Comparison of miRNA Expression Profiles between HIV-1 and HIV-2 Infected Monocyte-Derived Macrophages (MDMs) and Peripheral Blood Mononuclear Cells (PBMCs)
Abstract
1. Introduction
2. Results
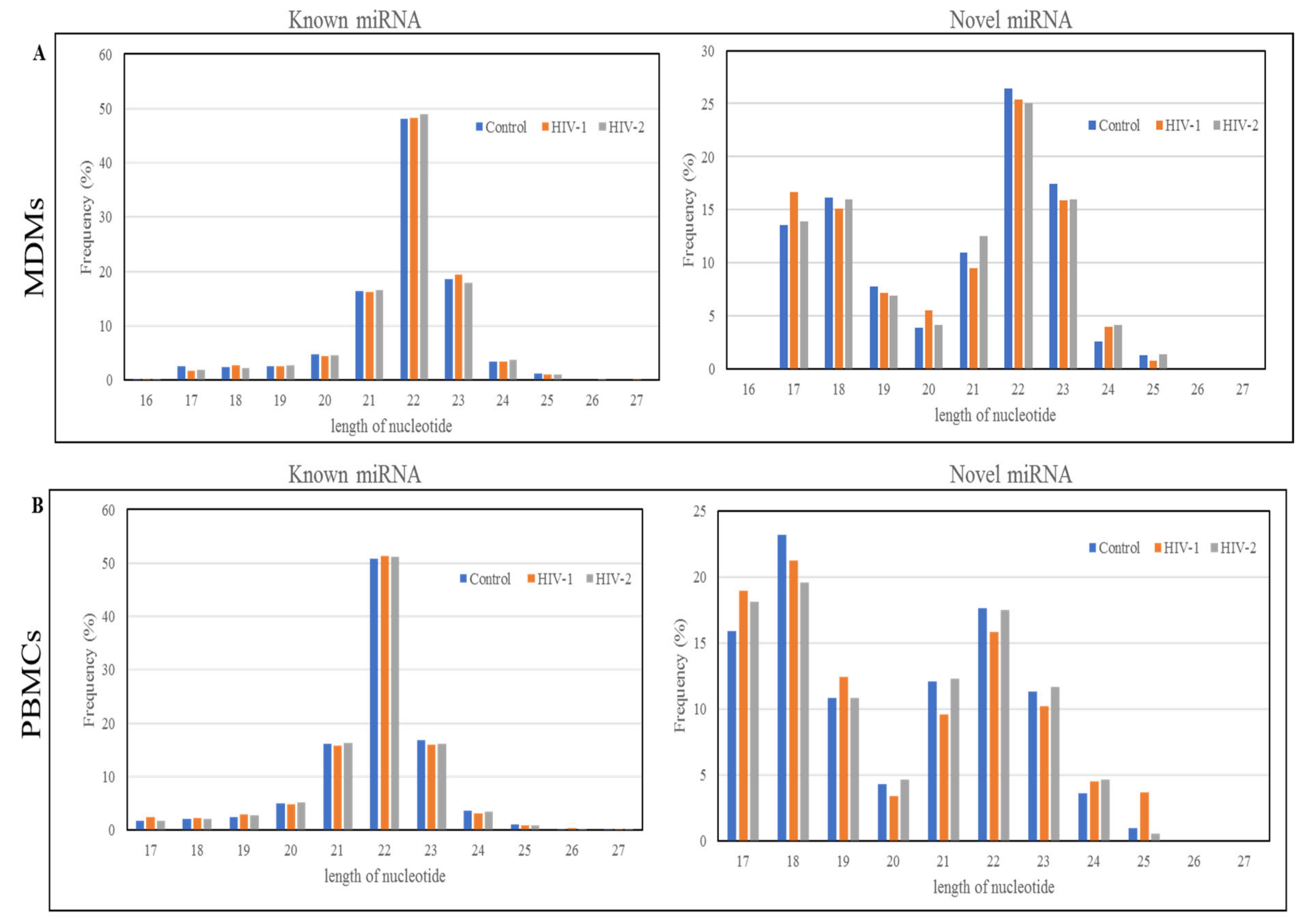
2.1. Length Distribution and Annotation of Small RNAs
2.2. Identification of miRNAs
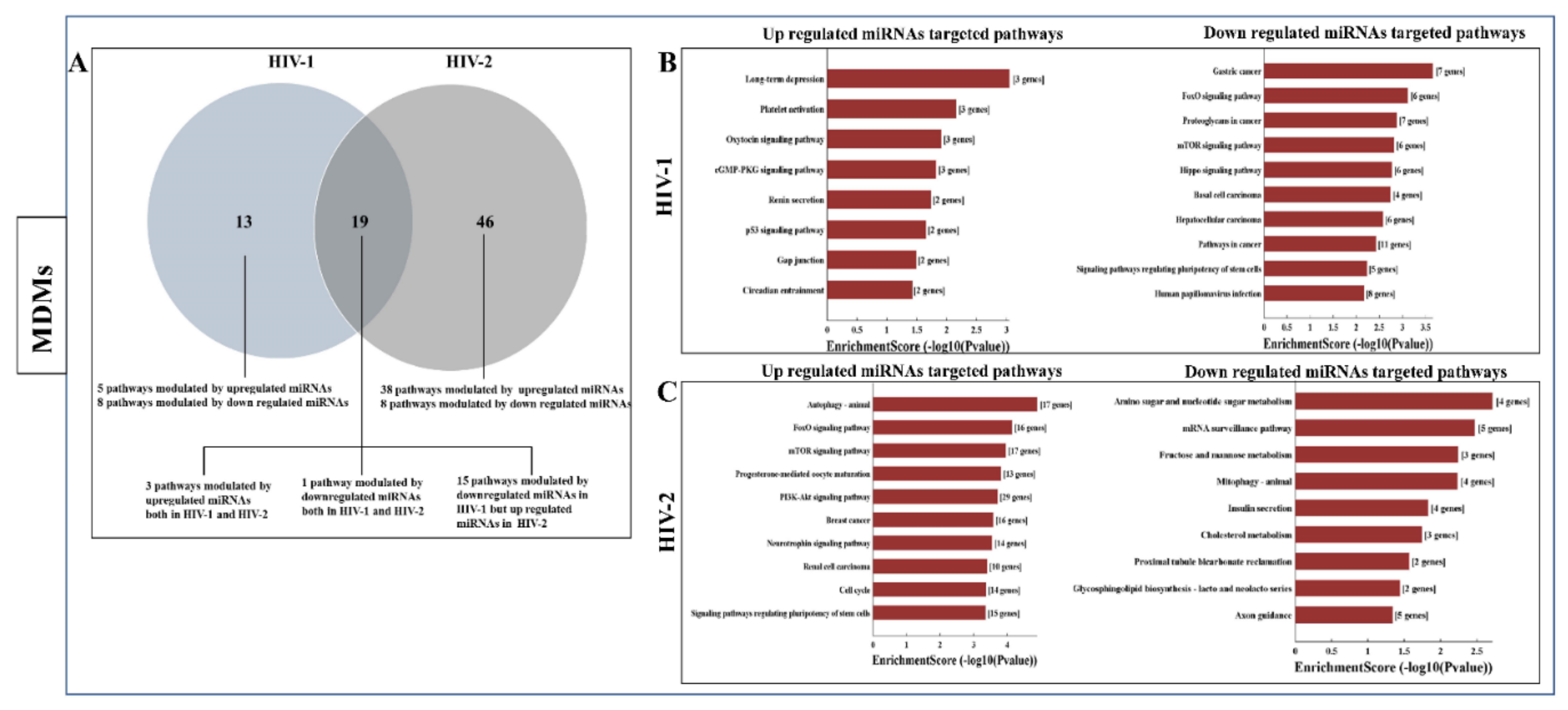
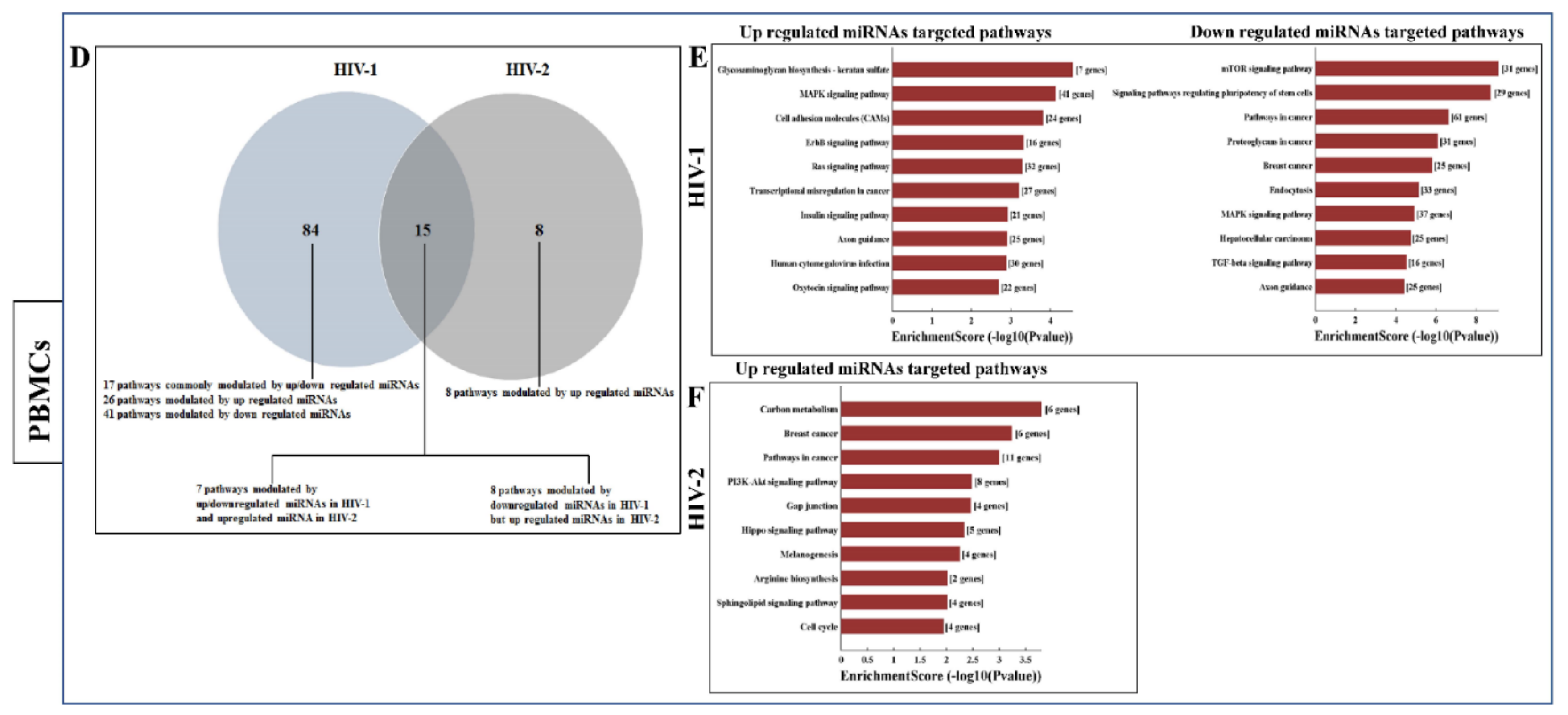
2.3. Differentially Expressed miRNA between HIV-1- and HIV-2-Infected MDMs and PBMCs
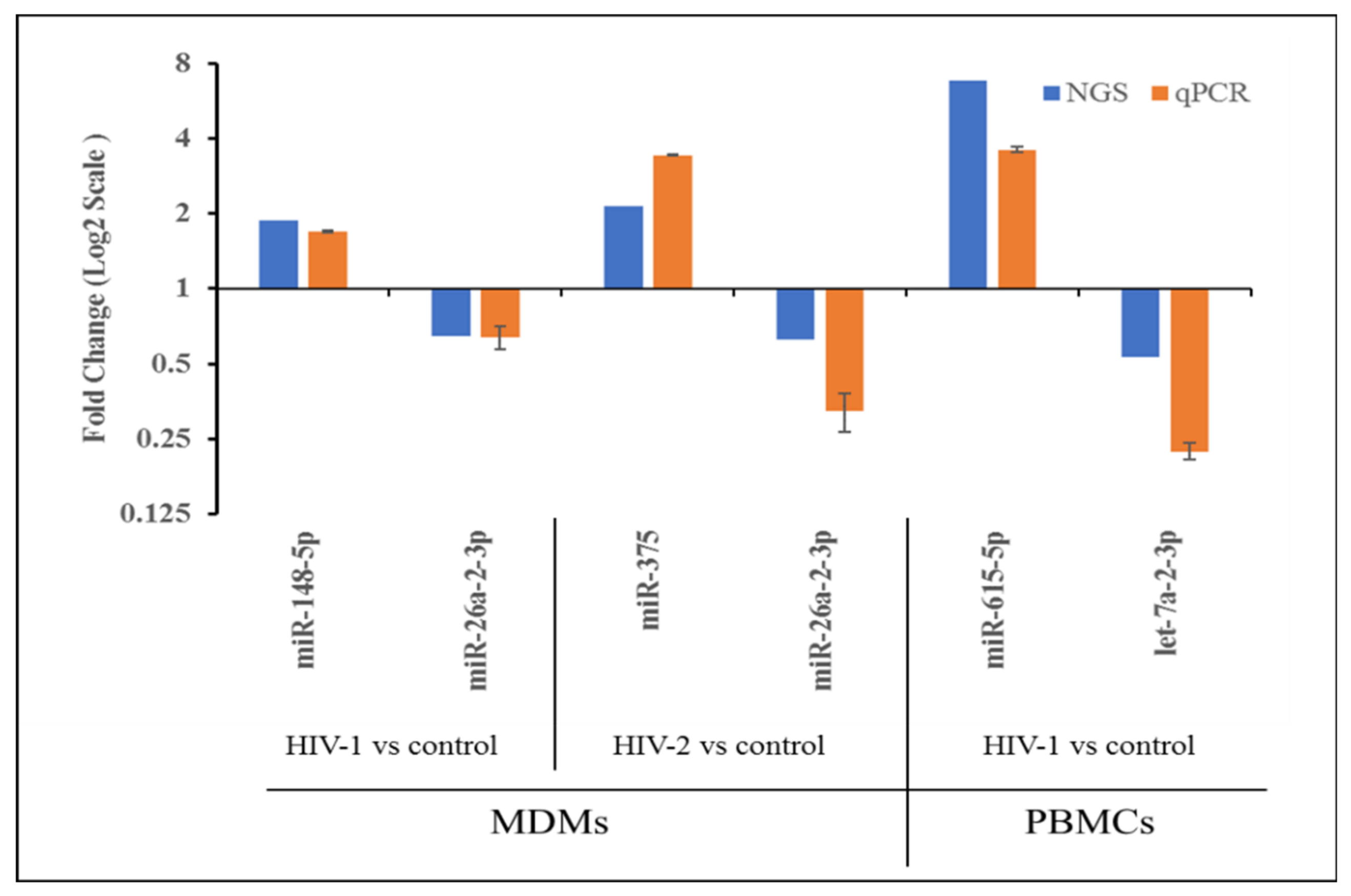
2.4. Validation of miRNA Expression by qRT-PCR
2.5. Functional Annotation and KEGG Pathway Enrichment Analysis of Differentially Expressed miRNA Target Genes
3. Discussion
4. Materials and Methods
4.1. Peripheral Blood Mononuclear Cells (PBMCs) and Monocyte-Derived Macrophages (MDMs) Cell Culture
4.2. Virus Infection in MDMs and PBMCs
4.3. Total RNA Isolation
4.4. Small RNA Library Preparation and Sequencing
4.5. Initial Processing of Reads
4.6. Normalization of miRNA Expression
4.7. Known miRNA Expression Analysis
4.8. Novel miRNA Prediction
4.9. Validation of Mature miRNA Expression by qPCR
4.10. MiRNA Target Prediction, Gene Ontology, and Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Additional Information
References
- Fackler, O.T.; Murooka, T.T.; Imle, A.; Mempel, T.R. Adding new dimensions: Towards an integrative understanding of HIV-1 spread. Nat. Rev. Microbiol. 2014, 12, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Shiramizu, B.; Gartner, S.; Williams, A.; Shikuma, C.; Ratto-Kim, S.; Watters, M.; Aguon, J.; Valcour, V. Circulating proviral HIV DNA and HIV-associated dementia. AIDS 2005, 19, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Parrish, N.F.; Gao, F.; Li, H.; Giorgi, E.E.; Barbian, H.J.; Parrish, E.H.; Zajic, L.; Iyer, S.S.; Decker, J.M.; Kumar, A.; et al. Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. USA 2013, 110, 6626–6633. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Richter, H.E.; Clements, R.H.; Novak, L.; Huff, K.; Bimczok, D.; Sankaran-Walters, S.; Dandekar, S.; Clapham, P.R.; Smythies, L.E.; et al. Macrophages in vaginal but not intestinal mucosa are monocyte-like and permissive to human immunodeficiency virus type 1 infection. J. Virol. 2009, 83, 3258–3267. [Google Scholar] [CrossRef]
- Coleman, C.M.; Wu, L. HIV interactions with monocytes and dendritic cells: Viral latency and reservoirs. Retrovirology 2009, 6, 51. [Google Scholar] [CrossRef]
- Philpott, S.M. HIV-1 coreceptor usage, transmission, and disease progression. Curr. HIV Res. 2003, 1, 217–227. [Google Scholar] [CrossRef]
- Waki, K.; Freed, E.O. Macrophages and Cell-Cell Spread of HIV-1. Viruses 2010, 2, 1603–1620. [Google Scholar] [CrossRef]
- Murooka, T.T.; Deruaz, M.; Marangoni, F.; Vrbanac, V.D.; Seung, E.; von Andrian, U.H.; Tager, A.M.; Luster, A.D.; Mempel, T.R. HIV-infected T cells are migratory vehicles for viral dissemination. Nature 2012, 490, 283–287. [Google Scholar] [CrossRef]
- Baxter, A.E.; Russell, R.A.; Duncan, C.J.; Moore, M.D.; Willberg, C.B.; Pablos, J.L.; Finzi, A.; Kaufmann, D.E.; Ochsenbauer, C.; Kappes, J.C.; et al. Macrophage infection via selective capture of HIV-1-infected CD4+ T cells. Cell Host Microbe 2014, 16, 711–721. [Google Scholar] [CrossRef]
- Costiniuk, C.T.; Jenabian, M.A. Cell-to-cell transfer of HIV infection: Implications for HIV viral persistence. J. Gen. Virol. 2014, 95, 2346–2355. [Google Scholar] [CrossRef]
- Malbec, M.; Porrot, F.; Rua, R.; Horwitz, J.; Klein, F.; Halper-Stromberg, A.; Scheid, J.F.; Eden, C.; Mouquet, H.; Nussenzweig, M.C.; et al. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J. Exp. Med. 2013, 210, 2813–2821. [Google Scholar] [CrossRef] [PubMed]
