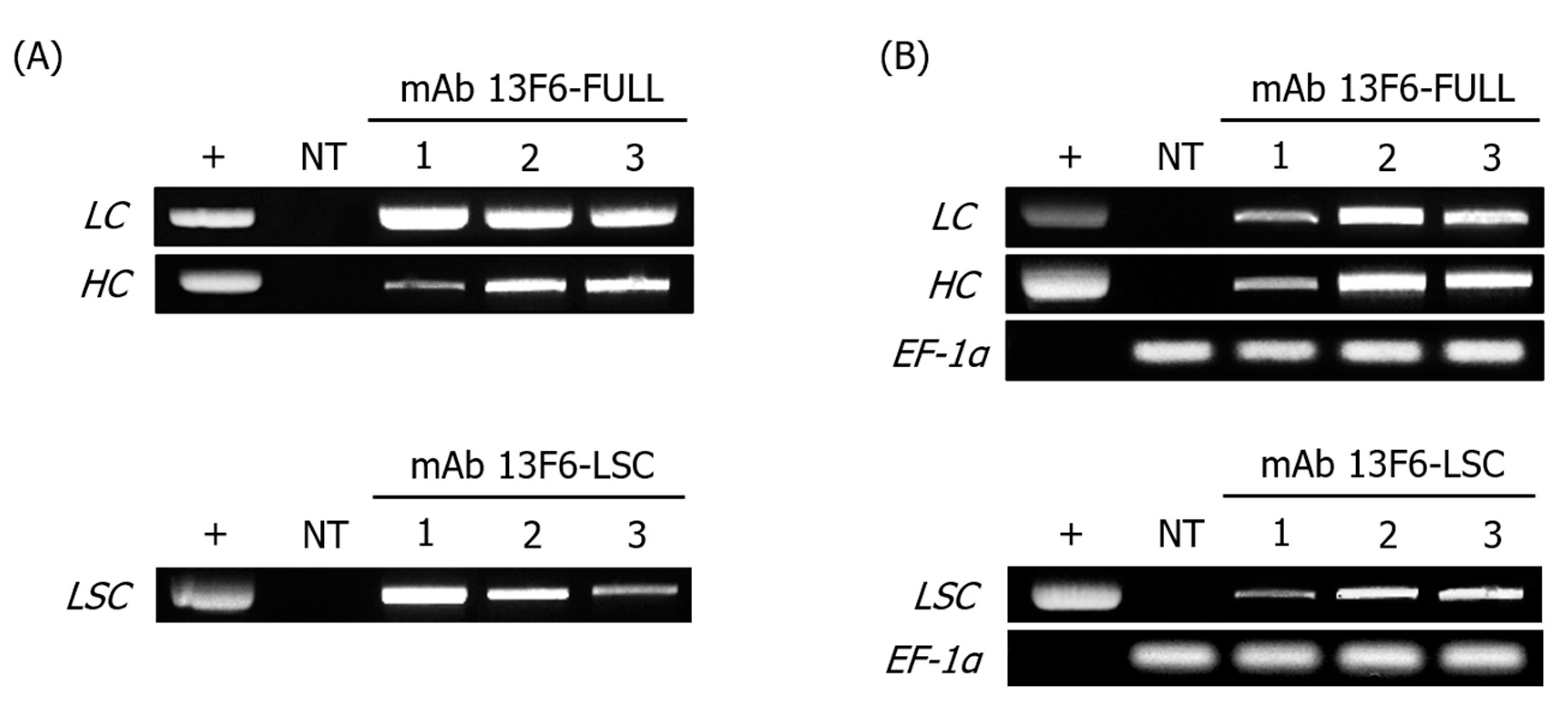
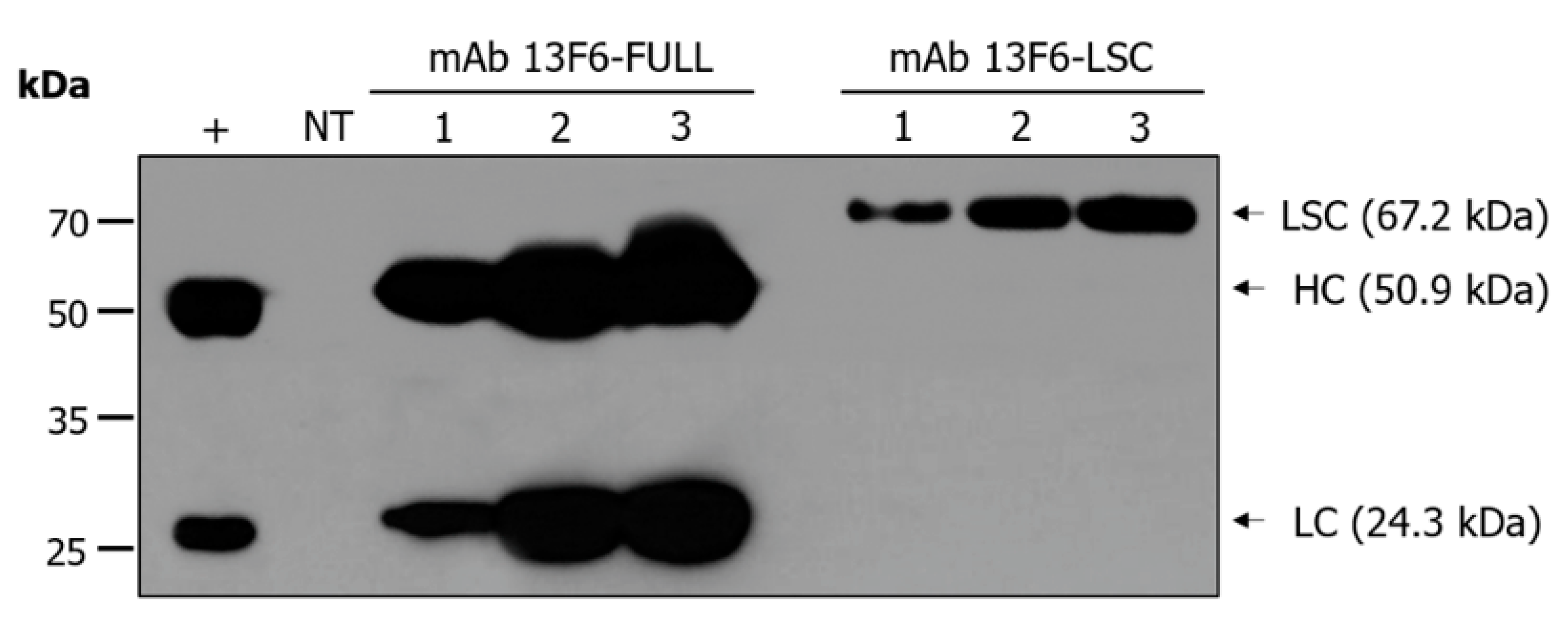
2.1. Expression of mAb 13F6-FULL and mAb 13F6-LSC in Transgenic N. tabacum Plants
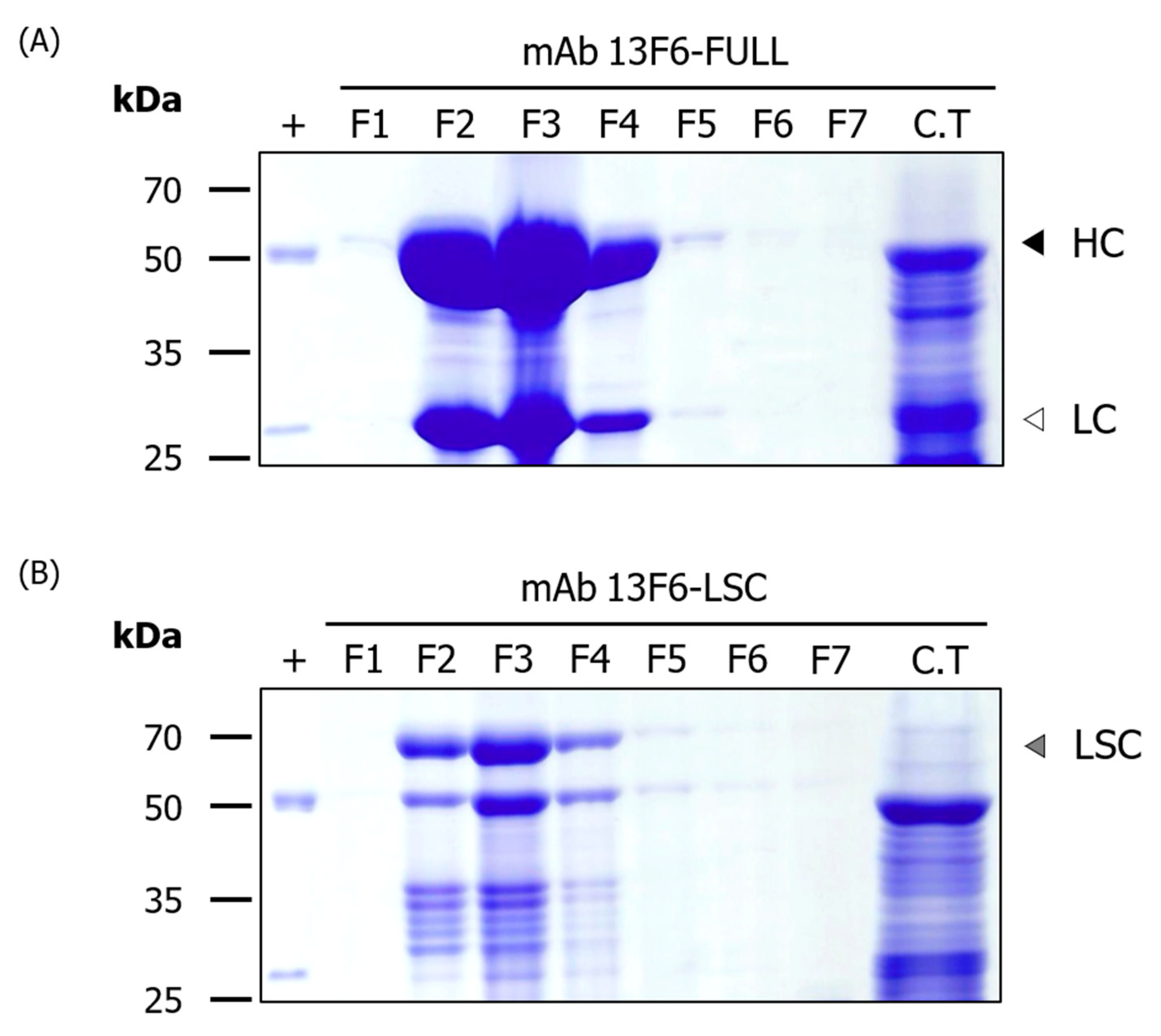
2.2. Purification of mAb 13F6-FULL and 13F6-LSC Antibodies from the Transgenic N. tabacum Plant
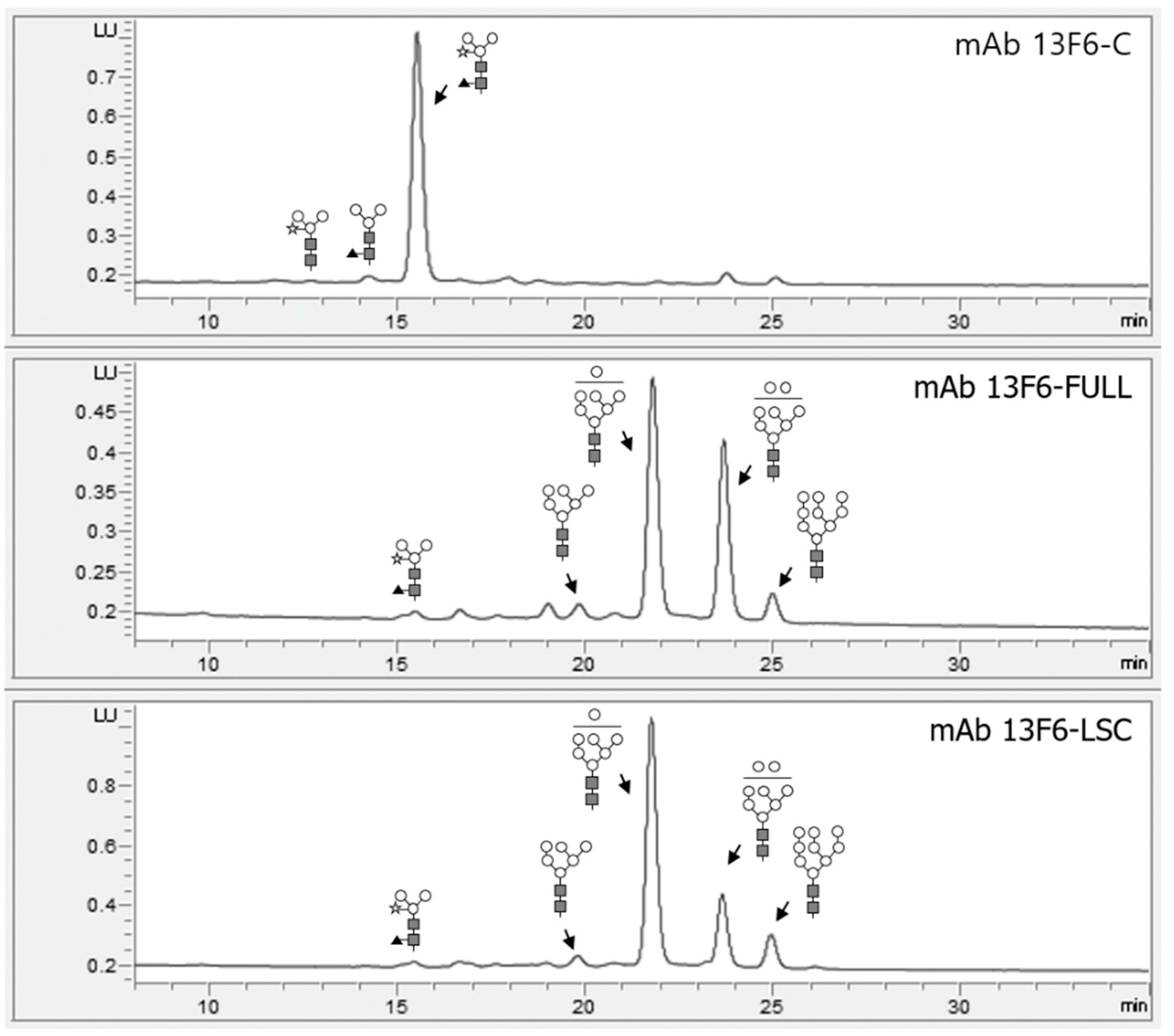
2.3. N-glycan Profile Analysis of mAb 13F6-FULL and mAb 13F6-LSC Using HPLC
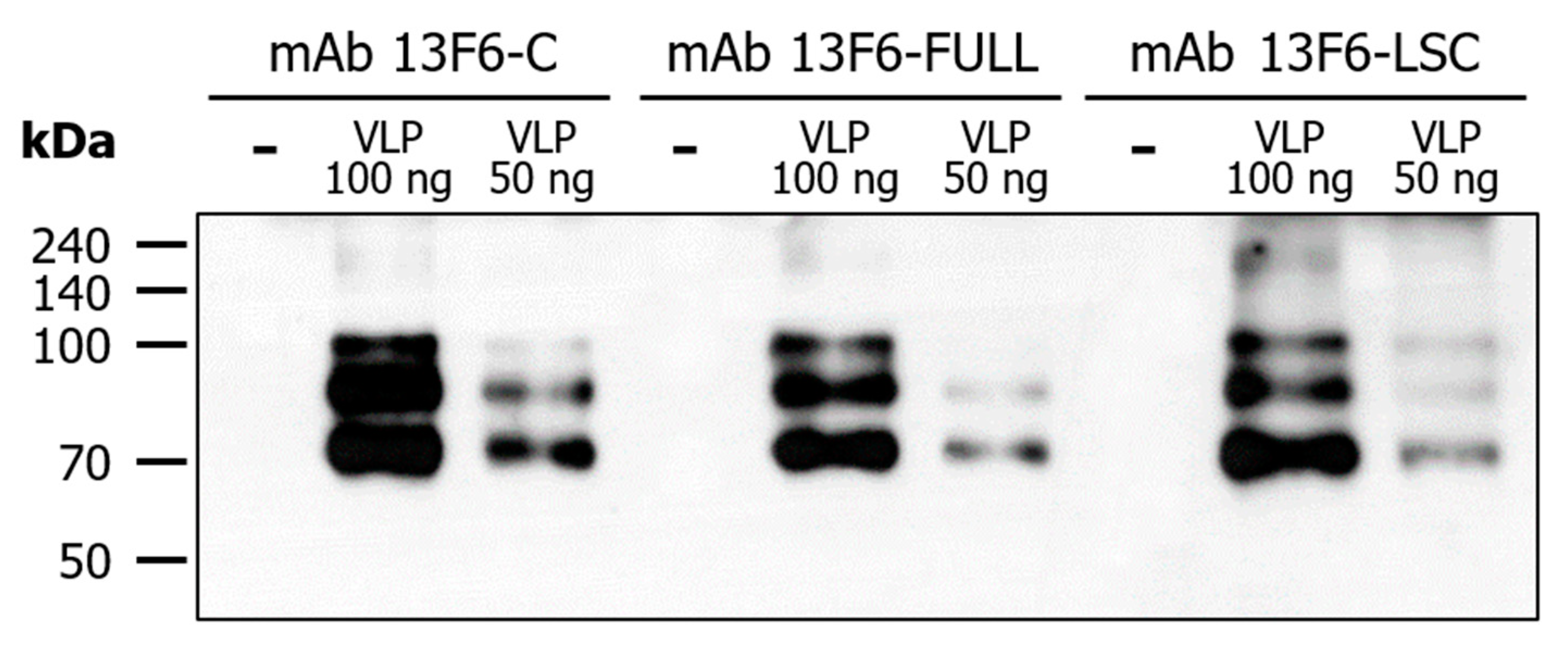
2.4. Binding of mAb 13F6-FULL and mAb 13F6-LSC to Ebola VLP
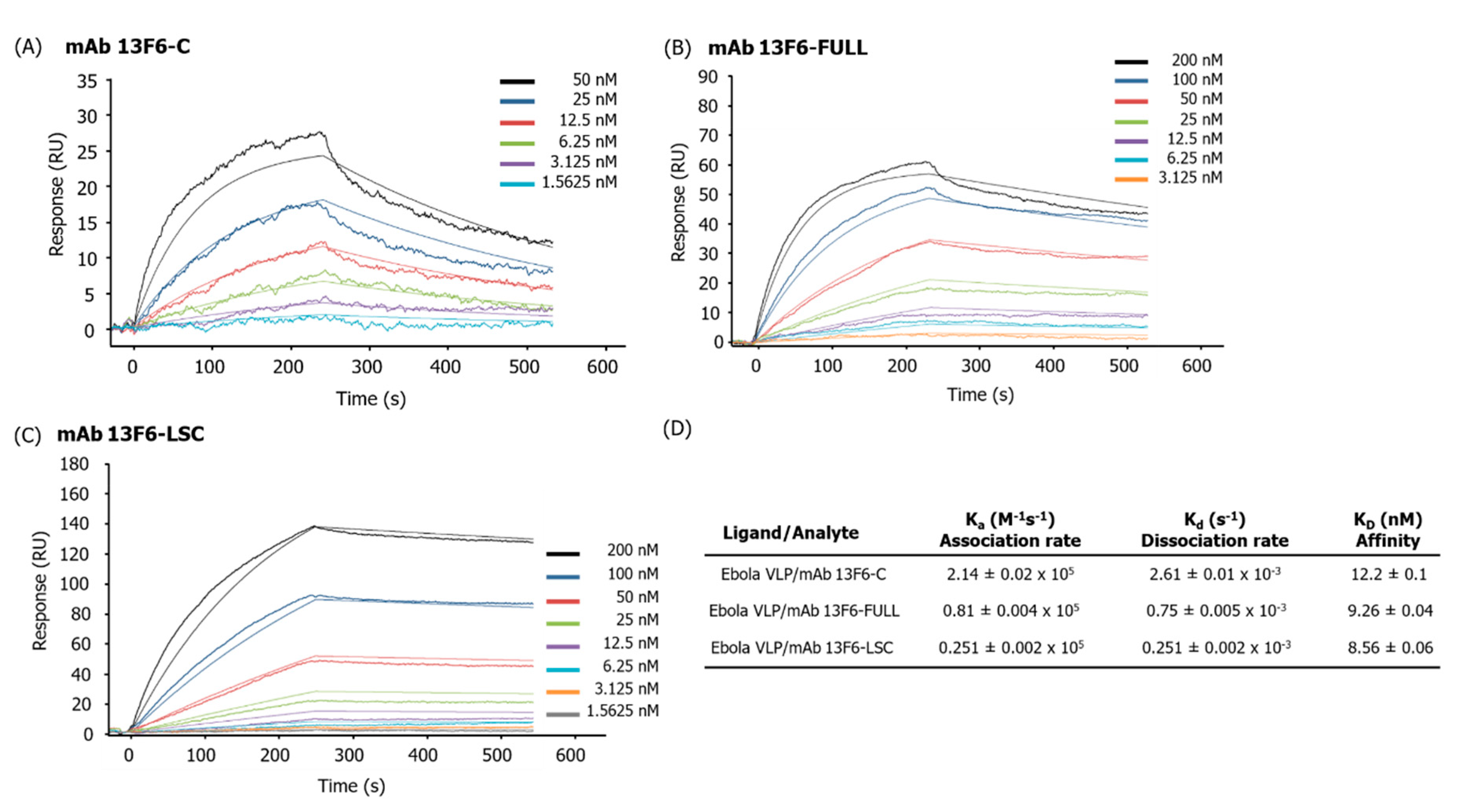
2.5. Comparison of the Binding Affinity of mAb 13F6-C, mAb 13F6-FULL, and mAb 13F6-LSC to the Ebola VLP Using Surface Plasmon Resonance (SPR)
3. Discussion
4. Materials and Methods
4.1. Construction of Expression Vectors for Plant Transformation
4.2. Generation of Transgenic Plants and Growth Conditions
4.3. Plant Genomic DNA, RNA Isolation, PCR, and RT-PCR Analyses
4.4. SDS-PAGE and Immunoblot Analysis
4.5. Purification and Dialysis of mAb 13F6-FULL and mAb 13F6-LSC
4.6. HPLC Analysis
4.7. ELISA Analysis
4.8. SPR Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Agrobacterium | Agrobacterium tumefaciens |
| COVID-19 | Coronavirus disease |
| DRC | Democratic republic of the Congo |
| EBOV | Ebola virus |
| ECL | Enhanced chemiluminescence |
| EDTA | Ethylenediaminetetra acetic acid |
| ELISA | Enzyme-linked immunosorbent assay |
| ER | Endoplasmic reticulum |
| EVD | Ebolavirus disease |
| FULL | Full size antibody |
| gDNA | Genomic deoxyribonucleic acid |
| GP | Glycoprotein |
| HC | Heavy chain |
| HPLC | High-performance liquid chromatography |
| HRP | Horseradish peroxidase |
| IgG | Immunoglobulin G |
| KD | Binding affinity |
| KDEL | Endoplasmic reticulum retention motif |
| LC | Light chain |
| LSC | Large single-chain |
| mAb | Monoclonal antibody |
| MERS | Middle east respiratory syndrome |
| MLD | Mucin-like domain |
| mRNA | Messenger ribonucleic acid |
| NT | Non-transgenic Nicotiana tabacum plant |
| N. tabacum | Nicotiana tabacum |
| PBS | Phosphate-buffer saline |
| PCR | Polymerase chain reaction |
| RNA | Ribonucleic acid |
| RT | Room temperature |
| RT-PCR | Reverse transcription polymerase chain reaction |
| RU | Response units |
| SARS | Acute respiratory syndrome |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide |
| SPR | Surface plasmon resonance |
