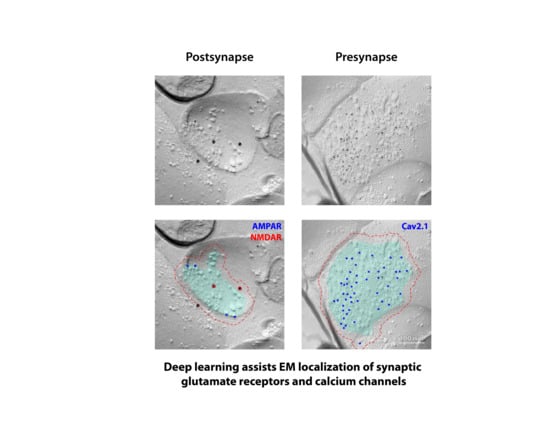
Deep Learning-Assisted High-Throughput Analysis of Freeze-Fracture Replica Images Applied to Glutamate Receptors and Calcium Channels at Hippocampal Synapses
Abstract
1. Introduction
2. Results
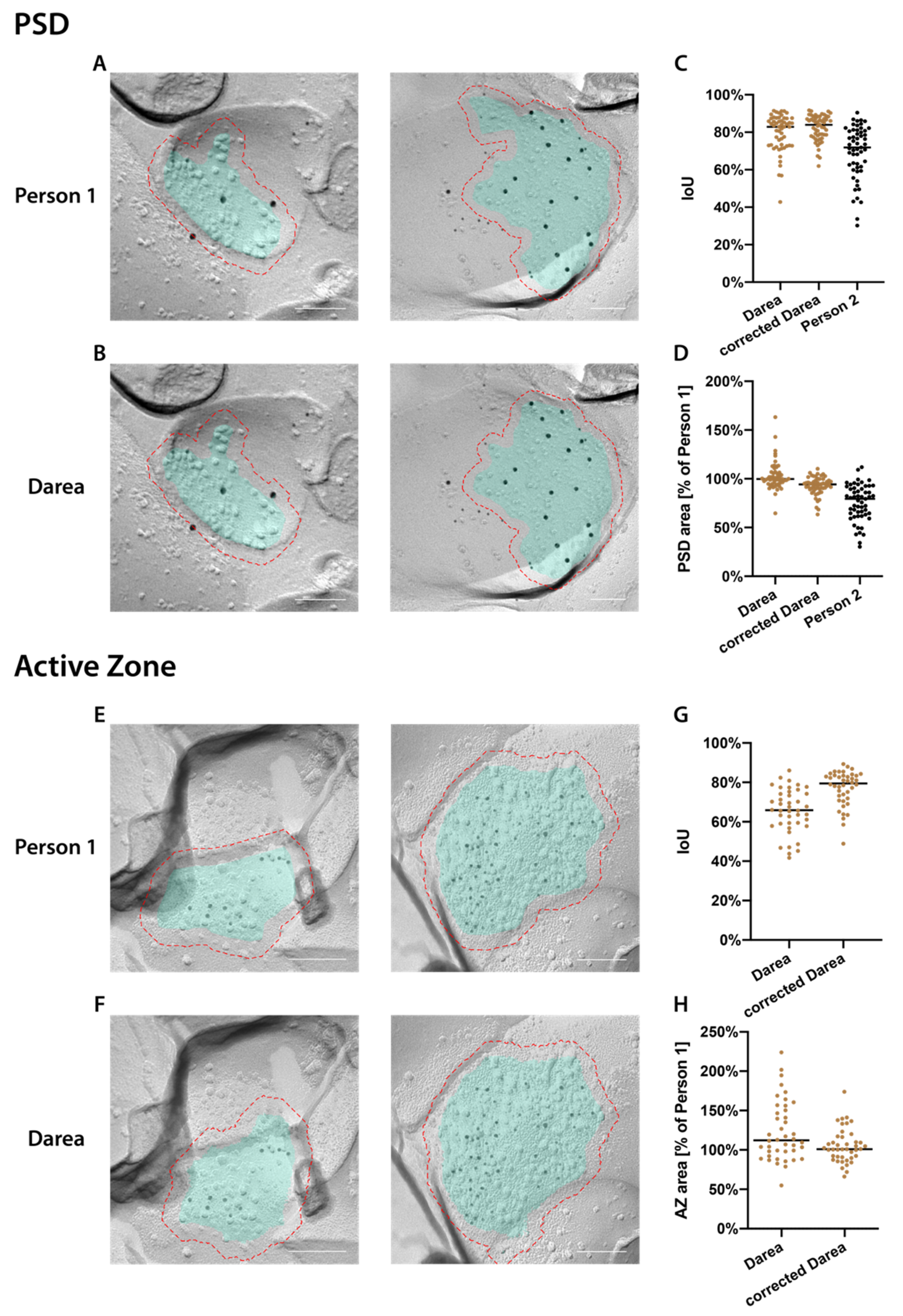
2.1. Darea Software
2.2. Automated Demarcation of Synaptic Membranes and Annotation of Gold Particles
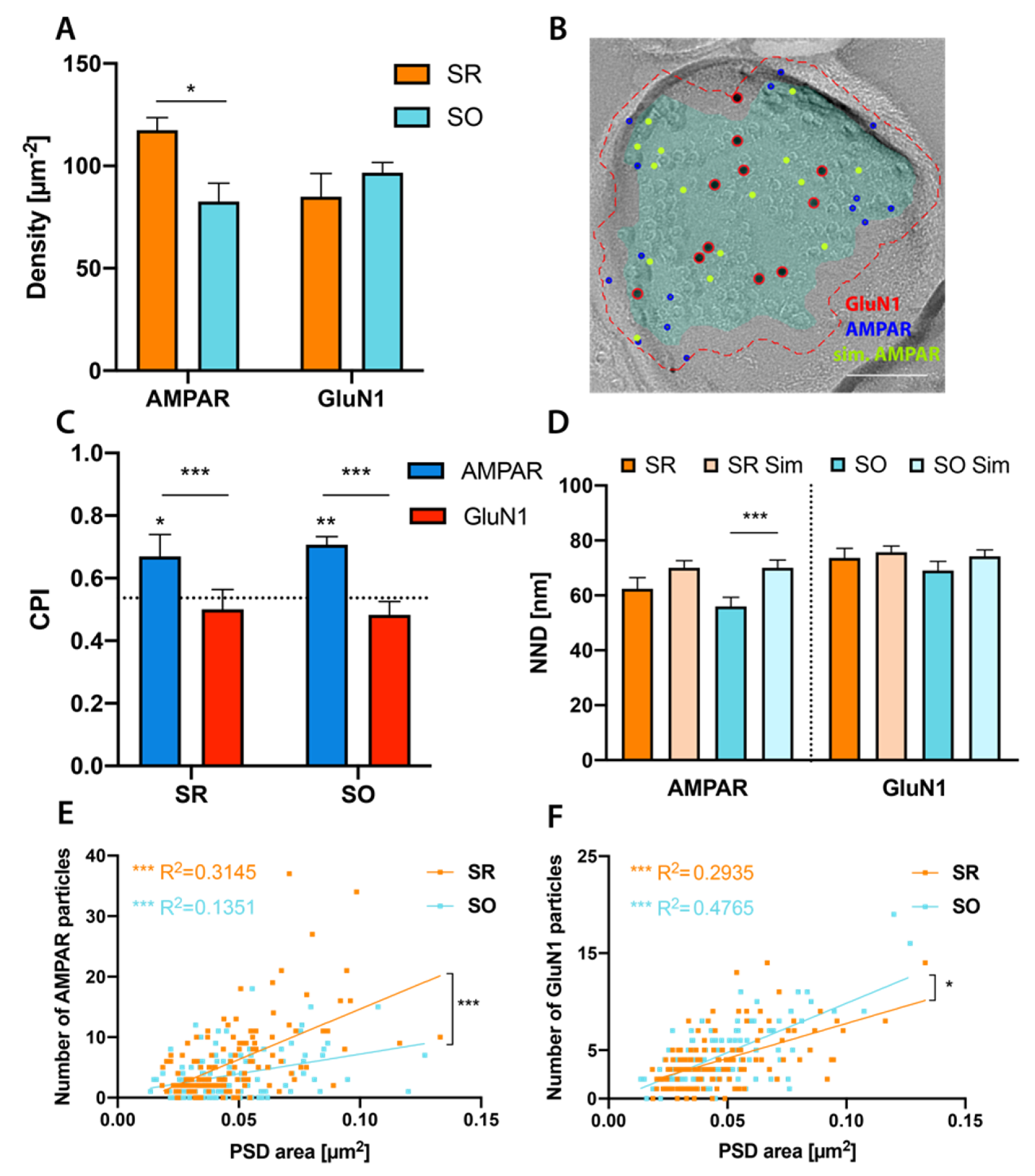
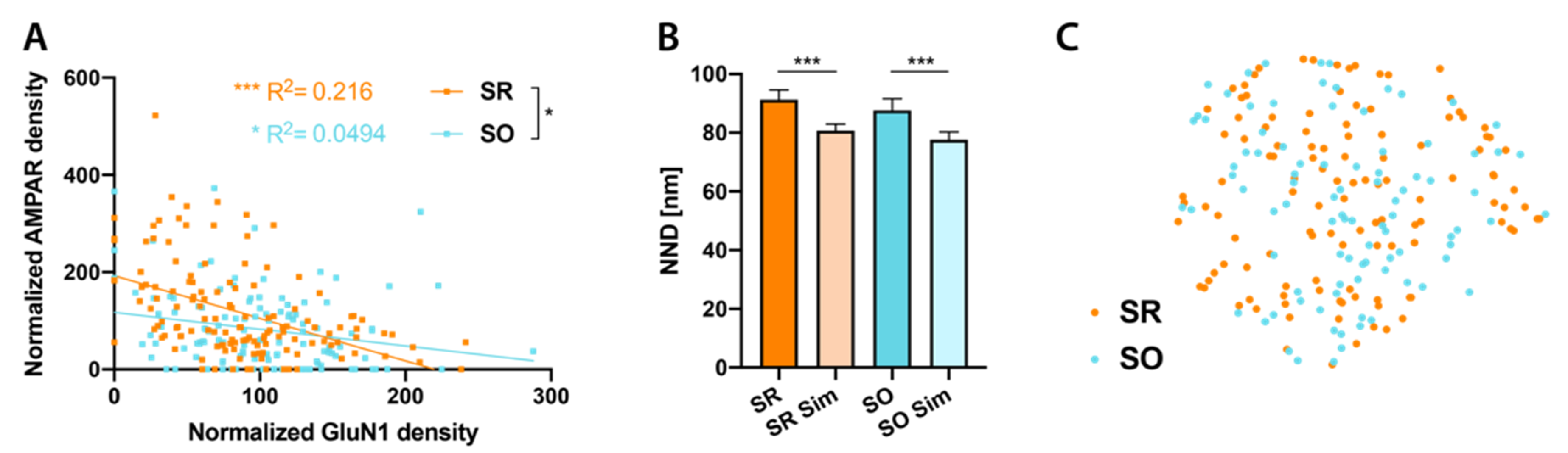
2.3. Glutamate Receptor Distributions in CA1 Pyramidal Cell Spine Synapses
2.4. Cav2.1 Channel Distribution in SR and SO
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Antibodies
4.3. SDS Freeze-Fracture Replica Labeling
4.4. Imaging and Manual Demarcation
4.5. Darea Software
4.5.1. Deep Learning for Automated Segmentation of Synaptic Areas
4.5.2. Gold Particle Detection
4.5.3. Monte-Carlo Simulations
4.5.4. Center Periphery Index (CPI)
4.5.5. Particle Cluster Analysis
4.6. Serial Section EM Analysis of PSD Area
4.7. Statistics and Visualization
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPA | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| AMPAR | AMPA receptor |
| ANOVA | Analysis of variance |
| AZ | Active zone |
| CPI | Center periphery index |
| CPU | Central processing unit |
| EM | Electron microscopy |
| EPSC | excitatory postsynaptic current |
| fEPSP | field excitatory postsynaptic potential |
| gp | Guinea pig |
| GPU | Graphics processing unit |
| gt | Goat |
| IMP | Intramembrane particle |
| IoU | Intersect over union |
| IPN | Interpeduncular nucleus |
| LTP | Long-term potentiation |
| ms | Mouse |
| NMDA | N-methyl-D-aspartate |
| NMDAR | NMDAR receptor |
| NND | Nearest-neighbor distance |
| PFA | Paraformaldehyde |
