Classifications of Neurodegenerative Disorders Using a Multiplex Blood Biomarkers-Based Machine Learning Model
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics
2.2. Plasma Biomarker Levels in Different Disease Groups
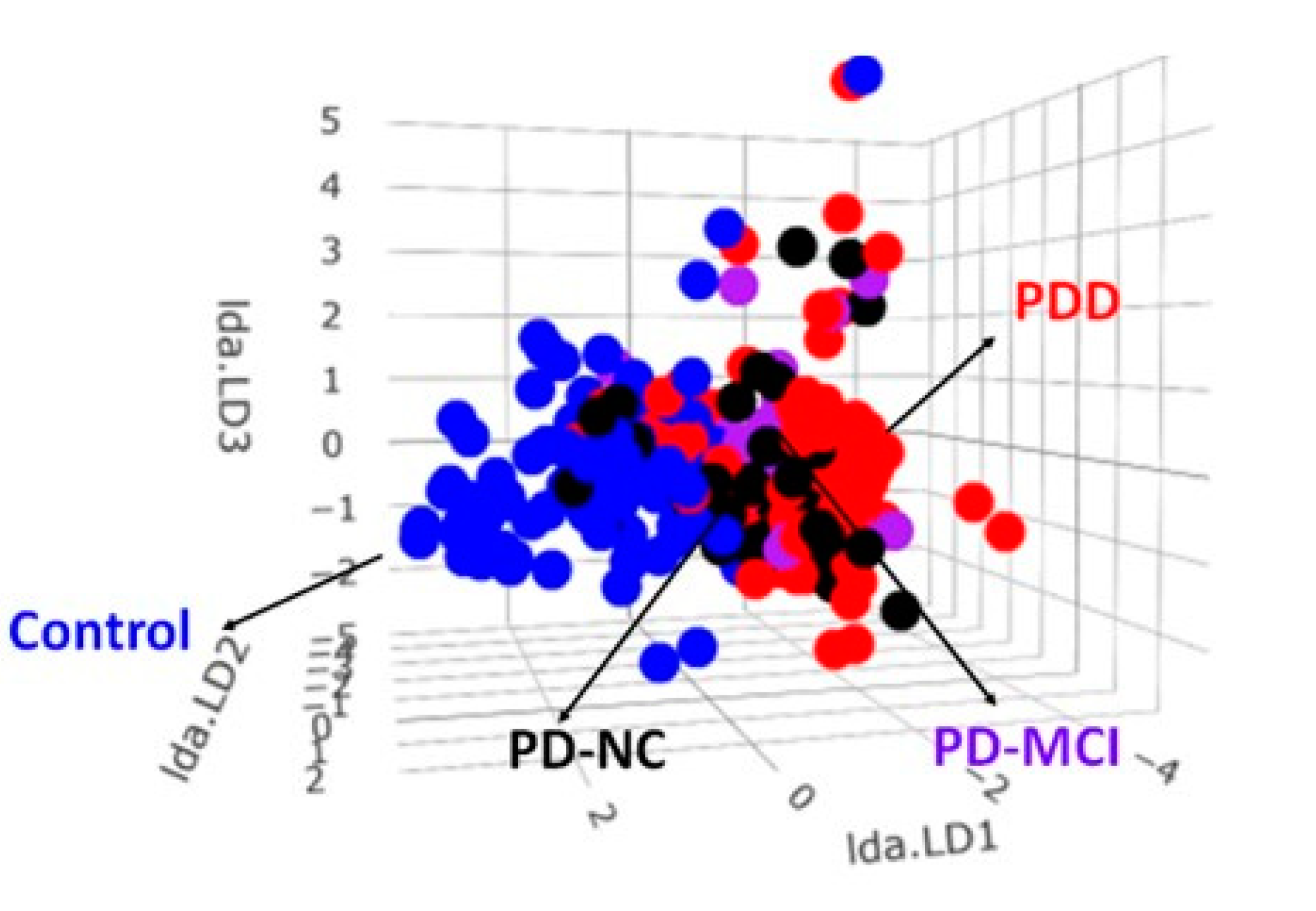
2.3. The LDA Model for Classifying Controls, AD Spectrum, PD Spectrum, and FTD
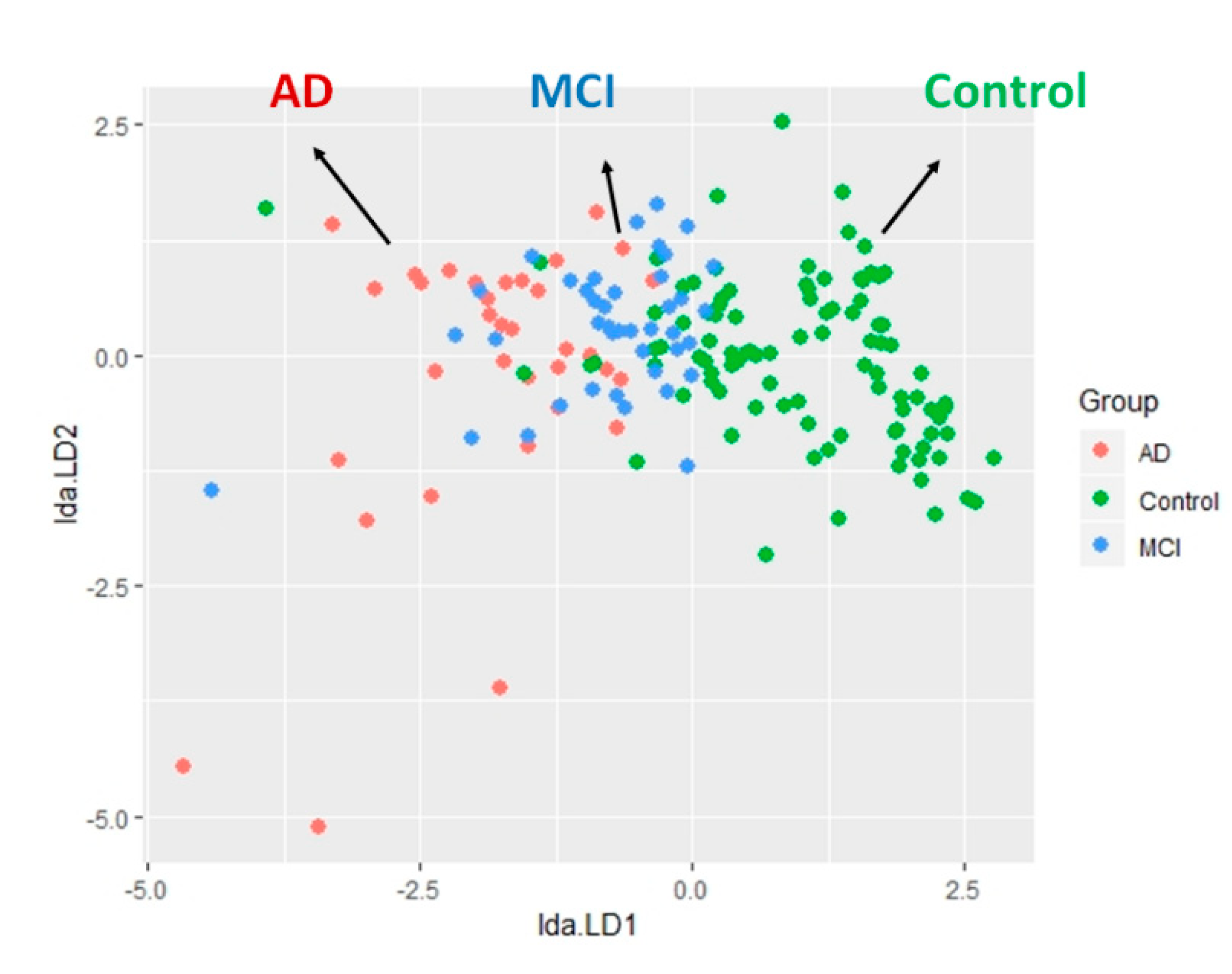
2.4. The LDA Model for Classifying Controls and AD Spectrum
2.5. The LDA Model for Classifying Controls and PD Spectrum
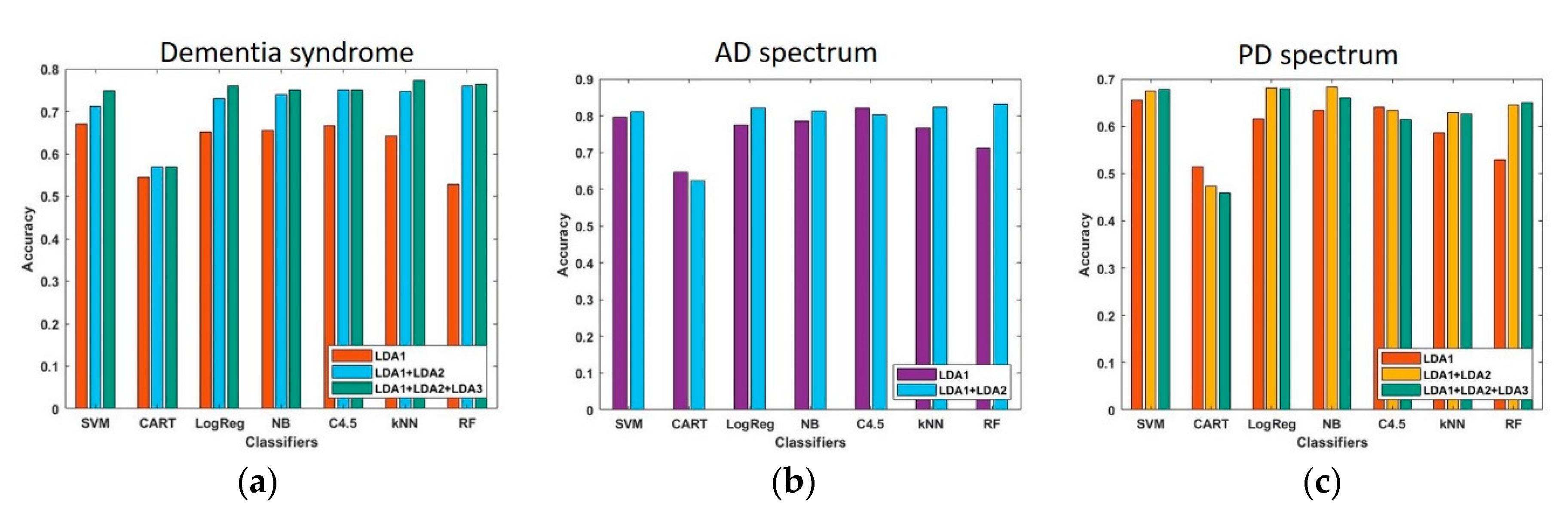
2.6. Measure the Performance of the Established Models by LDA
3. Discussion
4. Materials and Methods
4.1. Study Participants
4.2. Biomarker Assessments
4.3. Statistical Analyses for Clinical and Biomarker Characteristics
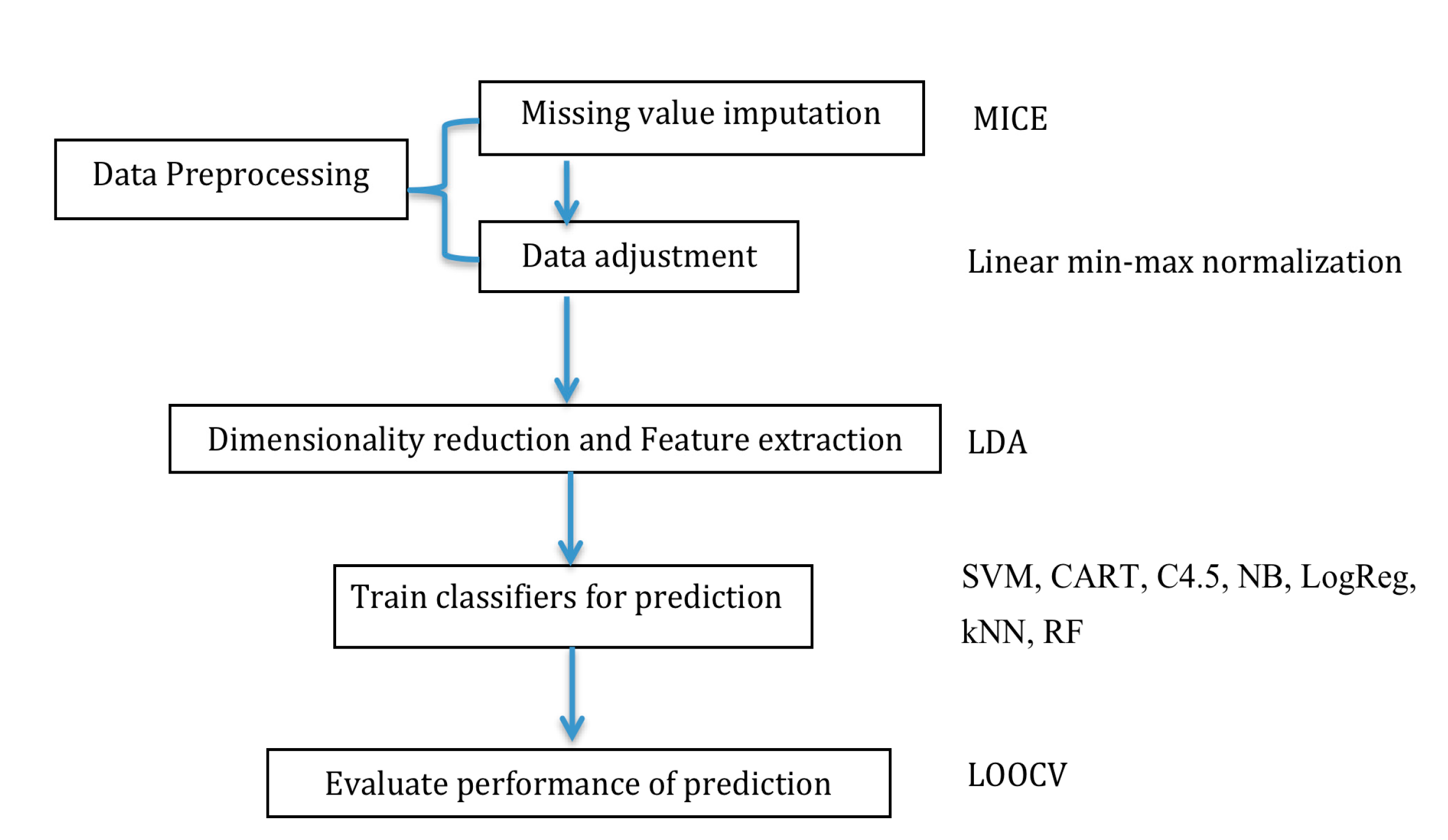
4.4. Data Processing and Dimensionality Reduction
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MCI | Mild cognitive impairment |
| AD | Alzheimer’s disease |
| PD | Parkinson’s disease |
| PD-NC | Parkinson’s disease with normal cognition |
| PD-MCI | Parkinson’s disease with mild cognitive impairment |
| FTD | Frontotemporal dementia |
| LDA | Linear discriminant analysis |
| MICE | Multivariate imputation by chained equation |
| LOOCV | Leave-one-out cross-validation |
| NB | Naïve Bayes |
| kNN | k-nearest neighbor |
| SVM | Support vector machine |
| C4.5 | C4.5 decision tree |
| CART | Classification and regression trees |
| RF | Random forest |
| LogReg | Logistic regression. |
References
- Johnson, I.P. Age-related neurodegenerative disease research needs aging models. Front. Aging Neurosci. 2015, 7, 168. [Google Scholar] [CrossRef]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Del Tredici, K.; Rub, U.; de Vos, R.A.; Jansen Steur, E.N.; Braak, E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 2003, 24, 197–211. [Google Scholar] [CrossRef]