| TEV | Untranslated leader sequence of RNA4 of an Alfalfa mosaic virus |
| VH | Heavy chain variable region |
| VL | Light chain variable region |
| VLP | Virus-like particle |
| WHO | World health organization |
| ZEBOV | Zaire ebolavirus species |
| 2-AB | 2-Aminobenzamide |
References
- Audet, J.; Wong, G.; Wang, H.; Lu, G.; Gao, G.F.; Kobinger, G.; Qiu, X. Molecular characterization of the monoclonal antibodies composing ZMAb: A protective cocktail against Ebola virus. Sci. Rep. 2014, 4, 6881. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, U.; Phillips, J.; Artsaenko, O.; Conrad, U. Optimization of scFv antibody production in transgenic plants. Immunotechnology 1997, 3, 205–216. [Google Scholar] [CrossRef]
- WHO. Ebola Virus Disease. Available online: https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease (accessed on 22 August 2020).
- Team, W.E.R.; Aylward, B.; Barboza, P.; Bawo, L.; Bertherat, E.; Bilivogui, P.; Blake, I.; Brennan, R.; Briand, S.; Chakauya, J.M.; et al. Ebola virus disease in West Africa-the first 9 months of the epidemic and forward projections. N. Engl. J. Med. 2014, 371, 1481–1495. [Google Scholar] [CrossRef] [PubMed]
- Beniac, D.R.; Booth, T.F. Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 A resolution. Sci. Rep. 2017, 7, 46374. [Google Scholar] [CrossRef]
- WHO. Ebola in the Democratic Republic of the Congo-Health Emergency Update. Available online: https://www.who.int/emergencies/diseases/ebola/drc-2019 (accessed on 22 August 2020).
- Malvy, D.; McElroy, A.K.; Clerck, H.; Günther, S.; Griensven, J.V. Ebola virus disease. Lancet 2019, 393, 936–948. [Google Scholar] [CrossRef]
- Broadhurst, M.J.; Brooks, T.J.; Pollock, N.R. Diagnosis of Ebola Virus Disease: Past, Present, and Future. Clin. Microbiol. Rev. 2016, 29, 773–793. [Google Scholar] [CrossRef] [PubMed]
- Towner, J.S.; Rollin, P.E.; Bausch, D.G.; Sanchez, A.; Crary, S.M.; Vincent, M.; Lee, W.F.; Spiropoulou, C.F.; Ksiazek, T.G.; Lukwiya, M.; et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J. Virol. 2004, 78, 4330–4341. [Google Scholar] [CrossRef] [PubMed]
- Fujihira, H.; Usami, K.; Matsuno, K.; Takeuchi, H.; Denda-Nagai, K.; Furukawa, J.I.; Shinohara, Y.; Takada, A.; Kawaoka, Y.; Irimura, T. A critical domain of Ebolavirus envelope glycoprotein determines glycoform and infectivity. Sci. Rep. 2018, 8, 5495. [Google Scholar] [CrossRef]