- Sattentau, Q. Avoiding the void: Cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 2008, 6, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Santini, P.A.; Sullivan, J.S.; He, B.; Shan, M.; Ball, S.C.; Dyer, W.B.; Ketas, T.J.; Chadburn, A.; Cohen-Gould, L.; et al. HIV-1 evades virus-specific IgG2 and IgA responses by targeting systemic and intestinal B cells via long-range intercellular conduits. Nat. Immunol. 2009, 10, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Embretson, J.; Zupancic, M.; Ribas, J.L.; Burke, A.; Racz, P.; Tenner-Racz, K.; Haase, A.T. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature 1993, 362, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Streeck, H.; Nixon, D.F. T cell immunity in acute HIV-1 infection. J. Infect. Dis. 2010, 202 (Suppl. 2), S302–S308. [Google Scholar] [CrossRef] [PubMed]
- Goulder, P.J.; Walker, B.D. The great escape—AIDS viruses and immune control. Nat. Med. 1999, 5, 1233–1235. [Google Scholar] [CrossRef]
- Fujiwara, M.; Takiguchi, M. HIV-1-specific CTLs effectively suppress replication of HIV-1 in HIV-1-infected macrophages. Blood 2007, 109, 4832–4838. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Severino, M.E.; Sipsas, N.V.; Nguyen, P.T.; Kalams, S.A.; Walker, B.D.; Johnson, R.P.; Yang, O.O. Inhibition of human immunodeficiency virus type 1 replication in primary CD4(+) T lymphocytes, monocytes, and dendritic cells by cytotoxic T lymphocytes. J. Virol. 2000, 74, 6695–6699. [Google Scholar] [CrossRef]
- Walker-Sperling, V.E.; Buckheit, R.W., 3rd; Blankson, J.N. Comparative analysis of the capacity of elite suppressor CD4+ and CD8+ T cells to inhibit HIV-1 replication in monocyte-derived macrophages. J. Virol. 2014, 88, 9789–9798. [Google Scholar] [CrossRef]
- Walker-Sperling, V.E.; Merlo, C.A.; Buckheit, R.W., 3rd; Lambert, A.; Tarwater, P.; Kirk, G.D.; Drummond, M.B.; Blankson, J.N. Short Communication: HIV Controller T Cells Effectively Inhibit Viral Replication in Alveolar Macrophages. AIDS Res. Hum. Retrovir. 2016, 32, 1097–1099. [Google Scholar] [CrossRef]
- Kawamura, T.; Gulden, F.O.; Sugaya, M.; McNamara, D.T.; Borris, D.L.; Lederman, M.M.; Orenstein, J.M.; Zimmerman, P.A.; Blauvelt, A. R5 HIV productively infects Langerhans cells, and infection levels are regulated by compound CCR5 polymorphisms. Proc. Natl. Acad. Sci. USA 2003, 100, 8401–8406. [Google Scholar] [CrossRef] [PubMed]
- Jekle, A.; Keppler, O.T.; De Clercq, E.; Schols, D.; Weinstein, M.; Goldsmith, M.A. In vivo evolution of human immunodeficiency virus type 1 toward increased pathogenicity through CXCR4-mediated killing of uninfected CD4 T cells. J. Virol. 2003, 77, 5846–5854. [Google Scholar] [CrossRef] [PubMed]
- An, P.; Winkler, C.A. Host genes associated with HIV/AIDS: Advances in gene discovery. Trends Genet. 2010, 26, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Brass, A.L.; Dykxhoorn, D.M.; Benita, Y.; Yan, N.; Engelman, A.; Xavier, R.J.; Lieberman, J.; Elledge, S.J. Identification of host proteins required for HIV infection through a functional genomic screen. Science 2008, 319, 921–926. [Google Scholar] [CrossRef]
- Corbeil, J.; Sheeter, D.; Genini, D.; Rought, S.; Leoni, L.; Du, P.; Ferguson, M.; Masys, D.R.; Welsh, J.B.; Fink, J.L.; et al. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 2001, 11, 1198–1204. [Google Scholar] [CrossRef]
- McLaren, P.J.; Pulit, S.L.; Gurdasani, D.; Bartha, I.; Shea, P.R.; Pomilla, C.; Gupta, N.; Gkrania-Klotsas, E.; Young, E.H.; Bannert, N.; et al. Evaluating the Impact of Functional Genetic Variation on HIV-1 Control. J. Infect. Dis. 2017, 216, 1063–1069. [Google Scholar] [CrossRef]