- Emre, M.; Aarsland, D.; Brown, R.; Burn, D.J.; Duyckaerts, C.; Mizuno, Y.; Broe, G.A.; Cummings, J.; Dickson, D.W.; Gauthier, S.; et al. Clinical diagnostic criteria for dementia associated with Parkinson’s disease. Mov. Disord. 2007, 22, 1689–1707, quiz 1837. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, E.; Wenning, G.; Poewe, W. The diagnosis of Parkinson’s disease. Lancet Neurol. 2006, 5, 75–86. [Google Scholar] [CrossRef]
- Dubois, B.; Feldman, H.H.; Jacova, C.; Dekosky, S.T.; Barberger-Gateau, P.; Cummings, J.; Delacourte, A.; Galasko, D.; Gauthier, S.; Jicha, G.; et al. Research criteria for the diagnosis of Alzheimer’s disease: Revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007, 6, 734–746. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Volc, D.; Poewe, W.; Kutzelnigg, A.; Luhrs, P.; Thun-Hohenstein, C.; Schneeberger, A.; Galabova, G.; Majbour, N.; Vaikath, N.; El-Agnaf, O.; et al. Safety and immunogenicity of the alpha-synuclein active immunotherapeutic PD01A in patients with Parkinson’s disease: A randomised, single-blinded, phase 1 trial. Lancet Neurol. 2020, 19, 591–600. [Google Scholar] [CrossRef]
- Marciani, D.J. Promising Results from Alzheimer’s Disease Passive Immunotherapy Support the Development of a Preventive Vaccine. Research 2019, 2019, 5341375. [Google Scholar] [CrossRef]
- Chiu, M.J.; Yang, S.Y.; Horng, H.E.; Yang, C.C.; Chen, T.F.; Chieh, J.J.; Chen, H.H.; Chen, T.C.; Ho, C.S.; Chang, S.F.; et al. Combined plasma biomarkers for diagnosing mild cognition impairment and Alzheimer’s disease. ACS Chem. Neurosci. 2013, 4, 1530–1536. [Google Scholar] [CrossRef]
