Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application
Abstract
1. Introduction
2. Short History of Cell-Free Nucleic Acids
3. Extracellular Vesicles
4. Cell-Free DNA
4.1. Cell-Free Nuclear DNA (cf-nDNA)
4.2. Cell-Free DNA Fragments
4.3. Vesicle-Bound DNA
4.4. DNA-Macromolecule Complexes
4.5. Cell-Free mtDNA
5. Cell-Free Microbial and Viral DNA
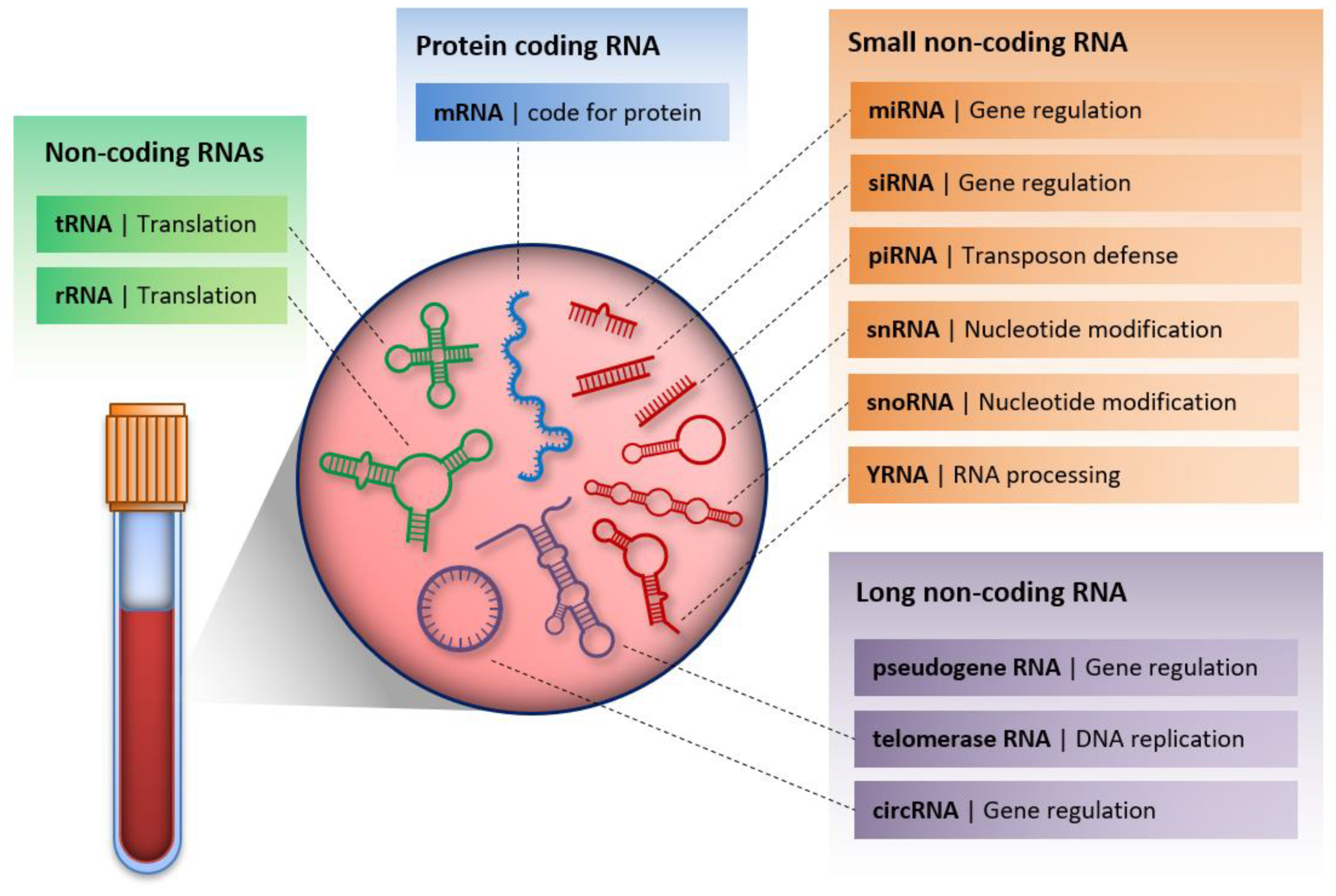
6. Cell-Free RNA
6.1. MicroRNAs
6.2. Long Non-Coding RNAs
6.3. Circular RNAs
6.4. PIWI-Interacting RNAs
6.5. YRNA
6.6. Vault RNA
6.7. Other Non-Coding RNAs
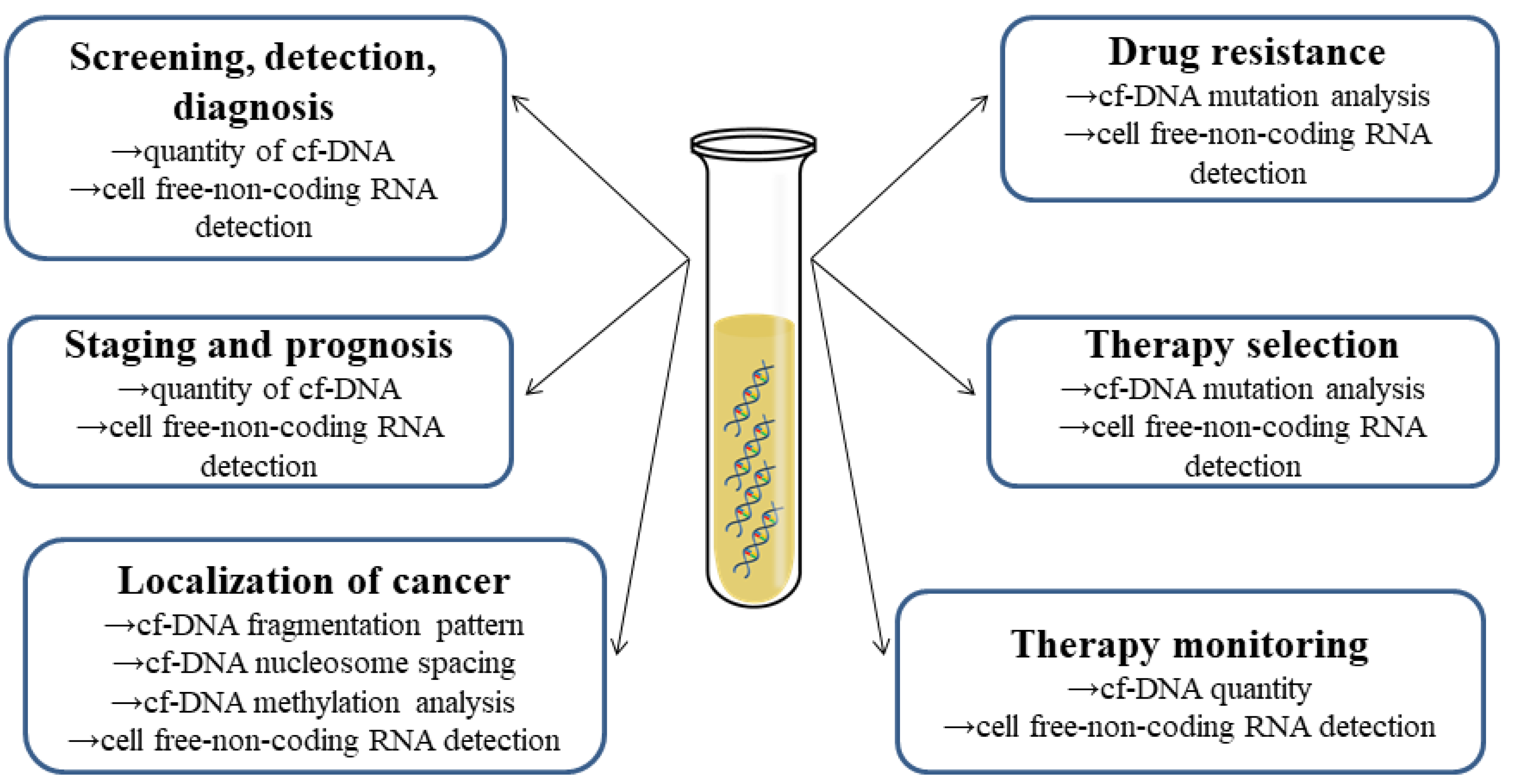
7. Cf-NA Detection
8. Current Perspectives on the Role of cfNAs as Biomarkers
Funding
Conflicts of Interest
References
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2018, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, R.; Van Roosbroeck, K.; Calin, G.A. Cell-to-cell communication: MicroRNAs as hormones. Mol. Oncol. 2017, 11, 1673–1686. [Google Scholar] [CrossRef] [PubMed]
- Alimirzaie, S.; Bagherzadeh, M.; Akbari, M.R. Liquid biopsy in breast cancer: A comprehensive review. Clin. Genet. 2019, 95, 643–660. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; O Fackelmayer, F.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Pös, O.; Bíró, O.; Szemes, T.; Nagy, B. Circulating cell-free nucleic acids: Characteristics and applications. Eur. J. Hum. Genet. 2018, 26, 937–945. [Google Scholar] [CrossRef]
- Pös, O.; Budiš, J.; Szemes, T. Recent trends in prenatal genetic screening and testing. F1000Research 2019, 8, 764. [Google Scholar] [CrossRef]
- Nagy, B. Cell-free nucleic acids. Int. J. Mol. Sci. 2019, 20, 5645. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez lʹhomme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Tan, E.M.; Schur, P.H.; Carr, R.I.; Kunkel, H.G. Deoxyribonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J. Clin. Investig. 1966, 45, 1732–1740. [Google Scholar] [CrossRef]
- Steinman, C.R. Free DNA in serum and plasma from normal adults. J. Clin. Investig. 1975, 56, 512–515. [Google Scholar] [CrossRef]
- Leon, S.; Green, A.; Yaros, M.; Shapiro, B. Radioimmunoassay for nanogram quantities of DNA. J. Immunol. Methods 1975, 9, 157–164. [Google Scholar] [CrossRef]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomark. Prev. 1994, 3, 67–71. [Google Scholar]
- Anker, P.; Stroun, M. Circulating nucleic acids and evolution. Expert Opin. Boil. 2012, 12, 113–117. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef]
- Palomaki, G.E.; Kloza, E.M.; Lambert-Messerlian, G.; Haddow, J.E.; Neveux, L.M.; Ehrich, M.; Boom, D.V.D.; Bombard, A.T.; Deciu, C.; Grody, W.W.; et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet. Med. 2011, 13, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Boil. Chem. 1946, 166, 189–197. [Google Scholar]
- Kawamura, Y.; Yamamoto, Y.; Sato, T.; Ochiya, T. Extracellular vesicles as trans-genomic agents: Emerging roles in disease and evolution. Cancer Sci. 2017, 108, 824–830. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Lui, Y.Y.; Chik, K.-W.; Chiu, R.W.; Ho, C.-Y.; Lam, C.W.; Lo, Y.M.D. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 2002, 48, 421–427. [Google Scholar] [CrossRef]
- Tong, Y.-K.; Lo, Y.M.D. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin. Chim. Acta 2006, 363, 187–196. [Google Scholar] [CrossRef]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [PubMed]
- Volik, S.; Alcaide, M.; Morin, R.D.; Collins, C. Cell-free DNA (cfDNA): Clinical significance and utility in cancer shaped by emerging technologies. Mol. Cancer Res. 2016, 14, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Duque-Afonso, J.; Waterhouse, M.; Pfeifer, D.; Follo, M.; Duyster, J.; Bertz, H.; Finke, J. Cell-free DNA characteristics and chimerism analysis in patients after allogeneic cell transplantation. Clin. Biochem. 2018, 52, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Di Renzo, G.C.; Bartha, J.L.; Bilardo, C.M. Expanding the indications for cell-free DNA in the maternal circulation: Clinical considerations and implications. Am. J. Obs. Gynecol. 2019, 220, 537–542. [Google Scholar] [CrossRef]