- Ockenhouse, C.F.; Bernstein, W.B.; Wang, Z.; Vahey, M.T. Functional genomic relationships in HIV-1 disease revealed by gene-expression profiling of primary human peripheral blood mononuclear cells. J. Infect. Dis. 2005, 191, 2064–2074. [Google Scholar] [CrossRef]
- Devadas, K.; Biswas, S.; Haleyurgirisetty, M.; Wood, O.; Ragupathy, V.; Lee, S.; Hewlett, I. Analysis of Host Gene Expression Profile in HIV-1 and HIV-2 Infected T-Cells. PLoS ONE 2016, 11, e0147421. [Google Scholar] [CrossRef]
- Biswas, S.; Haleyurgirisetty, M.; Ragupathy, V.; Wang, X.; Lee, S.; Hewlett, I.; Devadas, K. Differentially expressed host long intergenic noncoding RNA and mRNA in HIV-1 and HIV-2 infection. Sci. Rep. 2018, 8, 2546. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, F.; Argyris, E.; Chen, K.; Liang, Z.; Tian, H.; Huang, W.; Squires, K.; Verlinghieri, G.; Zhang, H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007, 13, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Lecellier, C.H.; Dunoyer, P.; Arar, K.; Lehmann-Che, J.; Eyquem, S.; Himber, C.; Saib, A.; Voinnet, O. A cellular microRNA mediates antiviral defense in human cells. Science 2005, 308, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, M.; Jing, Q.; Georgel, P.; New, L.; Chen, J.; Mols, J.; Kang, Y.J.; Jiang, Z.; Du, X.; Cook, R.; et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 2007, 27, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Bignami, F.; Pilotti, E.; Bertoncelli, L.; Ronzi, P.; Gulli, M.; Marmiroli, N.; Magnani, G.; Pinti, M.; Lopalco, L.; Mussini, C.; et al. Stable changes in CD4+ T lymphocyte miRNA expression after exposure to HIV-1. Blood 2012, 119, 6259–6267. [Google Scholar] [CrossRef]
- Wang, X.; Ye, L.; Hou, W.; Zhou, Y.; Wang, Y.J.; Metzger, D.S.; Ho, W.Z. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood 2009, 113, 671–674. [Google Scholar] [CrossRef]
- Witwer, K.W.; Watson, A.K.; Blankson, J.N.; Clements, J.E. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology 2012, 9, 5. [Google Scholar] [CrossRef]
- Wu, J.Q.; Dwyer, D.E.; Dyer, W.B.; Yang, Y.H.; Wang, B.; Saksena, N.K. Genome-wide analysis of primary CD4+ and CD8+ T cell transcriptomes shows evidence for a network of enriched pathways associated with HIV disease. Retrovirology 2011, 8, 18. [Google Scholar] [CrossRef]
- Ahluwalia, J.K.; Khan, S.Z.; Soni, K.; Rawat, P.; Gupta, A.; Hariharan, M.; Scaria, V.; Lalwani, M.; Pillai, B.; Mitra, D.; et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology 2008, 5, 117. [Google Scholar] [CrossRef]
- Gavegnano, C.; Detorio, M.A.; Bassit, L.; Hurwitz, S.J.; North, T.W.; Schinazi, R.F. Cellular pharmacology and potency of HIV-1 nucleoside analogs in primary human macrophages. Antimicrob. Agents Chemother. 2013, 57, 1262–1269. [Google Scholar] [CrossRef] [PubMed]
- Crowe, S.M.; Westhorpe, C.L.; Mukhamedova, N.; Jaworowski, A.; Sviridov, D.; Bukrinsky, M. The macrophage: The intersection between HIV infection and atherosclerosis. J. Leukoc. Biol. 2010, 87, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Garden, G.A. Microglia in human immunodeficiency virus-associated neurodegeneration. Glia 2002, 40, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Lamers, S.L.; Fogel, G.B.; Singer, E.J.; Salemi, M.; Nolan, D.J.; Huysentruyt, L.C.; McGrath, M.S. HIV-1 Nef in macrophage-mediated disease pathogenesis. Int. Rev. Immunol. 2012, 31, 432–450. [Google Scholar] [CrossRef] [PubMed]
- Hsu, D.C.; Sereti, I.; Ananworanich, J. Serious Non-AIDS events: Immunopathogenesis and interventional strategies. AIDS Res. Ther. 2013, 10, 29. [Google Scholar] [CrossRef]