- Lin, C.H.; Yang, S.Y.; Horng, H.E.; Yang, C.C.; Chieh, J.J.; Chen, H.H.; Liu, B.H.; Chiu, M.J. Plasma alpha-synuclein predicts cognitive decline in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2017, 88, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Compta, Y.; Parkkinen, L.; Kempster, P.; Selikhova, M.; Lashley, T.; Holton, J.L.; Lees, A.J.; Revesz, T. The significance of alpha-synuclein, amyloid-beta and tau pathologies in Parkinson’s disease progression and related dementia. Neurodegener. Dis. 2014, 13, 154–156. [Google Scholar] [CrossRef]
- Kertesz, A.; McMonagle, P.; Blair, M.; Davidson, W.; Munoz, D.G. The evolution and pathology of frontotemporal dementia. Brain 2005, 128, 1996–2005. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.C.; Yang, S.Y.; Chieh, J.J.; Horng, H.E.; Hong, C.Y.; Yang, H.C.; Chen, K.H.; Shih, B.Y.; Chen, T.F.; Chiu, M.J. Biofunctionalized magnetic nanoparticles for specifically detecting biomarkers of Alzheimer’s disease in vitro. ACS Chem. Neurosci. 2011, 2, 500–505. [Google Scholar] [CrossRef]
- Zhang, H. The optimality of naive Bayes. In Proceedings of the Seventeenth International Florida Artificial Intelligence Research Society Conference, Miami Beach, FL, USA, 12–14 May 2004. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 2006, 13, 21–27. [Google Scholar] [CrossRef]
- Siac, A. Support Vector Machines, 1st ed.; Springer: Berlin, Germany, 2008. [Google Scholar]
- Quinlan, J. C4.5: Programs for Machine Learning; Morgan Kaufmann Publisher: Burlington, MA, USA, 1993. [Google Scholar]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and regression trees. WadsworthBelmont 1984, 40, 874. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–21. [Google Scholar] [CrossRef]
- Molinaro, A.M.; Simon, R.; Pfeiffer, R.M. Prediction error estimation: A comparison of resampling methods. Bioinformatics 2005, 21, 3301–3307. [Google Scholar] [CrossRef]
- Lin, C.H.; Liu, H.C.; Yang, S.Y.; Yang, K.C.; Wu, C.C.; Chiu, M.J. Plasma pS129-alpha-Synuclein Is a Surrogate Biofluid Marker of Motor Severity and Progression in Parkinson’s Disease. J. Clin. Med. 2019, 8, 1601. [Google Scholar] [CrossRef]