| PSD | Postsynaptic density |
| rb | Rabbit |
| RIM | Rab3-interacting molecule |
| SD | Standard deviation |
| SDS-FRL | SDS freeze-fracture replica immunolabeling electron microscopy |
| SO | Stratum oriens |
| SR | Stratum radiatum |
| tSNE | t-distributed stochastic neighbor embedding |
References
- Scheefhals, N.; MacGillavry, H.D. Functional organization of postsynaptic glutamate receptors. Mol. Cell. Neurosci. 2018, 91, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Shipton, O.A.; Paulsen, O. GluN2A and GluN2B subunit-containing NMDA receptors in hippocampal plasticity. Philos. Trans. R. Soc. B Biol. Sci. 2013, 369, 20130163. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Shang, Y.; Zhang, M. Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat. Rev. Neurosci. 2016, 17, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Eggermann, E.; Bucurenciu, I.; Goswami, S.P.; Jonas, P. Nanodomain coupling between Ca2+ channels and sensors of exocytosis at fast mammalian synapses. Nat. Rev. Neurosci. 2012, 13, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Leenders, A.; Sheng, Z. Modulation of neurotransmitter release by the second messenger-activated protein kinases: Implications for presynaptic plasticity. Pharmacol. Ther. 2005, 105, 69–84. [Google Scholar] [CrossRef]
- Südhof, T.C. Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 2017, 171, 745–769. [Google Scholar] [CrossRef] [PubMed]
- Savtchenko, L.P.; Rusakov, D.A. Moderate AMPA receptor clustering on the nanoscale can efficiently potentiate synaptic current. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130167. [Google Scholar] [CrossRef]
- Compans, B.; Choquet, D.; Hosy, E. Review on the role of AMPA receptor nano-organization and dynamic in the properties of synaptic transmission. Neurophotonics 2016, 3, 041811. [Google Scholar] [CrossRef]