- Nagy, B. Cell-free nucleic acids in prenatal diagnosis and pregnancy-associated diseases. EJIFCC 2019, 30, 215–223. [Google Scholar]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Sun, K.; Jiang, P.; Chan, K.C.A.; Wong, J.; Cheng, Y.K.Y.; Liang, R.H.S.; Chan, W.-K.; Ma, E.S.K.; Chan, S.L.; Cheng, S.H.; et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. USA 2015, 112, E5503–E5512. [Google Scholar] [CrossRef]
- Nuzzo, P.V.; Berchuck, J.E.; Korthauer, K.; Spisak, S.; Nassar, A.H.; Alaiwi, S.A.; Chakravarthy, A.; Shen, S.Y.; Bakouny, Z.; Boccardo, F.; et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat. Med. 2020, 26, 1041–1043. [Google Scholar] [CrossRef]
- Ou, Z.; Li, K.; Yang, T.; Dai, Y.; Chandra, M.; Ning, J.; Wang, Y.; Xu, R.; Gao, T.; Xie, Y.; et al. Detection of bladder cancer using urinary cell-free DNA and cellular DNA. Clin. Transl. Med. 2020, 9, 1–12. [Google Scholar] [CrossRef]
- Frattini, M.; Gallino, G.; Signoroni, S.; Balestra, D.; Lusa, L.; Battaglia, L.; Sozzi, G.; Bertario, L.; Leo, E.; Pilotti, S.; et al. Quantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancer. Cancer Lett. 2008, 263, 170–181. [Google Scholar] [CrossRef]
- Heitzer, E.; Auer, M.; Ulz, P.; Geigl, J.B.; Speicher, M.R. Circulating tumor cells and DNA as liquid biopsies. Genome Med. 2013, 5, 73. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.A.; Hovinga, J.A.K.; Schatzberg, D.; Wagner, D.D.; Lämmle, B. Circulating DNA and myeloperoxidase indicate disease activity in patients with thrombotic microangiopathies. Blood 2012, 120, 1157–1164. [Google Scholar] [CrossRef]
- Mouliere, F.; Thierry, A.R. The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert Opin. Boil. 2012, 12, 209–215. [Google Scholar] [CrossRef]
- Contreras-Naranjo, J.C.; Wu, H.-J.; Ugaz, V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab. A Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef]
- Di Vizio, L.; Kim, J.; Hager, M.H.; Morello, M.; Yang, W.; LaFargue, C.J.; True, L.D.; Rubin, M.A.; Adam, R.M.; Beroukhim, R.; et al. Oncosome formation in prostate cancer: Association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009, 69, 5601–5609. [Google Scholar] [CrossRef]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef]
- Ma, X.; Zhu, L.; Wu, X.; Bao, H.; Wang, X.; Chang, Z.; Shao, Y.W.; Wang, Z. Cell-free DNA provides a good representation of the tumor genome despite its biased fragmentation patterns. PLoS ONE 2017, 12, e0169231. [Google Scholar] [CrossRef]
- Thierry, A.R.; Rabinovich, P.; Peng, B.; Mahan, L.C.; Bryant, J.L.; Gallo, R.C. Characterization of liposome-mediated gene delivery: Expression, stability and pharmacokinetics of plasmid DNA. Gene 1997, 4, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; Norris, V.; Molina, F.; Schmutz, M. Lipoplex nanostructures reveal a general self-organization of nucleic acids. Biochim. Et Biophys. Acta BBA Gen. Subj. 2009, 1790, 385–394. [Google Scholar] [CrossRef]
- Veltri, K.L.; Espiritu, M.; Singh, G. Distinct genomic copy number in mitochondria of different mammalian organs. J. Cell. Physiol. 1990, 143, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Thurairajah, K.; Briggs, G.D.; Balogh, Z.J. The source of cell-free mitochondrial DNA in trauma and potential therapeutic strategies. Eur. J. Trauma Emerg. Surg. 2018, 44, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Nakahira, K.; Guo, X.; Choi, A.M.; Gu, Z. Very short mitochondrial DNA fragments and heteroplasmy in human plasma. Sci. Rep. 2016, 6, 36097. [Google Scholar] [CrossRef] [PubMed]