- Chiu, M.J.; Chen, T.F.; Hu, C.J.; Yan, S.H.; Sun, Y.; Liu, B.H.; Chang, Y.T.; Yang, C.C.; Yang, S.Y. Nanoparticle-based immunomagnetic assay of plasma biomarkers for differentiating dementia and prodromal states of Alzheimer’s disease—A cross-validation study. Nanomedicine 2020, 28, 102182. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Knopman, D.S.; Jagust, W.J.; Shaw, L.M.; Aisen, P.S.; Weiner, M.W.; Petersen, R.C.; Trojanowski, J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010, 9, 119–128. [Google Scholar] [CrossRef]
- Nakamura, A.; Kaneko, N.; Villemagne, V.L.; Kato, T.; Doecke, J.; Dore, V.; Fowler, C.; Li, Q.X.; Martins, R.; Rowe, C.; et al. High performance plasma amyloid-beta biomarkers for Alzheimer’s disease. Nature 2018, 554, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Teunissen, C.E.; Chiu, M.J.; Yang, C.C.; Yang, S.Y.; Scheltens, P.; Zetterberg, H.; Blennow, K. Plasma Amyloid-beta (Abeta42) Correlates with Cerebrospinal Fluid Abeta42 in Alzheimer’s Disease. J. Alzheimers Dis. 2018, 62, 1857–1863. [Google Scholar] [CrossRef]
- Fan, L.Y.; Tzen, K.Y.; Chen, Y.F.; Chen, T.F.; Lai, Y.M.; Yen, R.F.; Huang, Y.Y.; Shiue, C.Y.; Yang, S.Y.; Chiu, M.J. The Relation Between Brain Amyloid Deposition, Cortical Atrophy, and Plasma Biomarkers in Amnesic Mild Cognitive Impairment and Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Bitan, G.; Kirkitadze, M.D.; Lomakin, A.; Vollers, S.S.; Benedek, G.B.; Teplow, D.B. Amyloid beta -protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. USA 2003, 100, 330–335. [Google Scholar] [CrossRef]
- Duering, M.; Grimm, M.O.; Grimm, H.S.; Schroder, J.; Hartmann, T. Mean age of onset in familial Alzheimer’s disease is determined by amyloid beta 42. Neurobiol. Aging 2005, 26, 785–788. [Google Scholar] [CrossRef]