- Qiu, X.; Fernando, L.; Alimonti, J.B.; Melito, P.L.; Feldmann, F.; Dick, D.; Stroher, U.; Feldmann, H.; Jones, S.M. Mucosal immunization of cynomolgus macaques with the VSVDeltaG/ZEBOVGP vaccine stimulates strong ebola GP-specific immune responses. PLoS ONE 2009, 4, e5547. [Google Scholar] [CrossRef]
- Wong, G.; Richardson, J.S.; Pillet, S.; Patel, A.; Qiu, X.; Alimonti, J.; Hogan, J.; Zhang, Y.; Takada, A.; Feldmann, H.; et al. Immune parameters correlate with protection against ebola virus infection in rodents and nonhuman primates. Sci. Transl. Med. 2012, 4, 158ra146. [Google Scholar] [CrossRef]
- Hood, C.J.; Abraham, J.; Boyington, J.C.; Leung, K.; Kwong, P.D.; Nabel, G.J. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: Implications for viral entry and immunogenicity. J. Virol. 2010, 84, 2972–2982. [Google Scholar] [CrossRef] [PubMed]
- Takada, A.; Robison, C.; Goto, H.; Sanchez, A.; Murti, K.G.; Whitt, M.A.; Kawaoka, Y. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 1997, 94, 14764–14769. [Google Scholar] [CrossRef] [PubMed]
- Holtsberg, F.W.; Shulenin, S.; Vu, H.; Howell, K.A.; Patel, S.J.; Gunn, B.; Karim, M.; Lai, J.R.; Frei, J.C.; Nyakatura, E.K.; et al. Pan-ebolavirus and Pan-filovirus Mouse Monoclonal Antibodies: Protection against Ebola and Sudan Viruses. J. Virol. 2016, 90, 266–278. [Google Scholar] [CrossRef]
- Flyak, A.I.; Shen, X.; Murin, C.D.; Turner, H.L.; David, J.A.; Fusco, M.L.; Lampley, R.; Kose, N.; Ilinykh, P.A.; Kuzmina, N.; et al. Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection. Cell 2016, 164, 392–405. [Google Scholar] [CrossRef]
- Wilson, J.A.; Hevey, M.; Bakken, R.; Guest, S.; Bray, M.; Schmaljohn, A.L.; Hart, M.K. Epitopes involved in antibody-mediated protection from Ebola virus. Science 2000, 287, 1664–1666. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, L.; Pettitt, J.; Scully, C.; Bohorova, N.; Kim, D.; Pauly, M.; Hiatt, A.; Ngo, L.; Steinkellner, H.; Whaley, K.J.; et al. Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proc. Natl. Acad. Sci. USA 2011, 108, 20690–20694. [Google Scholar] [CrossRef] [PubMed]
- Gunn, B.M.; Yu, W.H.; Karim, M.M.; Brannan, J.M.; Herbert, A.S.; Wec, A.Z.; Halfmann, P.J.; Fusco, M.L.; Schendel, S.L.; Gangavarapu, K.; et al. A Role for Fc Function in Therapeutic Monoclonal Antibody-Mediated Protection against Ebola Virus. Cell Host Microbe 2018, 24, 221–233.e5. [Google Scholar] [CrossRef]