- McIlroy, D.J.; Bigland, M.; White, A.E.; Hardy, B.M.; Lott, N.; Smith, D.W.; Balogh, Z.J. Cell necrosis–independent sustained mitochondrial and nuclear DNA release following trauma surgery. J. Trauma Acute Care Surg. 2015, 78, 282–288. [Google Scholar] [CrossRef]
- Burnham, P.; Kim, M.S.; Agbor-Enoh, S.; Luikart, H.; Valantine, H.A.; Khush, K.K.; De Vlaminck, I. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci. Rep. 2016, 6, 27859. [Google Scholar] [CrossRef]
- Ellinger, J.; Albers, P.; Müller, S.C.; Von Ruecker, A.; Bastian, P.J. Circulating mitochondrial DNA in the serum of patients with testicular germ cell cancer as a novel noninvasive diagnostic biomarker. BJU Int. 2009, 104, 48–52. [Google Scholar] [CrossRef]
- Lazaro-Ibanez, E.; Lässer, C.; Shelke, G.V.; Crescitelli, R.; Jang, S.C.; Cvjetkovic, A.; García-Rodríguez, A.; Lötvall, J. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J. Extracell. Vesicles 2019, 8, 1656993. [Google Scholar] [CrossRef]
- Kohler, C.; Radpour, R.; Barekati, Z.; Asadollahi, R.; Bitzer, J.; Wight, E.; Bürki, N.; Diesch, C.; Holzgreve, W.; Zhong, X.Y. Levels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumors. Mol. Cancer 2009, 8, 1–8. [Google Scholar] [CrossRef]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef]
- Dache, Z.A.A.; Otandault, A.; Tanos, R.; Pastor, B.; Meddeb, R.; Sanchez, C.; Arena, G.; Lasorsa, L.; Bennett, A.; Grange, T.; et al. Blood contains circulating cell-free respiratory competent mitochondria. FASEB J. 2020, 34, 3616–3630. [Google Scholar] [CrossRef]
- Soltesz, B.; Urbancsek, R.; Pös, O.; Hajas, O.; Forgács, I.N.; Szilágyi, E.; Nagy-Baló, E.; Szemes, T.; Csanádi, Z.; Nagy, B. Quantification of peripheral whole blood, cell-free plasma and exosome encapsulated mitochondrial DNA copy numbers in patients with atrial fibrillation. J. Biotechnol. 2019, 299, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Lowes, H.; Pyle, A.; Duddy, M.; Hudson, G. Cell-free mitochondrial DNA in progressive multiple sclerosis. Mitochondrion 2018, 46, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Lowes, H.; Pyle, A.; Santibanez-Koref, M.; Hudson, G. Circulating cell-free mitochondrial DNA levels in Parkinson’s disease are influenced by treatment. Mol. Neurodegener. 2020, 15, 1–8. [Google Scholar] [CrossRef]
- An, Q.; Hu, Y.; Li, Q.; Chen, X.; Huang, J.; Pellegrini, M.; Zhou, X.J.; Rettig, M.; Fan, G. The size of cell-free mitochondrial DNA in blood is inversely correlated with tumor burden in cancer patients. Precis. Clin. Med. 2019, 2, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Newell, C.; Hume, S.; Greenway, S.C.; Podemski, L.; Shearer, J.; Khan, A. Plasma-derived cell-free mitochondrial DNA: A novel non-invasive methodology to identify mitochondrial DNA haplogroups in humans. Mol. Genet. Metab. 2018, 125, 332–337. [Google Scholar] [CrossRef]
- Huang, Y.-F.; Chen, Y.-J.; Fan, T.-C.; Chang, N.-C.; Chen, Y.-J.; Midha, M.K.; Chen, T.-H.; Yang, H.-H.; Wang, Y.-T.; Yu, A.L.; et al. Analysis of microbial sequences in plasma cell-free DNA for early-onset breast cancer patients and healthy females. BMC Med. Genom. 2018, 11, 16. [Google Scholar] [CrossRef]
- Castillo, D.J.; Rifkin, R.F.; Cowan, D.A.; Potgieter, M. The healthy human blood microbiome: Fact or fiction? Front. Microbiol. 2019, 9, 148. [Google Scholar] [CrossRef]
- Gosiewski, T.; Ludwig-Galezowska, A.H.; Huminska-Lisowska, K.; Sroka-Oleksiak, A.; Radkowski, P.; Salamon, D.; Wojciechowicz, J.; Kus-Slowinska, M.; Bulanda, M.; Wołkow, P.P. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method—The observation of DNAemia. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 36, 329–336. [Google Scholar] [CrossRef]
- Roy, S.; Trinchieri, G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef]