- Molinuevo, J.L.; Ayton, S.; Batrla, R.; Bednar, M.M.; Bittner, T.; Cummings, J.; Fagan, A.M.; Hampel, H.; Mielke, M.M.; Mikulskis, A.; et al. Current state of Alzheimer’s fluid biomarkers. Acta Neuropathol. 2018, 136, 821–853. [Google Scholar] [CrossRef]
- Mielke, M.M.; Hagen, C.E.; Xu, J.; Chai, X.; Vemuri, P.; Lowe, V.J.; Airey, D.C.; Knopman, D.S.; Roberts, R.O.; Machulda, M.M.; et al. Plasma phospho-tau181 increases with Alzheimer’s disease clinical severity and is associated with tau- and amyloid-positron emission tomography. Alzheimers Dement. 2018, 14, 989–997. [Google Scholar] [CrossRef]
- Palmqvist, S.; Insel, P.S.; Stomrud, E.; Janelidze, S.; Zetterberg, H.; Brix, B.; Eichenlaub, U.; Dage, J.L.; Chai, X.; Blennow, K.; et al. Cerebrospinal fluid and plasma biomarker trajectories with increasing amyloid deposition in Alzheimer’s disease. EMBO Mol. Med. 2019, 11, e11170. [Google Scholar] [CrossRef]
- Lin, C.H.; Yang, S.Y.; Horng, H.E.; Yang, C.C.; Chieh, J.J.; Chen, H.H.; Liu, B.H.; Chiu, M.J. Plasma Biomarkers Differentiate Parkinson’s Disease From Atypical Parkinsonism Syndromes. Front. Aging Neurosci. 2018, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, K.A.; Attems, J. Neuropathological evaluation of mixed dementia. J. Neurol. Sci. 2007, 257, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Caudle, W.M.; Zhang, J. Biomarker discovery in neurodegenerative diseases: A proteomic approach. Neurobiol. Dis. 2009, 35, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, M.; Matson, W.R.; Wang, L.; Matson, T.; Saunders-Pullman, R.; Bressman, S.S.; Flint Beal, M. Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain 2008, 131, 389–396. [Google Scholar] [CrossRef]
- Scherzer, C.R.; Eklund, A.C.; Morse, L.J.; Liao, Z.; Locascio, J.J.; Fefer, D.; Schwarzschild, M.A.; Schlossmacher, M.G.; Hauser, M.A.; Vance, J.M.; et al. Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc. Natl. Acad. Sci. USA 2007, 104, 955–960. [Google Scholar] [CrossRef]