- Olinger, G.G., Jr.; Pettitt, J.; Kim, D.; Working, C.; Bohorov, O.; Bratcher, B.; Hiatt, E.; Hume, S.D.; Johnson, A.K.; Morton, J.; et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc. Natl. Acad. Sci. USA 2012, 109, 18030–18035. [Google Scholar] [CrossRef]
- Pettitt, J.; Zeitlin, L.; Kim, D.H.; Working, C.; Johnson, J.C.; Bohorov, O.; Bratcher, B.; Hiatt, E.; Hume, S.D.; Johnson, A.K.; et al. Therapeutic Intervention of Ebola Virus Infection in Rhesus Macaques with the MB-003 Monoclonal Antibody Cocktail. Sci. Transl. Med. 2013, 5, 199ra113. [Google Scholar] [CrossRef]
- Ko, K. Expression of recombinant vaccines and antibodies in plants. Monoclon. Antib. Immunodiagn. Immunother. 2014, 33, 192–198. [Google Scholar] [CrossRef]
- Lee, J.H.; Ko, K. Production of Recombinant Anti-Cancer Vaccines in Plants. Biomol. Ther. 2017, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Lee, K.J.; Shao, Y.; Lee, J.H.; So, Y.; Choo, Y.K.; Oh, D.B.; Hwang, K.A.; Oh, S.H.; Han, Y.S.; et al. Expression of GA733-Fc Fusion Protein as a Vaccine Candidate for Colorectal Cancer in Transgenic Plants. J. Biomed. Biotechnol. 2012, 2012, 364240. [Google Scholar] [CrossRef] [PubMed]
- Shin, C.; Kang, Y.; Kim, H.S.; Shin, Y.K.; Ko, K. Immune response of heterologous recombinant antigenic protein of viral hemorrhagic septicemia virus (VHSV) in mice. Anim. Cells Syst. (Seoul) 2019, 23, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Song, I.; Kang, Y.; Lee, Y.K.; Myung, S.C.; Ko, K. Endoplasmic reticulum retention motif fused to recombinant anti-cancer monoclonal antibody (mAb) CO17-1A affects mAb expression and plant stress response. PLoS ONE 2018, 13, e0198978. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Shin, Y.K.; Park, S.W.; Ko, K. Effect of nitrogen deficiency on recombinant protein production and dimerization and growth in transgenic plants. Hortic. Environ. Biotechnol. 2016, 57, 299–307. [Google Scholar] [CrossRef]
- Lim, C.Y.; Lee, K.J.; Oh, D.B.; Ko, K. Effect of the developmental stage and tissue position on the expression and glycosylation of recombinant glycoprotein GA733-FcK in transgenic plants. Front. Plant Sci. 2015, 5, 778. [Google Scholar] [CrossRef]