- Dzutsev, A.; Trinchieri, G. Microbial DNA signature in plasma enables cancer diagnosis. Nat. Rev. Clin. Oncol. 2020, 17, 453–454. [Google Scholar] [CrossRef] [PubMed]
- Blauwkamp, T.A.; Thair, S.; Rosen, M.J.; Blair, L.; Lindner, M.S.; Vilfan, I.D.; Kawli, T.; Christians, F.C.; Venkatasubrahmanyam, S.; Wall, G.D.; et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 2019, 4, 663–674. [Google Scholar] [CrossRef]
- Hong, D.K.; Blauwkamp, T.A.; Kertesz, M.; Bercovici, S.; Truong, C.; Banaei, N. Liquid biopsy for infectious diseases: Sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn. Microbiol. Infect. Dis. 2018, 92, 210–213. [Google Scholar] [CrossRef]
- Witt, R.G.; Blair, L.; Frascoli, M.; Rosen, M.J.; Nguyen, Q.-H.; Bercovici, S.; Zompi, S.; Romero, R.; MacKenzie, T. Detection of microbial cell-free DNA in maternal and umbilical cord plasma in patients with chorioamnionitis using next generation sequencing. PLoS ONE 2020, 15, e0231239. [Google Scholar] [CrossRef]
- Burnham, P.; Dadhania, D.; Heyang, M.; Chen, F.; Westblade, L.F.; Suthanthiran, M.; Lee, J.R.; De Vlaminck, I. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 2018, 9, 2412. [Google Scholar] [CrossRef] [PubMed]
- Chiu, K.-P.; Yu, A.L. Application of cell-free DNA sequencing in characterization of bloodborne microbes and the study of microbe-disease interactions. PeerJ 2019, 7, e7426. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, X.; Li, L. Biogenesis and function of extracellular miRNAs. ExRNA 2019, 1, 38. [Google Scholar] [CrossRef]
- Umu, S.U.; Langseth, H.; Bucher-Johannessen, C.; Fromm, B.; Keller, A.; Meese, E.; Lauritzen, M.; Leithaug, M.; Lyle, R.; Rounge, T.B. A comprehensive profile of circulating RNAs in human serum. RNA Biol. 2017, 15, 242–250. [Google Scholar] [CrossRef]
- Xue, V.W.; Cheung, M.T.; Chan, P.T.; Luk, L.L.Y.; Lee, V.H.; Au, T.C.; Yu, A.C.; Cho, W.C.S.; Tsang, H.F.A.; Chan, A.K.; et al. Non-invasive potential circulating mRNA markers for colorectal adenoma using targeted sequencing. Sci. Rep. 2019, 9, 12943. [Google Scholar] [CrossRef]
- De Souza, M.F.; Kuasne, H.; Barros-Filho, M.D.C.; Cilião, H.L.; Marchi, F.A.; Fuganti, P.E.; Paschoal, A.R.; Rogatto, S.R.; Cólus, I.M.D.S. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS ONE 2017, 12, e0184094. [Google Scholar] [CrossRef]
- Larrea, E.; Solé, C.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.M.; Araujo, A.M.; Araiz, M.; Fernandez-Mercado, M.; et al. New concepts in cancer biomarkers: Circulating miRNAs in liquid biopsies. Int. J. Mol. Sci. 2016, 17, 627. [Google Scholar] [CrossRef] [PubMed]
- Penyige, A.; Márton, É.; Soltesz, B.; Szilágyi, M.; Poka, R.; Lukács, J.; Széles, L.; Nagy, B. Circulating miRNA profiling in plasma samples of ovarian cancer patients. Int. J. Mol. Sci. 2019, 20, 4533. [Google Scholar] [CrossRef] [PubMed]
- Márton, É.; Lukács, J.; Penyige, A.; Janka, E.; Hegedüs, L.; Soltész, B.; Méhes, G.; Poka, R.; Nagy, B.; Szilágyi, M. Circulating epithelial-mesenchymal transition-associated miRNAs are promising biomarkers in ovarian cancer. J. Biotechnol. 2019, 297, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.-Y.; Zou, D.; Koirala, P. Long non-coding RNAs as key regulators of cancer metastasis. J. Cancer Metastasis Treat. 2015, 2, 1–10. [Google Scholar] [CrossRef]
- Boon, R.A.; Jaé, N.; Holdt, L.; Dimmeler, S. Long noncoding RNAs: From clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 2016, 67, 1214–1226. [Google Scholar] [CrossRef]
- Lu, M. Circular RNA: Functions, applications and prospects. ExRNA 2020, 2, 1–7. [Google Scholar] [CrossRef]
- Creugny, A.; Fender, A.; Pfeffer, S. Regulation of primary microRNA processing. FEBS Lett. 2018, 592, 1980–1996. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Bracken, C.P.; Khew-Goodall, Y.; Goodall, G.J. Network-based approaches to understand the roles of miR-200 and other microRNAs in cancer. Cancer Res. 2015, 75, 2594–2599. [Google Scholar] [CrossRef]