- Chahine, L.M.; Stern, M.B.; Chen-Plotkin, A. Blood-based biomarkers for Parkinson’s disease. Parkinsonism Relat. Disord. 2014, 20 (Suppl. 1), S99–S103. [Google Scholar] [CrossRef]
- Hunter, P. The advent of AI and deep learning in diagnostics and imaging: Machine learning systems have potential to improve diagnostics in healthcare and imaging systems in research. EMBO Rep. 2019, 20, e48559. [Google Scholar] [CrossRef]
- Barber, I.; Nevado-Holgado, A.; Lovestone, S. A Parkinson’S Disease Protein Biomarker Panel Using the Somamer Assay And Machine Learning. Alzheimer’s Dement. 2017, 13, P1027. [Google Scholar] [CrossRef]
- Patel, H.; Iniesta, R.; Stahl, D.; Dobson, R.J.B.; Newhouse, S.J. Working Towards a Blood-Derived Gene Expression Biomarker Specific for Alzheimer’s Disease. J. Alzheimers Dis. 2020, 74, 545–561. [Google Scholar] [CrossRef]
- Huang, H.; Nguyen, T.; Ibrahim, S.; Shantharam, S.; Yue, Z.; Chen, J.Y. DMAP: A connectivity map database to enable identification of novel drug repositioning candidates. BMC Bioinform. 2015, 16 (Suppl. 13), S4. [Google Scholar] [CrossRef]
- Lue, L.F.; Pai, M.C.; Chen, T.F.; Hu, C.J.; Huang, L.K.; Lin, W.C.; Wu, C.C.; Jeng, J.S.; Blennow, K.; Sabbagh, M.N.; et al. Age-Dependent Relationship Between Plasma Abeta40 and Abeta42 and Total Tau Levels in Cognitively Normal Subjects. Front. Aging Neurosci. 2019, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.J.; Cairns, N.J.; Grossman, M.; McMillan, C.T.; Lee, E.B.; Van Deerlin, V.M.; Lee, V.M.; Trojanowski, J.Q. Frontotemporal lobar degeneration: Defining phenotypic diversity through personalized medicine. Acta Neuropathol. 2015, 129, 469–491. [Google Scholar] [CrossRef]