- Rigano, M.M.; Walmsley, A.M. Expression systems and developments in plant-made vaccines. Immunol. Cell Biol. 2005, 83, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Arcalis, E.; Stadlmann, J.; Rademacher, T.; Marcel, S.; Sack, M.; Altmann, F.; Stoger, E. Plant species and organ influence the structure and subcellular localization of recombinant glycoproteins. Plant Mol. Biol. 2013, 83, 105–117. [Google Scholar] [CrossRef]
- Fischer, R.; Stoger, E.; Schillberg, S.; Christou, P.; Twyman, R.M. Plant-based production of biopharmaceuticals. Curr. Opin. Plant Biol. 2004, 7, 152–158. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, S.R.; Phoolcharoen, W.; Ko, K. Expression, function, and glycosylation of anti-colorectal cancer large single-chain antibody (LSC) in plant. Plant Biotechnol. Rep. 2020, 14, 363–371. [Google Scholar] [CrossRef]
- Bates, A.; Power, C.A. David vs. Goliath: The Structure, Function, and Clinical Prospects of Antibody Fragments. Antibodies 2019, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Spadiut, O.; Capone, S.; Krainer, F.; Glieder, A.; Herwig, C. Microbials for the production of monoclonal antibodies and antibody fragments. Trends Biotechnol. 2014, 32, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Jamal, A.; Lee, J.H.; Lee, K.J.; Oh, D.B.; Kim, D.S.; Lee, K.K.; Choo, Y.K.; Hwang, K.A.; Ko, K. Chimerism of multiple monoclonal antibodies expressed in a single plant. Hortic. Environ. Biotechnol. 2012, 53, 544–551. [Google Scholar] [CrossRef]
- González-González, E.; Alvarez, M.M.; Márquez-Ipiña, A.R.; Trujillo-de Santiago, G.; Rodríguez-Martínez, L.M.; Annabi, N.; Khademhosseini, A. Anti-Ebola therapies based on monoclonal antibodies: Current state and challenges ahead. Crit. Rev. Biotechnol. 2017, 37, 53–68. [Google Scholar] [CrossRef]
- Angelo, K.M.; Kozarsky, P.E.; Ryan, E.T.; Chen, L.H.; Sotir, M.J. What proportion of international travellers acquire a travel-related illness? A review of the literature. J. Travel Med. 2017, 24, 1–13. [Google Scholar] [CrossRef]
- Harvey, K.; Esposito, D.H.; Han, P.; Kozarsky, P.; Freedman, D.O.; Plier, D.A.; Sotir, M.J. Surveillance for travel-related disease-GeoSentinel Surveillance System, United States, 1997–2011. MMWR Surveill. Summ. 2013, 62, 1–23. [Google Scholar]
- Stoney, R.J.; Esposito, D.H.; Kozarsky, P.; Hamer, D.H.; Grobusch, M.P.; Gkrania-Klotsas, E.; Libman, M.; Gautret, P.; Lim, P.L.; Leder, K.; et al. Infectious diseases acquired by international travellers visiting the USA. J. Travel Med. 2018, 25, 1–7. [Google Scholar] [CrossRef]