- Tang, A.-H.; Chen, H.; Li, T.P.; Metzbower, S.R.; MacGillavry, H.D.; Blanpied, T.A. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 2016, 536, 210–214. [Google Scholar] [CrossRef]
- Burnashev, N.; Monyer, H.; Seeburg, P.H.; Sakmann, B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron 1992, 8, 189–198. [Google Scholar] [CrossRef]
- Soto, D.; Coombs, I.D.; Kelly, L.; Farrant, M.; Cull-Candy, S.G. Stargazin attenuates intracellular polyamine block of calcium-permeable AMPA receptors. Nat. Neurosci. 2007, 10, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Diering, G.H.; Huganir, R.L. The AMPA Receptor Code of Synaptic Plasticity. Neuron 2018, 100, 314–329. [Google Scholar] [CrossRef]
- Barria, A.; Malinow, R. NMDA Receptor Subunit Composition Controls Synaptic Plasticity by Regulating Binding to CaMKII. Neuron 2005, 48, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, R.-Q.; Gu, Q.-H.; Yan, J.-Z.; Wang, S.-H.; Liu, S.-Y.; Lu, W. Metaplastic Regulation of Long-Term Potentiation/Long-Term Depression Threshold by Activity-Dependent Changes of NR2A/NR2B Ratio. J. Neurosci. 2009, 29, 8764–8773. [Google Scholar] [CrossRef] [PubMed]
- Yashiro, K.; Philpot, B.D. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology 2008, 55, 1081–1094. [Google Scholar] [CrossRef]
- Henley, J.M.; Wilkinson, K.A. Synaptic AMPA receptor composition in development, plasticity and disease. Nat. Rev. Neurosci. 2016, 17, 337–350. [Google Scholar] [CrossRef]
- Shinohara, Y.; Hosoya, A.; Yahagi, K.; Ferecskó, A.S.; Yaguchi, K.; Sík, A.; Itakura, M.; Takahashi, M.; Hirase, H. Hippocampal CA3 and CA2 have distinct bilateral innervation patterns to CA1 in rodents: Rodent hippocampal CA3 and CA2 projection to CA1. Eur. J. Neurosci. 2012, 35, 702–710. [Google Scholar] [CrossRef]