- Baborie, A.; Griffiths, T.D.; Jaros, E.; McKeith, I.G.; Burn, D.J.; Richardson, A.; Ferrari, R.; Moreno, J.; Momeni, P.; Duplessis, D.; et al. Pathological correlates of frontotemporal lobar degeneration in the elderly. Acta Neuropathol. 2011, 121, 365–371. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.J.; Daniel, S.E.; Kilford, L.; Lees, A.J. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry 1992, 55, 181–184. [Google Scholar] [CrossRef]
- Litvan, I.; Goldman, J.G.; Troster, A.I.; Schmand, B.A.; Weintraub, D.; Petersen, R.C.; Mollenhauer, B.; Adler, C.H.; Marder, K.; Williams-Gray, C.H.; et al. Diagnostic criteria for mild cognitive impairment in Parkinson’s disease: Movement Disorder Society Task Force guidelines. Mov. Disord. 2012, 27, 349–356. [Google Scholar] [CrossRef]
- Van Buuren, S.; Groothuis-Oudshoorn, K. MICE: Multivariate imputation by chained equations in R. J. Stat. Softw. 2010, 45, 1–68. [Google Scholar] [CrossRef]
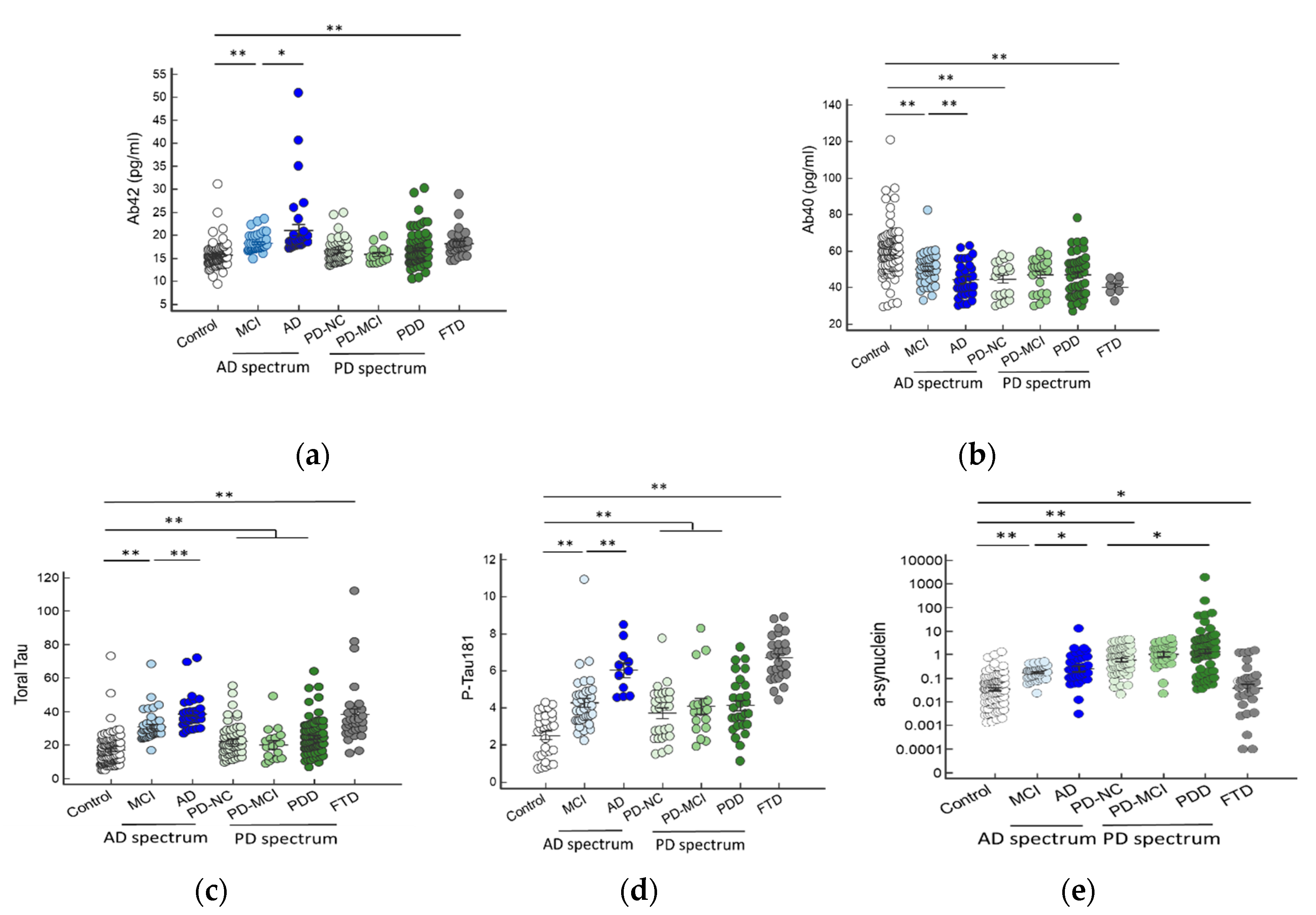
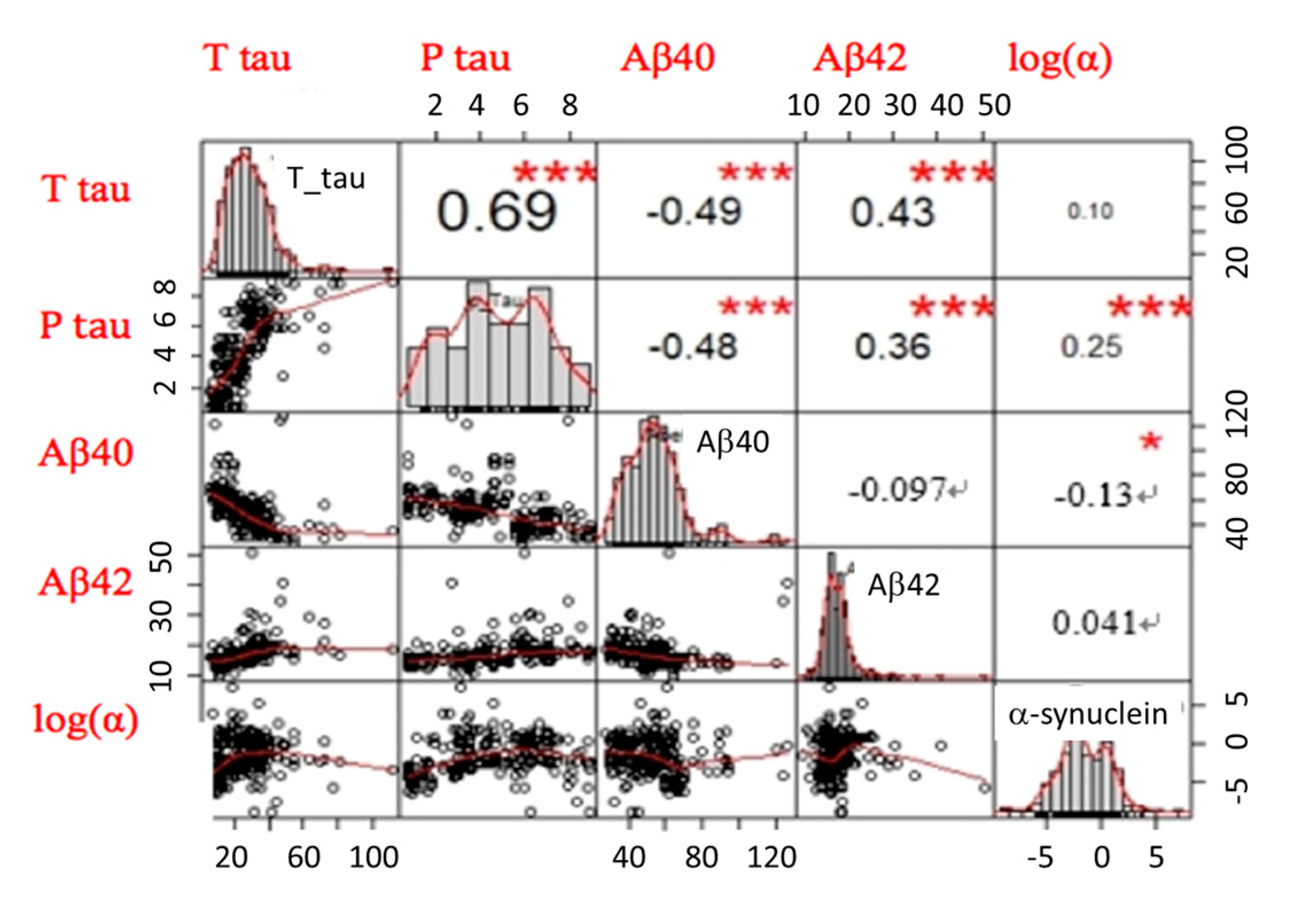
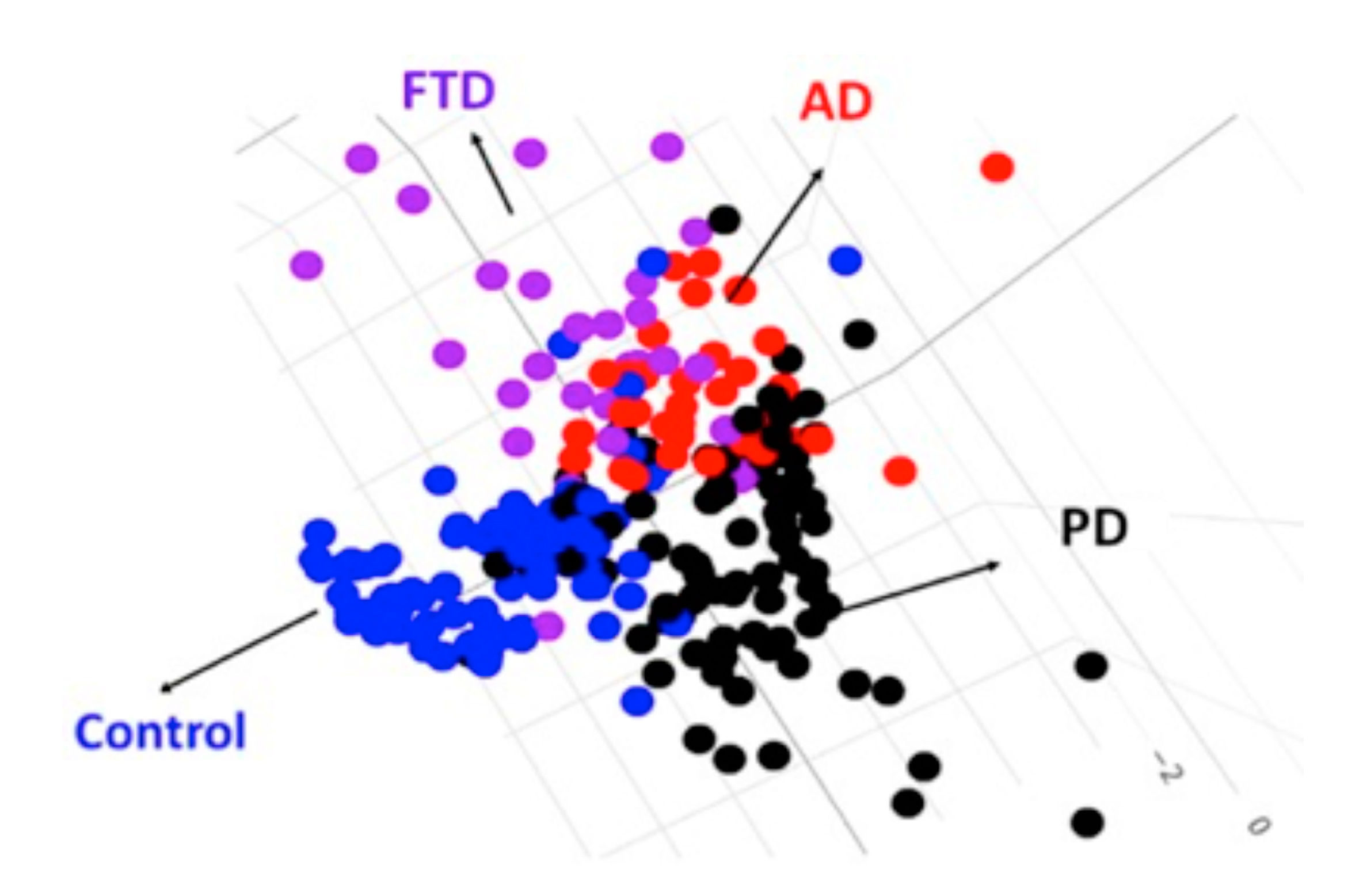
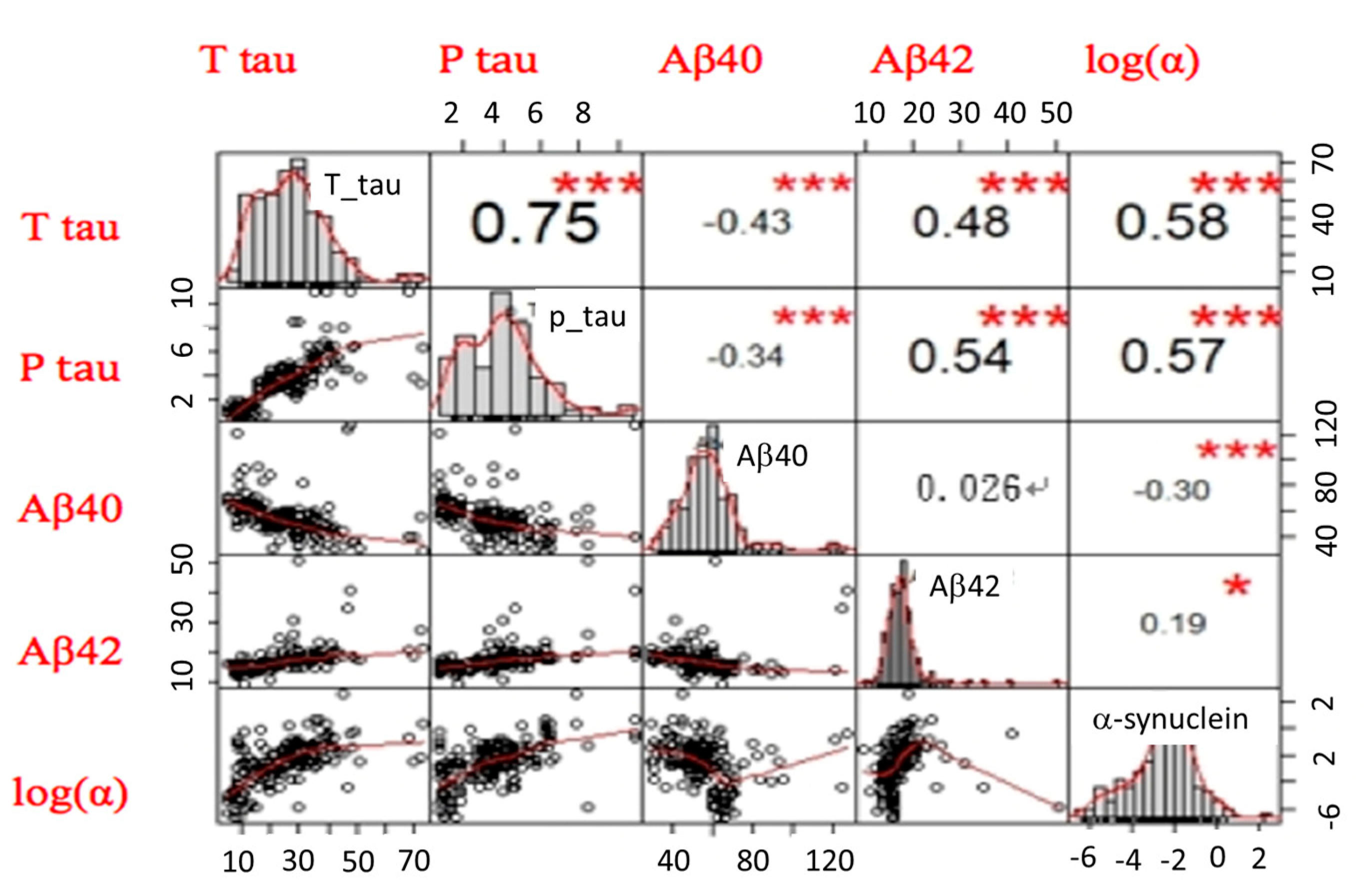

| Controls (n = 97) | AD Spectrum | PD Spectrum | FTD (n = 31) | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| (n = 76) | (n = 173) | |||||||
| MCI | AD | PD-NC | PD-MCI | PDD | ||||
| (n = 41) | (n = 35) | (n = 57) | (n = 29) | (n = 87) | ||||
| Age (years) | 64.0 ± 7.8 | 72.9 ± 7.9 | 75.2 ± 11.6 | 62.4 ± 11.2 | 66.5 ± 11.8 | 72.8 ± 8.9 | 60.7 ± 7.1 | p < 0.01 ** |
| Gender (M, n, %) | 31 (31.9%) | 16 (39.0) | 14 (40.04%) | 31 (54.4) | 20 (68.9) | 53 (60.9) | 6 (19.4) | p < 0.01 ** |
| Disease duration (y) | N.A. | 2.7 ± 0.9 | 6.2 ± 2.9 | 2.6 ± 1.1 | 5.6 ± 1.6 | 9.6 ± 3.2 | 5.4 ± 2.1 | p < 0.01 ** |
| MMSE | 28.9 ± 0.9 | 26.8 ± 1.2 | 18.2 ± 5.8 | 28.3 ± 0.9 | 26.8 ± 1.1 | 20.8 ± 4.3 | 18.6 ± 4.8 | p < 0.01 ** |
| Hoehn-Yahr stage | N.A. | N.A. | N.A. | 1.7 ± 0.9 | 2.0 ± 0.8 | 2.5 ± 1.1 | N.A. | p < 0.01 ** |