- Dragomir, M.P.; Mafra, A.C.P.; Dias, S.M.G.; Vasilescu, C.; Calin, G.A. Using microRNA networks to understand cancer. Int. J. Mol. Sci. 2018, 19, 1871. [Google Scholar] [CrossRef]
- Hayes, J.L.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lu, Z.; Unruh, A.K.; Ivan, C.; Baggerly, K.A.; Calin, G.A.; Li, Z.; Bast, R.C.; Le, X.-F. Clinically relevant microRNAs in ovarian cancer. Mol. Cancer Res. 2014, 13, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 1–24. [Google Scholar] [CrossRef]
- Balacescu, O.; Visan, S.; Baldasici, O.; Balacescu, L.; Vlad, C.; Achimas-Cadariu, P. MiRNA-based therapeutics in oncology, realities, and challenges. In Antisense Therapy; IntechOpen: London, UK, 2019. [Google Scholar]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in cancer: Potential and challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef]
- Liang, J.; Wang, S.; Wang, Z. Role of microRNAs in embryo implantation. Reprod. Boil. Endocrinol. 2017, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- Cuman, C.; Van Sinderen, M.; Gantier, M.P.; Rainczuk, K.; Sorby, K.; Rombauts, L.; Osianlis, T.; Dimitriadis, E. Human blastocyst secreted microRNA regulate endometrial epithelial cell adhesion. EBioMedicine 2015, 2, 1528–1535. [Google Scholar] [CrossRef]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating cancer with microRNA replacement therapy: A literature review. J. Cell. Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef]
- Lodes, M.J.; Caraballo, M.; Suciu, D.; Munro, S.; Kumar, A.; Anderson, B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS ONE 2009, 4, e6229. [Google Scholar] [CrossRef]
- Roth, C.; Rack, B.; Müller, V.; Janni, W.; Pantel, K.; Schwarzenbach, H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010, 12, R90. [Google Scholar] [CrossRef]
- Roth, C.; Kasimir-Bauer, S.; Pantel, K.; Schwarzenbach, H. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol. Oncol. 2011, 5, 281–291. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H. MicroRNA-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014, 5, e1327. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Sawada, K.; Yoshimura, A.; Kinose, Y.; Nakatsuka, E.; Kimura, T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol. Cancer 2016, 15, 48. [Google Scholar] [CrossRef] [PubMed]
- Elias, K.M.; Fendler, W.; Stawiski, K.; Fiascone, S.J.; Vitonis, A.F.; Berkowitz, R.S.; Frendl, G.; Konstantinopoulos, P.A.; Crum, C.P.; Kedzierska, M.; et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. eLife 2017, 6, 28932. [Google Scholar] [CrossRef] [PubMed]
- Bratulic, S.; Gatto, F.; Nielsen, J. The translational status of cancer liquid biopsies. Regen. Eng. Transl. Med. 2019, 1–41. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sunwoo, H.; Spector, D. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef]
- Cajal, S.R.Y.; Segura, M.F.; Hümmer, S. Interplay between ncRNAs and cellular communication: A proposal for understanding cell-specific signaling pathways. Front. Genet. 2019, 10, 281. [Google Scholar] [CrossRef]
- Dragomir, M.; Chen, B.; Calin, G.A. Exosomal lncRNAs as new players in cell-to-cell communication. Transl. Cancer Res. 2018, 7, S243–S252. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, M.; Du, S.; Feng, W.; Zhang, K.; Zhang, L.; Liu, H.; Jia, G.; Wu, L.; Hu, X.; et al. Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat. Commun. 2019, 10, 1637. [Google Scholar] [CrossRef]