- Gannon, P.; Khan, M.Z.; Kolson, D.L. Current understanding of HIV-associated neurocognitive disorders pathogenesis. Curr. Opin. Neurol. 2011, 24, 275–283. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Popper, S.J.; Sarr, A.D.; Gueye-Ndiaye, A.; Mboup, S.; Essex, M.E.; Kanki, P.J. Low plasma human immunodeficiency virus type 2 viral load is independent of proviral load: Low virus production in vivo. J. Virol. 2000, 74, 1554–1557. [Google Scholar] [CrossRef]
- Popper, S.J.; Sarr, A.D.; Travers, K.U.; Gueye-Ndiaye, A.; Mboup, S.; Essex, M.E.; Kanki, P.J. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 1999, 180, 1116–1121. [Google Scholar] [CrossRef]
- Jin, W.; Ibeagha-Awemu, E.M.; Liang, G.; Beaudoin, F.; Zhao, X.; Guan le, L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genom. 2014, 15, 181. [Google Scholar] [CrossRef]
- Lawless, N.; Foroushani, A.B.; McCabe, M.S.; O’Farrelly, C.; Lynn, D.J. Next generation sequencing reveals the expression of a unique miRNA profile in response to a gram-positive bacterial infection. PLoS ONE 2013, 8, e57543. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; Gabriely, G. miR-21: A small multi-faceted RNA. J. Cell. Mol. Med. 2009, 13, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Verollet, C.; Souriant, S.; Bonnaud, E.; Jolicoeur, P.; Raynaud-Messina, B.; Kinnaer, C.; Fourquaux, I.; Imle, A.; Benichou, S.; Fackler, O.T.; et al. HIV-1 reprograms the migration of macrophages. Blood 2015, 125, 1611–1622. [Google Scholar] [CrossRef]
- Devadas, K.; Biswas, S.; Haleyurgirisetty, M.; Ragupathy, V.; Wang, X.; Lee, S.; Hewlett, I. Identification of Host Micro RNAs That Differentiate HIV-1 and HIV-2 Infection Using Genome Expression Profiling Techniques. Viruses 2016, 8, 121. [Google Scholar] [CrossRef]
- Narla, V.; Bhakta, N.; Freedman, J.E.; Tanriverdi, K.; Maka, K.; Deeks, S.G.; Ganz, P.; Hsue, P. Unique Circulating MicroRNA Profiles in HIV Infection. J. Acquir. Immune Defic. Syndr. 2018, 79, 644–650. [Google Scholar] [CrossRef]
- Qi, Y.; Hu, H.; Guo, H.; Xu, P.; Shi, Z.; Huan, X.; Zhu, Z.; Zhou, M.; Cui, L. MicroRNA profiling in plasma of HIV-1 infected patients: Potential markers of infection and immune status. J. Public Health Emerg. 2017, 1, 65. [Google Scholar] [CrossRef]
- De Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Di Leva, G.; Piovan, C.; Gasparini, P.; Ngankeu, A.; Taccioli, C.; Briskin, D.; Cheung, D.G.; Bolon, B.; Anderlucci, L.; Alder, H.; et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013, 9, e1003311. [Google Scholar] [CrossRef]
- Kang, S.M.; Lee, H.J.; Cho, J.Y. MicroRNA-365 regulates NKX2-1, a key mediator of lung cancer. Cancer Lett. 2013, 335, 487–494. [Google Scholar] [CrossRef]
- Kefas, B.; Comeau, L.; Floyd, D.H.; Seleverstov, O.; Godlewski, J.; Schmittgen, T.; Jiang, J.; di Pierro, C.G.; Li, Y.; Chiocca, E.A.; et al. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J. Neurosci. 2009, 29, 15161–15168. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, N.; Ahmad, H.M.; Molparia, B.; Kulshreshtha, R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis 2013, 34, 1889–1899. [Google Scholar] [CrossRef] [PubMed]
- Oster, B.; Linnet, L.; Christensen, L.L.; Thorsen, K.; Ongen, H.; Dermitzakis, E.T.; Sandoval, J.; Moran, S.; Esteller, M.; Hansen, T.F.; et al. Non-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancer. Int. J. Cancer 2013, 132, 2303–2315. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Cui, X.; Hong, Y.; Wang, J.; Li, Y.; Chen, L.; Liu, Y.; Gao, Y.; Xu, D.; Wang, Q. MicroRNA-185 suppresses proliferation, invasion, migration, and tumorigenicity of human prostate cancer cells through targeting androgen receptor. Mol. Cell. Biochem. 2013, 377, 121–130. [Google Scholar] [CrossRef]