- Mangili, A.; Vindenes, T.; Gendreau, M. Infectious Risks of Air Travel. Microbiol. Spectr. 2015, 3, 1–10. [Google Scholar] [CrossRef]
- Geisbert, T.W.; Jahrling, P.B. Exotic emerging viral diseases: Progress and challenges. Nat. Med. 2004, 10, S110–S121. [Google Scholar] [CrossRef]
- CDC. Ebola (Ebola Virus Disease). Available online: https://www.cdc.gov/vhf/ebola/treatment/index.html (accessed on 22 August 2020).
- Ebadat, S.; Ahmadi, S.; Ahmadi, M.; Nematpour, F.; Barkhordari, F.; Mahdian, R.; Davami, F.; Mahboudi, F. Evaluating the efficiency of CHEF and CMV promoter with IRES and Furin/2A linker sequences for monoclonal antibody expression in CHO cells. PLoS ONE 2017, 12, e0185967. [Google Scholar] [CrossRef]
- Klimtchuk, E.S.; Prokaeva, T.B.; Spencer, B.H.; Gursky, O.; Connors, L.H. In vitro co-expression of human amyloidogenic immunoglobulin light and heavy chain proteins: A relevant cell-based model of AL amyloidosis. Amyloid 2017, 24, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.Y.; Zhao, J.; Park, J.H.; Choi, G.W.; Park, K.; Lee, Y.; Chung, I. Human colorectal cancer antigen GA733-2-Fc fused to endoplasmic reticulum retention motif KDEL enhances its immunotherapeutic effects. J. Cancer Res. Ther. 2018, 14, 748–757. [Google Scholar]
- Triguero, A.; Cabrera, G.; Cremata, J.A.; Yuen, C.T.; Wheeler, J.; Ramírez, N.I. Plant-derived mouse IgG monoclonal antibody fused to KDEL endoplasmic reticulum-retention signal is N-glycosylated homogeneously throughout the plant with mostly high-mannose-type N-glycans. Plant Biotechnol. J. 2005, 3, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Park, D.Y.; Lee, K.J.; Kim, Y.K.; So, Y.K.; Ryu, J.S.; Oh, S.H.; Han, Y.S.; Ko, K.; Choo, Y.K.; et al. Intracellular reprogramming of expression, glycosylation, and function of a plant-derived antiviral therapeutic monoclonal antibody. PLoS ONE 2013, 8, e68772. [Google Scholar] [CrossRef]
- Park, S.R.; Ko, K.; Lim, S.; Cha, S.Y.; Chung, H.J.; Park, S.J.; Myung, S.C.; Kim, M.K. In vitro wound healing: Inhibition activity of insect-derived mAb CO17-1A in human colorectal cancer cell migration. Entomol. Res. 2020, 50, 199–204. [Google Scholar] [CrossRef]