- Chow, J.T.-S.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, K.; Wilczek, A.L.; De Gonzalo-Calvo, D.; Pfanne, A.; Derda, A.A.; Zwadlo, C.; Bavendiek, U.; Bauersachs, J.; Fiedler, J.; Thum, T. Serum circular RNAs act as blood-based biomarkers for hypertrophic obstructive cardiomyopathy. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Xie, Y.; Li, J.; Li, P.; Li, N.; Zhang, Y.; Binang, H.; Zhao, Y.; Duan, W.; Chen, Y.; Wang, Y.; et al. RNA-Seq profiling of serum exosomal circular RNAs reveals Circ-PNN as a potential biomarker for human colorectal cancer. Front. Oncol. 2020, 10, 982. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wei, Y.; Yu, L.; Xiao, Y. Identification of altered circular RNA expression in serum exosomes from patients with papillary thyroid carcinoma by high-throughput sequencing. Med. Sci. Monit. 2019, 25, 2785–2791. [Google Scholar] [CrossRef]
- Xu, H.; Gong, Z.; Shen, Y.; Fang, Y.; Zhong, S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics 2018, 10, 187–197. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2018, 20, 89–108. [Google Scholar] [CrossRef]
- Yu, Y.; Xiao, J.; Hann, S.S. The emerging roles of PIWI-interacting RNA in human cancers. Cancer Manag. Res. 2019, 11, 5895–5909. [Google Scholar] [CrossRef]
- Iliev, R.; Fedorko, M.; Machackova, T.; Mlcochova, H.; Svoboda, M.; Pacik, D.; Dolezel, J.; Stanik, M.; Slaby, O. Expression levels of PIWI-interacting RNA, piR-823, are deregulated in tumor tissue, blood serum and urine of patients with renal cell carcinoma. Anticancer. Res. 2016, 36, 6419–6424. [Google Scholar] [CrossRef]
- Coenen-Stass, A.M.; Sork, H.; Gatto, S.; Godfrey, C.; Bhomra, A.; Krjutškov, K.; Hart, J.R.; Westholm, J.O.; O’Donovan, L.; Roos, A.; et al. Comprehensive RNA-sequencing analysis in serum and muscle reveals novel small RNA signatures with biomarker potential for DMD. Mol. Nucleic Acids 2018, 13, 1–15. [Google Scholar] [CrossRef]
- Kowalski, M.P.; Krude, T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Boil. 2015, 66, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Nientiedt, M.; Schmidt, D.; Kristiansen, G.; Müller, S.; Ellinger, J. YRNA expression profiles are altered in clear cell renal cell carcinoma. Eur. Urol. Focus 2018, 4, 260–266. [Google Scholar] [CrossRef]
- Tolkach, Y.; Niehoff, E.-M.; Stahl, A.F.; Zhao, C.; Kristiansen, G.; Müller, S.C.; Ellinger, J. YRNA expression in prostate cancer patients: Diagnostic and prognostic implications. World J. Urol. 2018, 36, 1073–1078. [Google Scholar] [CrossRef]
- Hizir, Z.; Bottini, S.; Grandjean, V.; Trabucchi, M.; Repetto, E. RNY (YRNA)-derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death Dis. 2017, 8, e2530. [Google Scholar] [CrossRef]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Boffelli, D.; Mote, P.; Martin, D.I.K. 5′-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol. Genom. 2013, 45, 990–998. [Google Scholar] [CrossRef]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Boffelli, D.; Martin, D.I.K. Deep sequencing of serum small RNAs identifies patterns of 5′ tRNA half and YRNA fragment expression associated with breast cancer. Biomark. Cancer 2014, 6, 37–47. [Google Scholar] [CrossRef]
- Dhahbi, J.; Nunez Lopez, Y.O.; Schneider, A.; Victoria, B.; Saccon, T.; Bharat, K.; McClatchey, T.; Atamna, H.; Scierski, W.; Golusinski, P.; et al. Profiling of tRNA halves and YRNA fragments in serum and tissue from oral squamous cell carcinoma patients identify key role of 5′ tRNA-Val-CAC-2-1 half. Front. Oncol. 2019, 9, 959. [Google Scholar] [CrossRef] [PubMed]
- Horos, R.; Büscher, M.; Kleinendorst, R.; Alleaume, A.-M.; Tarafder, A.K.; Schwarzl, T.; Dziuba, D.; Tischer, C.; Zielonka, E.M.; Adak, A.; et al. The Small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell 2019, 176, 1054–1067.e12. [Google Scholar] [CrossRef]
- Gopinath, S.C.; Wadhwa, R.; Kumar, P.K. Expression of noncoding vault RNA in human malignant cells and its importance in mitoxantrone resistance. Mol. Cancer Res. 2010, 8, 1536–1546. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Yamakawa, A.; Boffelli, D.; Mote, P.; Martin, D.I.K. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom. 2013, 14, 298. [Google Scholar] [CrossRef]