- Xu, Y.Y.; Wu, H.J.; Ma, H.D.; Xu, L.P.; Huo, Y.; Yin, L.R. MicroRNA-503 suppresses proliferation and cell-cycle progression of endometrioid endometrial cancer by negatively regulating cyclin D1. FEBS J. 2013, 280, 3768–3779. [Google Scholar] [CrossRef]
- Reynoso, R.; Laufer, N.; Hackl, M.; Skalicky, S.; Monteforte, R.; Turk, G.; Carobene, M.; Quarleri, J.; Cahn, P.; Werner, R.; et al. MicroRNAs differentially present in the plasma of HIV elite controllers reduce HIV infection in vitro. Sci. Rep. 2014, 4, 5915. [Google Scholar] [CrossRef]
- Qased, A.B.; Yi, H.; Liang, N.; Ma, S.; Qiao, S.; Liu, X. MicroRNA-18a upregulates autophagy and ataxia telangiectasia mutated gene expression in HCT116 colon cancer cells. Mol. Med. Rep. 2013, 7, 559–564. [Google Scholar] [CrossRef]
- Cao, P.; Zhang, M.; Wang, L.; Sai, B.; Tang, J.; Luo, Z.; Shuai, C.; Zhang, L.; Li, Z.; Wang, Y.; et al. miR-18a reactivates the Epstein-Barr virus through defective DNA damage response and promotes genomic instability in EBV-associated lymphomas. BMC Cancer 2018, 18, 1293. [Google Scholar] [CrossRef]
- Ariumi, Y.; Trono, D. Ataxia-telangiectasia-mutated (ATM) protein can enhance human immunodeficiency virus type 1 replication by stimulating Rev function. J. Virol. 2006, 80, 2445–2452. [Google Scholar] [CrossRef]
- Song, L.; Lin, C.; Wu, Z.; Gong, H.; Zeng, Y.; Wu, J.; Li, M.; Li, J. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS ONE 2011, 6, e25454. [Google Scholar] [CrossRef]
- Symmans, W.F.; Volm, M.D.; Shapiro, R.L.; Perkins, A.B.; Kim, A.Y.; Demaria, S.; Yee, H.T.; McMullen, H.; Oratz, R.; Klein, P.; et al. Paclitaxel-induced apoptosis and mitotic arrest assessed by serial fine-needle aspiration: Implications for early prediction of breast cancer response to neoadjuvant treatment. Clin. Cancer Res. 2000, 6, 4610–4617. [Google Scholar]
- Lehtinen, M.K.; Yuan, Z.; Boag, P.R.; Yang, Y.; Villen, J.; Becker, E.B.; DiBacco, S.; de la Iglesia, N.; Gygi, S.; Blackwell, T.K.; et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 2006, 125, 987–1001. [Google Scholar] [CrossRef] [PubMed]
- Skurk, C.; Maatz, H.; Kim, H.S.; Yang, J.; Abid, M.R.; Aird, W.C.; Walsh, K. The Akt-regulated forkhead transcription factor FOXO3a controls endothelial cell viability through modulation of the caspase-8 inhibitor FLIP. J. Biol. Chem. 2004, 279, 1513–1525. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, I.; Zhu, X.; Kharas, M.G.; Chen, J.; Fruman, D.A. Optimal B-cell proliferation requires phosphoinositide 3-kinase-dependent inactivation of FOXO transcription factors. Blood 2004, 104, 784–787. [Google Scholar] [CrossRef]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Greer, E.L.; Brunet, A. FOXO transcription factors in ageing and cancer. Acta Physiol. 2008, 192, 19–28. [Google Scholar] [CrossRef]
- Huang, H.; Regan, K.M.; Wang, F.; Wang, D.; Smith, D.I.; van Deursen, J.M.; Tindall, D.J. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA 2005, 102, 1649–1654. [Google Scholar] [CrossRef]
- Plas, D.R.; Thompson, C.B. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J. Biol. Chem. 2003, 278, 12361–12366. [Google Scholar] [CrossRef]
- Tran, H.; Brunet, A.; Griffith, E.C.; Greenberg, M.E. The many forks in FOXO’s road. Sci. STKE 2003, 2003, RE5. [Google Scholar] [CrossRef]
- Zhao, J.; Brault, J.J.; Schild, A.; Goldberg, A.L. Coordinate activation of autophagy and the proteasome pathway by FoxO transcription factor. Autophagy 2008, 4, 378–380. [Google Scholar] [CrossRef]
- Cui, M.; Huang, Y.; Zhao, Y.; Zheng, J. New insights for FOXO and cell-fate decision in HIV infection and HIV associated neurocognitive disorder. Adv. Exp. Med. Biol. 2009, 665, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.E.; Carneiro, J.; Meier-Schellersheim, M.; Grossman, Z.; Victorino, R.M. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J. Immunol. 2002, 169, 3400–3406. [Google Scholar] [CrossRef] [PubMed]