| Aβ42 (pg/mL) | 15.66 ± 2.58 | 18.30 ± 1.85 | 21.15 ± 7.17 | 16.56 ± 2.44 | 15.80 ± 1.75 | 16.96 ± 3.54 | 18.26 ± 2.84 | p < 0.01 ** |
| Aβ40 (pg/mL) | 59.45 ± 13.94 | 50.28 ± 8.69 | 49.01 ± 21.10 | 44.73 ± 9.47 | 47.10 ± 9.21 | 46.96 ± 11.93 | 40.34 ± 10.69 | p < 0.01 ** |
| Total tau (pg/mL) | 16.95 ± 9.61 | 30.93 ± 8.67 | 38.70 ± 9.74 | 22.32 ± 9.91 | 20.27 ± 10.01 | 24.71 ± 10.41 | 38.16 ± 10.73 | p < 0.01 ** |
| Phospho-tau (pg/mL) | 2.52 ± 1.17 | 4.28 ± 1.51 | 6.04 ± 1.33 | 3.72 ± 1.43 | 4.09 ± 1.77 | 4.15 ± 1.51 | 6.69 ± 1.19 | p < 0.01 ** |
| a-synuclein (pg/mL) | 0.09 ± 0.05 | 0.20 ± 0.11 | 0.80 ± 0.32 | 1.15 ± 0.09 | 1.62 ± 0.28 | 25.17 ± 7.83 | 0.05 ± 0.02 | p < 0.01 ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-H.; Chiu, S.-I.; Chen, T.-F.; Jang, J.-S.R.; Chiu, M.-J. Classifications of Neurodegenerative Disorders Using a Multiplex Blood Biomarkers-Based Machine Learning Model. Int. J. Mol. Sci. 2020, 21, 6914. https://doi.org/10.3390/ijms21186914
Lin C-H, Chiu S-I, Chen T-F, Jang J-SR, Chiu M-J. Classifications of Neurodegenerative Disorders Using a Multiplex Blood Biomarkers-Based Machine Learning Model. International Journal of Molecular Sciences. 2020; 21(18):6914. https://doi.org/10.3390/ijms21186914
Chicago/Turabian StyleLin, Chin-Hsien, Shu-I Chiu, Ta-Fu Chen, Jyh-Shing Roger Jang, and Ming-Jang Chiu. 2020. "Classifications of Neurodegenerative Disorders Using a Multiplex Blood Biomarkers-Based Machine Learning Model" International Journal of Molecular Sciences 21, no. 18: 6914. https://doi.org/10.3390/ijms21186914
APA StyleLin, C.-H., Chiu, S.-I., Chen, T.-F., Jang, J.-S. R., & Chiu, M.-J. (2020). Classifications of Neurodegenerative Disorders Using a Multiplex Blood Biomarkers-Based Machine Learning Model. International Journal of Molecular Sciences, 21(18), 6914. https://doi.org/10.3390/ijms21186914