- Jehn, J.; Treml, J.; Wulsch, S.; Ottum, B.; Erb, V.; Hewel, C.; Kooijmans, R.N.; Wester, L.; Fast, I.; Rosenkranz, D. 5′ tRNA halves are highly expressed in the primate hippocampus and might sequence-specifically regulate gene expression. RNA 2020, 26, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Buono, G.; Gerratana, L.; Bulfoni, M.; Provinciali, N.; Basile, D.; Giuliano, M.; Corvaja, C.; Arpino, G.; Del Mastro, L.; De Placido, S.; et al. Circulating tumor DNA analysis in breast cancer: Is it ready for prime-time? Cancer Treat. Rev. 2019, 73, 73–83. [Google Scholar] [CrossRef]
- Gárcia, J.; Forestier, J.; Dusserre, E.; Wozny, A.-S.; Geiguer, F.; Merle, P.; Tissot, C.; Ferraro-Peyret, C.; Jones, F.S.; Edelstein, D.L.; et al. Cross-platform comparison for the detection of RAS mutations in cfDNA (ddPCR Biorad detection assay, BEAMing assay, and NGS strategy). Oncotarget 2018, 9, 21122–21131. [Google Scholar] [CrossRef]
- Xu, T.; Kang, X.; You, X.; Dai, L.; Tian, D.; Yan, W.; Yang, Y.; Xiong, H.; Liang, Z.; Zhao, G.Q.; et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor DNA from non-small cell lung carcinoma patient plasma. Theranostics 2017, 7, 1437–1446. [Google Scholar] [CrossRef]
- Volckmar, A.-L.; Sültmann, H.; Riediger, A.; Fioretos, T.; Schirmacher, P.; Endris, V.; Stenzinger, A.; Dietz, S. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosom. Cancer 2017, 57, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Ziemann, M.; Kaspi, A.; El-Osta, A. Evaluation of microRNA alignment techniques. RNA 2016, 22, 1120–1138. [Google Scholar] [CrossRef] [PubMed]
- Amanullah, M.; Yu, M.; Sun, X.; Luo, A.; Zhou, Q.; Zhou, L.; Hou, L.; Wang, W.; Lu, W.; Liu, P.; et al. MDEHT: A multivariate approach for detecting differential expression of microRNA isoform data in RNA-sequencing studies. Bioinformatics 2020, 36, 2657–2664. [Google Scholar] [CrossRef]
- Williams, Z.; Ben-Dov, I.Z.; Elias, R.; Mihailovic, A.; Brown, M.; Rosenwaks, Z.; Tuschl, T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA 2013, 110, 4255–4260. [Google Scholar] [CrossRef]
- Koh, W.; Pan, W.; Gawad, C.; Fan, H.C.; Kerchner, G.A.; Wyss-Coray, T.; Blumenfeld, Y.J.; El-Sayed, Y.Y.; Quake, S.R. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc. Natl. Acad. Sci. USA 2014, 111, 7361–7366. [Google Scholar] [CrossRef]
- Capková, M.; Sachova, J.; Strnad, H.; Kolar, M.; Hroudová, M.; Chovanec, M.; Čada, Z.; Šteffl, M.; Valach, J.; Kastner, J.; et al. Microarray analysis of serum mRNA in patients with head and neck squamous cell carcinoma at whole-genome scale. Biomed Res. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- Whitehead, C.L.; Walker, S.P.; Tong, S. Measuring circulating placental RNAs to non-invasively assess the placental transcriptome and to predict pregnancy complications. Prenat. Diagn. 2016, 36, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Beck, T.N.; Boumber, Y.A.; Aggarwal, C.; Pei, J.; Thrash-Bingham, C.; Fittipaldi, P.; Vlasenkova, R.; Rao, C.; Borghaei, H.; Cristofanilli, M.; et al. Circulating tumor cell and cell-free RNA capture and expression analysis identify platelet-associated genes in metastatic lung cancer. BMC Cancer 2019, 19, 1–13. [Google Scholar] [CrossRef]
- Forero, D.A.; González-Giraldo, Y.; Castro-Vega, L.J.; Barreto, G.E. qPCR-based methods for expression analysis of miRNAs. Biotechniques 2019, 67, 192–199. [Google Scholar] [CrossRef]
- Zaporozhchenko, I.A.; Ponomaryova, A.A.; Rykova, E.Y.; Laktionov, P.P. The potential of circulating cell-free RNA as a cancer biomarker: Challenges and opportunities. Expert Rev. Mol. Diagn. 2018, 18, 133–145. [Google Scholar] [CrossRef]
- Otandault, A.; Anker, P.; Dache, Z.A.A.; Guillaumon, V.; Meddeb, R.; Pastor, B.; Pisareva, E.; Sanchez, C.; Tanos, R.; Tousch, G.; et al. Recent advances in circulating nucleic acids in oncology. Ann. Oncol. 2019, 30, 374–384. [Google Scholar] [CrossRef]