- Miedema, F.; Hazenberg, M.D.; Tesselaar, K.; van Baarle, D.; de Boer, R.J.; Borghans, J.A. Immune activation and collateral damage in AIDS pathogenesis. Front. Immunol. 2013, 4, 298. [Google Scholar] [CrossRef] [PubMed]
- Younes, S.A.; Yassine-Diab, B.; Dumont, A.R.; Boulassel, M.R.; Grossman, Z.; Routy, J.P.; Sekaly, R.P. HIV-1 viremia prevents the establishment of interleukin 2-producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J. Exp. Med. 2003, 198, 1909–1922. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Oyaizu, N.; Than, S.; McCloskey, T.W.; Pahwa, S. IL-2 rescues in vitro lymphocyte apoptosis in patients with HIV infection: Correlation with its ability to block culture-induced down-modulation of Bcl-2. J. Immunol. 1996, 157, 4184–4193. [Google Scholar]
- Banki, K.; Hutter, E.; Gonchoroff, N.J.; Perl, A. Molecular ordering in HIV-induced apoptosis. Oxidative stress, activation of caspases, and cell survival are regulated by transaldolase. J. Biol. Chem. 1998, 273, 11944–11953. [Google Scholar] [CrossRef]
- Bian, T.; Gibbs, J.D.; Orvell, C.; Imani, F. Respiratory syncytial virus matrix protein induces lung epithelial cell cycle arrest through a p53 dependent pathway. PLoS ONE 2012, 7, e38052. [Google Scholar] [CrossRef]
- Janicke, R.U.; Sohn, D.; Schulze-Osthoff, K. The dark side of a tumor suppressor: Anti-apoptotic p53. Cell Death Differ. 2008, 15, 959–976. [Google Scholar] [CrossRef]
- Genini, D.; Sheeter, D.; Rought, S.; Zaunders, J.J.; Susin, S.A.; Kroemer, G.; Richman, D.D.; Carson, D.A.; Corbeil, J.; Leoni, L.M. HIV induces lymphocyte apoptosis by a p53-initiated, mitochondrial-mediated mechanism. FASEB J. 2001, 15, 5–6. [Google Scholar] [CrossRef]
- Imbeault, M.; Lodge, R.; Ouellet, M.; Tremblay, M.J. Efficient magnetic bead-based separation of HIV-1-infected cells using an improved reporter virus system reveals that p53 up-regulation occurs exclusively in the virus-expressing cell population. Virology 2009, 393, 160–167. [Google Scholar] [CrossRef]
- Roshal, M.; Kim, B.; Zhu, Y.; Nghiem, P.; Planelles, V. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 2003, 278, 25879–25886. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Ali, A.; Arora, S.; Banerjea, A.C. Inhibition of beta-TrcP-dependent ubiquitination of p53 by HIV-1 Vpu promotes p53-mediated apoptosis in human T cells. Blood 2011, 117, 6600–6607. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.H.; Kim, S.Y.; Byeon, S.E.; Jeong, Y.; Lee, J.; Kim, K.P.; Park, J.; Bae, Y.S. p53-derived host restriction of HIV-1 replication by protein kinase R-mediated Tat phosphorylation and inactivation. J. Virol. 2015, 89, 4262–4280. [Google Scholar] [CrossRef] [PubMed]
- Breton, Y.; Desrosiers, V.; Ouellet, M.; Deshiere, A.; Torresilla, C.; Cohen, E.A.; Tremblay, M.J. Expression of MDM2 in Macrophages Promotes the Early Postentry Steps of HIV-1 Infection through Inhibition of p53. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Muthumani, K.; Shedlock, D.J.; Choo, D.K.; Fagone, P.; Kawalekar, O.U.; Goodman, J.; Bian, C.B.; Ramanathan, A.A.; Atman, P.; Tebas, P.; et al. HIV-mediated phosphatidylinositol 3-kinase/serine-threonine kinase activation in APCs leads to programmed death-1 ligand upregulation and suppression of HIV-specific CD8 T cells. J. Immunol. 2011, 187, 2932–2943. [Google Scholar] [CrossRef] [PubMed]
- Pasquereau, S.; Kumar, A.; Abbas, W.; Herbein, G. Counteracting Akt Activation by HIV Protease Inhibitors in Monocytes/Macrophages. Viruses 2018, 10, 190. [Google Scholar] [CrossRef]



| Sample Name | HIV-1 (pg/mL) | HIV-2 (pg/mL) |
|---|---|---|