- Song, I.; Kang, Y.J.; Kim, D.H.; Kim, M.K.; Ko, K. Expression and in vitro function of anti-cancer mAbs in transgenic Arabidopsis thaliana. BMB Rep. 2020, 53, 229–233. [Google Scholar] [CrossRef]
- Ko, K.; Tekoah, Y.; Rudd, P.M.; Harvey, D.J.; Dwek, R.A.; Spitsin, S.; Hanlon, C.A.; Rupprecht, C.; Dietzschold, B.; Golovkin, M.; et al. Function and glycosylation of plant-derived antiviral monoclonal antibody. Proc. Natl. Acad. Sci. USA 2003, 100, 8013–8018. [Google Scholar] [CrossRef]
- Park, S.R.; Lee, J.H.; Kim, K.; Kim, T.M.; Lee, S.H.; Choo, Y.K.; Kim, K.S.; Ko, K. Expression and In Vitro Function of Anti-Breast Cancer Llama-Based Single Domain Antibody VHH Expressed in Tobacco Plants. Int. J. Mol. Sci. 2020, 21, 1354. [Google Scholar] [CrossRef]
- Sharp, J.M.; Doran, P.M. Characterization of monoclonal antibody fragments produced by plant cells. Biotechnol. Bioeng. 2001, 73, 338–346. [Google Scholar] [CrossRef]
- Schouten, A.; Roosien, J.; van Engelen, F.A.; de Jong, G.A.; Borst-Vrenssen, A.W.; Zilverentant, J.F.; Bosch, D.; Stiekema, W.J.; Gommers, F.J.; Schots, A.; et al. The C-terminal KDEL sequence increases the expression level of a single-chain antibody designed to be targeted to both the cytosol and the secretory pathway in transgenic tobacco. Plant Mol. Biol. 1996, 30, 781–793. [Google Scholar] [CrossRef]
- Wandelt, C.I.; Khan, M.R.; Craig, S.; Schroeder, H.E.; Spencer, D.; Higgins, T. Vicilin with carboxy-terminal KDEL is retained in the endoplasmic reticulum and accumulates to high levels in the leaves of transgenic plants. Plant J. 1992, 2, 181–192. [Google Scholar] [PubMed]
- Bar-On, Y.M.; Milo, R. The global mass and average rate of rubisco. Proc. Natl. Acad. Sci. USA 2019, 116, 4738–4743. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.; Koprowski, H. Plant biopharming of monoclonal antibodies. Virus Res. 2005, 111, 93–100. [Google Scholar] [CrossRef]
- Gu, X.; Jia, X.; Feng, J.; Shen, B.; Huang, Y.; Geng, S.; Sun, Y.; Wang, Y.; Li, Y.; Long, M. Molecular modeling and affinity determination of scFv antibody: Proper linker peptide enhances its activity. Ann. Biomed. Eng. 2010, 38, 537–549. [Google Scholar] [CrossRef]