- Bachet, J.; Bouché, O.; Taıeb, J.; Dubreuil, O.; Garcia, M.; Meurisse, A.; Normand, C.; Gornet, J.; Artru, P.; Louafi, S.; et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: The AGEO RASANC prospective multicenter study. Ann. Oncol. 2018, 29, 1211–1219. [Google Scholar] [CrossRef]
- Keller, L.; Guibert, N.; Casanova, A.; Brayer, S.; Farella, M.; Delaunay, M.; Gilhodes, J.; Martin, E.; Balagué, G.; Favre, G.; et al. Early circulating tumour DNA variations predict tumour response in melanoma patients treated with immunotherapy. Acta Derm. Venereol. 2019, 99, 206–210. [Google Scholar] [CrossRef]
| Extracellular Vesicle Type | Size | Plasma/Serum Concentration | Origin | Content | Markers | Function | Morphology |
|---|---|---|---|---|---|---|---|
| Exosome | 40–100 nm | 5.3 particle/mL × 106 | Exocytosis from multivesicular bodies | Proteins, lipids, DNA, mRNA, miRNA, lncRNA, circRNA | Alix, Tsg101, tetraspanins (CD81, CD63, CD9), flotilin | Intracellular communication | Cup-shape |
| Microvesicle | 50–3000 nm | 5–50 µg/mL | Outward budding of plasma membrane | Proteins, lipids, mRNA, miRNA, ncRNAs | Phosphatidylserin, Integrins, selectins, CD40 | Intracellular communication | Cup-shape |
| Apoptotic body | 800–5000 nm | Much less compared to EVs and EXOs (Lazaro-Ibanez 2014) | Programmed cell death or apoptosis | Nuclear fractions, cell organelles, proteins, mRNA, ncRNA, DNA | Annexin V, phosphatidylserin | Facilitate phagocytosis | Heterogeneous |
| Full Name | Size | Concentration | Clinical Application |
|---|---|---|---|
| Genomic DNA | 166->10.000 bp | 13.9 ± 3.7 mg/L | Prenatal testing, diagnosis of cancer, mutation detection, cancer localization |
| Mitochondrial DNA | 20–100 bp; <1–21 kbp | 4.21 ± 0.38 copies/L | Diagnosis of cancer |
| Microbial DNA | Variable | 20–450.000 microbe specific cfDNA/µL | Diagnosis of microbial infections and cancer |
| Method | Platform | MAF | Specificity | Limitations |
|---|---|---|---|---|
| NGS | WGS/WES | 0.02% | 80–90% | High ctDNA input |
| CAPP-Seq | 0.00025% | >99.99% | High ctDNA input; detects only known mutations | |
| Digital PCR | ddPCR | 0.1% | 100% | Detects only known mutations; limited in multiplexing |
| BEAMing | 0.01% | 100% | Detects only known mutations | |
| Real-Time PCR | qPCR | 0.1% | 99% | Detects only known mutations |
| AS-PCR | 1% | 98% | Detects only known mutations | |
| PNA-LNA PCR clamp | 0.1–1% | 79% | Detects only known point mutations | |
| COLD-PCR | 0.1% | 94.9% | Detects limited genomic loci; limited in multiplexing |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szilágyi, M.; Pös, O.; Márton, É.; Buglyó, G.; Soltész, B.; Keserű, J.; Penyige, A.; Szemes, T.; Nagy, B. Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application. Int. J. Mol. Sci. 2020, 21, 6827. https://doi.org/10.3390/ijms21186827
Szilágyi M, Pös O, Márton É, Buglyó G, Soltész B, Keserű J, Penyige A, Szemes T, Nagy B. Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application. International Journal of Molecular Sciences. 2020; 21(18):6827. https://doi.org/10.3390/ijms21186827
Chicago/Turabian StyleSzilágyi, Melinda, Ondrej Pös, Éva Márton, Gergely Buglyó, Beáta Soltész, Judit Keserű, András Penyige, Tomas Szemes, and Bálint Nagy. 2020. "Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application" International Journal of Molecular Sciences 21, no. 18: 6827. https://doi.org/10.3390/ijms21186827
APA StyleSzilágyi, M., Pös, O., Márton, É., Buglyó, G., Soltész, B., Keserű, J., Penyige, A., Szemes, T., & Nagy, B. (2020). Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application. International Journal of Molecular Sciences, 21(18), 6827. https://doi.org/10.3390/ijms21186827