| Cell Type: Peripheral Blood Mononuclear Cells (PBMCs) | ||
| Donor 1 | 3148.13 | 28,011.78 |
| Donor 2 | 4019.29 | 27,121.72 |
| Donor 3 | 1653.06 | 24,534.10 |
| Cell Type: Monocytes-Derived Macrophages (MDMs) | ||
| Donor 1 | 1468.62 | 17,981.50 |
| Donor 2 | 5451.53 | 22,746.26 |
| Donor 3 | 2065.89 | 18,446.87 |
| Sample Name | Clean Reads | Adapter-Trimmed Reads (Length ≥ 15 nt) | Reads Aligned to Human Pre-miRNA in miRBase 21 |
|---|---|---|---|
| Cell Type: Peripheral Blood Mononuclear Cells (PBMCs) | |||
| Control | |||
| Donor 1 | 16,367,003 | 15,696,848 | 8,684,872 |
| Donor 2 | 4,495,971 | 4,366,075 | 2,816,056 |
| Donor 3 | 13,046,715 | 12,445,186 | 6,553,373 |
| HIV-1 | |||
| Donor 1 | 17,138,982 | 15,436,055 | 6,409,199 |
| Donor 2 | 20,145,570 | 18,802,590 | 8,531,490 |
| Donor 3 | 11,429,935 | 10,390,651 | 5,447,327 |
| HIV-2 | |||
| Donor 1 | 14,140,517 | 13,362,208 | 8,078,730 |
| Donor 2 | 18,455,281 | 18,073,452 | 11,485,496 |
| Donor 3 | 19,037,081 | 18,423,451 | 11,583,858 |
| Cell Type: Monocytes-Derived Macrophages (MDMs) | |||
| Control | |||
| Donor 1 | 1,284,807 | 919,078 | 295,694 |
| Donor 2 | 1,509,542 | 1,373,235 | 587,411 |
| Donor 3 | 1,730,255 | 1,687,860 | 1,005,415 |
| HIV-1 | |||
| Donor 1 | 1,246,998 | 949,827 | 466,372 |
| Donor 2 | 1,378,466 | 1,166,657 | 386,670 |
| Donor 3 | 1,782,736 | 1,706,479 | 1,008,880 |
| HIV-2 | |||
| Donor 1 | 2,278,656 | 1,898,057 | 711,737 |
| Donor 2 | 1,365,011 | 1,194,546 | 559,974 |
| Donor 3 | 1,993,830 | 1,856,716 | 1,045,156 |
| Samples | Known-miRNAs | Novel-miRNAs | Total |
|---|---|---|---|
| Cell Type: Peripheral Blood Mononuclear Cells (PBMCs) | |||
| Control | 848 | 414 | 1262 |
| HIV-1 | 737 | 353 | 1090 |
| HIV-2 | 772 | 342 | 1114 |
| Total | 918 | 532 | 1450 |
| Cell Type: Monocytes-Derived Macrophages (MDMs) | |||
| Control | 642 | 155 | 797 |
| HIV-1 | 585 | 126 | 709 |
| HIV-2 | 658 | 144 | 802 |
| Total | 780 | 210 | 990 |
| Pathway Name | HIV-1 | |||||
|---|---|---|---|---|---|---|
| MDMs | PBMCs | |||||
| Enrichment Score | p-Value | Pathways Modulated by Up/Downregulated miRNAs | Enrichment Score | p-Value | Pathways Modulated by Up/Downregulated miRNAs | |
| Oxytocin signaling pathway | 1.905063 | 0.01244335 | up | 2.692827 | 0.002028492 | up |
| Renin secretion | 1.734644 | 0.01842282 | up | 1.661208 | 0.02181686 | up |
| p53 signaling pathway | 1.651102 | 0.02233047 | up | 2.98958 | 0.001024284 | down |
| mTOR signaling pathway | 2.810829 | 0.001545864 | down | 9.093665 | 0.0000000008060005 | down |
| Signaling pathways regulating pluripotency of stem cells | 2.228881 | 0.005903624 | down | 8.711413 | 0.000000001943511 | down |
| Pathways in cancer | 2.421814 | 0.003786047 | down | 6.618831 | 0.0000002405298 | down |
| Hepatocellular carcinoma | 2.576053 | 0.002654281 | down | 4.732601 | 0.00001850968 | down |
| Hippo signaling pathway | 2.767124 | 0.001709528 | down | 4.424071 | 0.00003766423 | down |
| FoxO signaling pathway | 3.114274 | 0.0007686461 | down | 3.539176 | 0.0002889511 | down |
| Gastric cancer | 3.65153 | 0.0002230849 | down | 3.28796 | 0.0005152755 | down |
| HTLV-I infection | 1.71839 | 0.0191254 | down | 2.912919 | 0.001222027 | down |
| Wnt signaling pathway | 2.13995 | 0.0072452 | down | 2.590099 | 0.002569812 | down |
| Chronic myeloid leukemia | 1.595796 | 0.0253632 | down | 2.2901 | 0.00512743 | down |
| ErbB signaling pathway | 1.470936 | 0.03381145 | down | 3.314262 | 0.0004849954 | up |
| 2.361913 | 0.004345974 | down | ||||
| Ras signaling pathway | 1.358576 | 0.0437949 | down | 3.285602 | 0.0005180819 | up |
| 3.212785 | 0.000612653 | down | ||||
| Human papillomavirus infection | 2.165002 | 0.006839085 | down | 2.123619 | 0.007522818 | up |