- Crispin, M.; Bowden, T.A.; Coles, C.H.; Harlos, K.; Aricescu, A.R.; Harvey, D.J.; Stuart, D.I.; Jones, E.Y. Carbohydrate and domain architecture of an immature antibody glycoform exhibiting enhanced effector functions. J. Mol. Biol. 2009, 387, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Kugelman, J.R.; Kugelman-Tonos, J.; Ladner, J.T.; Pettit, J.; Keeton, C.M.; Nagle, E.R.; Garcia, K.Y.; Froude, J.W.; Kuehne, A.I.; Kuhn, J.H.; et al. Emergence of Ebola Virus Escape Variants in Infected Nonhuman Primates Treated with the MB-003 Antibody Cocktail. Cell Rep. 2015, 12, 2111–2120. [Google Scholar] [CrossRef]
- Mayfield, S.P.; Franklin, S.E.; Lerner, R.A. Expression and assembly of a fully active antibody in algae. Proc. Natl. Acad. Sci. USA 2003, 100, 438–442. [Google Scholar] [CrossRef]
- So, Y.; Lee, K.J.; Kim, D.S.; Lee, J.H.; Oh, D.B.; Hwang, K.A.; Ko, K.; Choo, Y.K.; Ko, K. Glycomodification and characterization of anti-colorectal cancer immunotherapeutic monoclonal antibodies in transgenic tobacco. Plant Cell Tissue Org. 2013, 113, 41–49. [Google Scholar] [CrossRef]
- Chomczynski, P.; Mackey, K. Short technical report. Modification of the TRIZOL reagent procedure for isolation of RNA from Polysaccharide-and proteoglycan-rich sources. Biotechniques 1995, 19, 942–945. [Google Scholar]
- Park, S.R.; Lim, C.Y.; Kim, D.S.; Ko, K. Optimization of Ammonium Sulfate Concentration for Purification of Colorectal Cancer Vaccine Candidate Recombinant Protein GA733-FcK Isolated from Plants. Front. Plant Sci. 2015, 6, 1040. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, S.; Kim, D.-S.; Ko, K. Expression of a Large Single-Chain 13F6 Antibody with Binding Activity against Ebola Virus-Like Particles in a Plant System. Int. J. Mol. Sci. 2020, 21, 7007. https://doi.org/10.3390/ijms21197007
Lim S, Kim D-S, Ko K. Expression of a Large Single-Chain 13F6 Antibody with Binding Activity against Ebola Virus-Like Particles in a Plant System. International Journal of Molecular Sciences. 2020; 21(19):7007. https://doi.org/10.3390/ijms21197007
Chicago/Turabian StyleLim, Sohee, Do-Sun Kim, and Kisung Ko. 2020. "Expression of a Large Single-Chain 13F6 Antibody with Binding Activity against Ebola Virus-Like Particles in a Plant System" International Journal of Molecular Sciences 21, no. 19: 7007. https://doi.org/10.3390/ijms21197007
APA StyleLim, S., Kim, D.-S., & Ko, K. (2020). Expression of a Large Single-Chain 13F6 Antibody with Binding Activity against Ebola Virus-Like Particles in a Plant System. International Journal of Molecular Sciences, 21(19), 7007. https://doi.org/10.3390/ijms21197007