| 3.956626 | 0.000110503 | down | ||||
| Renal cell carcinoma | 1.705678 | 0.01969345 | down | 1.890512 | 0.01286731 | up |
| 4.353166 | 0.00004434387 | down | ||||
| Colorectal cancer | 1.458046 | 0.03483006 | down | 1.586711 | 0.02589933 | up |
| 3.290669 | 0.0005120717 | down | ||||
| Breast cancer | 2.127675 | 0.007452897 | down | 1.586524 | 0.02591051 | up |
| 5.795785 | 0.000001600352 | down | ||||
| Proteoglycans in cancer | 2.872341 | 0.001341712 | down | 1.572002 | 0.02679158 | up |
| 6.080734 | 0.00000083036 | down | ||||
| Melanogenesis | 2.00586 | 0.009865974 | down | 1.423319 | 0.03772947 | up |
| 1.777427 | 0.0166945 | down | ||||
| Cushing syndrome | 2.044558 | 0.009024896 | down | 1.410542 | 0.03885604 | up |
| 2.338646 | 0.004585155 | down | ||||
| Basal cell carcinoma | 2.739614 | 0.001821318 | down | 1.358427 | 0.04381 | up |
| 2.95548 | 0.001107949 | down | ||||
| Pathway Name | HIV-2 | |||||
|---|---|---|---|---|---|---|
| MDMs | PBMCs | |||||
| Enrichment Score | p-Value | Pathways Modulated by Up/Downregulated miRNAs | Enrichment Score | p-Value | Pathways Modulated by Up/Downregulated miRNAs | |
| PI3K-Akt signaling pathway | 3.708008 | 0.0001958807 | up | 2.475812 | 0.003343395 | up |
| Breast cancer | 3.577508 | 0.0002645407 | up | 3.235154 | 0.0005818966 | up |
| Cell cycle | 3.358965 | 0.000437557 | up | 1.936352 | 0.01157838 | up |
| Pathways in cancer | 3.248617 | 0.0005641353 | up | 2.994257 | 0.001013313 | up |
| Fc gamma R-mediated phagocytosis | 2.996289 | 0.001008581 | up | 1.570297 | 0.02689695 | up |
| Ras signaling pathway | 2.981019 | 0.001044674 | up | 1.614705 | 0.02428257 | up |
| Hippo signaling pathway | 2.445519 | 0.003584934 | up | 2.333154 | 0.004643505 | up |
| Endocrine resistance | 2.229404 | 0.005896524 | up | 1.487751 | 0.03252735 | up |
| Sphingolipid signaling pathway | 1.691501 | 0.02034693 | up | 2.010001 | 0.009772351 | up |
| Cushing syndrome | 1.670783 | 0.02134112 | up | 1.623592 | 0.02379075 | up |
| Hepatocellular carcinoma | 1.415382 | 0.03842537 | up | 1.50253 | 0.03143911 | up |
| Amino sugar and nucleotide sugar metabolism | 2.710335 | 0.001948342 | down | 1.338931 | 0.04582143 | up |
| Fructose and mannose metabolism | 2.23792 | 0.005782019 | down | 1.639237 | 0.02294895 | up |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswas, S.; Chen, E.; Haleyurgirisetty, M.; Lee, S.; Hewlett, I.; Devadas, K. Comparison of miRNA Expression Profiles between HIV-1 and HIV-2 Infected Monocyte-Derived Macrophages (MDMs) and Peripheral Blood Mononuclear Cells (PBMCs). Int. J. Mol. Sci. 2020, 21, 6970. https://doi.org/10.3390/ijms21186970
Biswas S, Chen E, Haleyurgirisetty M, Lee S, Hewlett I, Devadas K. Comparison of miRNA Expression Profiles between HIV-1 and HIV-2 Infected Monocyte-Derived Macrophages (MDMs) and Peripheral Blood Mononuclear Cells (PBMCs). International Journal of Molecular Sciences. 2020; 21(18):6970. https://doi.org/10.3390/ijms21186970
Chicago/Turabian StyleBiswas, Santanu, Emily Chen, Mohan Haleyurgirisetty, Sherwin Lee, Indira Hewlett, and Krishnakumar Devadas. 2020. "Comparison of miRNA Expression Profiles between HIV-1 and HIV-2 Infected Monocyte-Derived Macrophages (MDMs) and Peripheral Blood Mononuclear Cells (PBMCs)" International Journal of Molecular Sciences 21, no. 18: 6970. https://doi.org/10.3390/ijms21186970
APA StyleBiswas, S., Chen, E., Haleyurgirisetty, M., Lee, S., Hewlett, I., & Devadas, K. (2020). Comparison of miRNA Expression Profiles between HIV-1 and HIV-2 Infected Monocyte-Derived Macrophages (MDMs) and Peripheral Blood Mononuclear Cells (PBMCs). International Journal of Molecular Sciences, 21(18), 6970. https://doi.org/10.3390/ijms21186970