- Klausberger, T.; Somogyi, P. Neuronal Diversity and Temporal Dynamics: The Unity of Hippocampal Circuit Operations. Science 2008, 321, 53–57. [Google Scholar] [CrossRef]
- Spruston, N. Pyramidal neurons: Dendritic structure and synaptic integration. Nat. Rev. Neurosci. 2008, 9, 206–221. [Google Scholar] [CrossRef]
- Haley, J.E.; Schaible, E.; Pavlidis, P.; Murdock, A.; Madison, D.V. Basal and apical synapses of CA1 pyramidal cells employ different LTP induction mechanisms. Learn. Mem. 1996, 3, 289–295. [Google Scholar] [CrossRef][Green Version]
- Brzdak, P.; Wójcicka, O.; Zareba-Koziol, M.; Minge, D.; Henneberger, C.; Wlodarczyk, J.; Mozrzymas, J.W.; Wójtowicz, T. Synaptic Potentiation at Basal and Apical Dendrites of Hippocampal Pyramidal Neurons Involves Activation of a Distinct Set of Extracellular and Intracellular Molecular Cues. Cereb. Cortex 2019, 29, 283–304. [Google Scholar] [CrossRef] [PubMed]
- Leung, L.S.; Shen, B. Long-term potentiation at the apical and basal dendritic synapses of CA1 after local stimulation in behaving rats. J. Neurophysiol. 1995, 73, 1938–1946. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-B.; Du, D.; Hasan, M.T.; Köhr, G. D4 Receptor Activation Differentially Modulates Hippocampal Basal and Apical Dendritic Synapses in Freely Moving Mice. Cereb. Cortex 2016, 26, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Herwerth, M.; Jensen, V.; Novak, M.; Konopka, W.; Hvalby, O.; Kohr, G. D4 Dopamine Receptors Modulate NR2B NMDA Receptors and LTP in Stratum Oriens of Hippocampal CA1. Cereb. Cortex 2012, 22, 1786–1798. [Google Scholar] [CrossRef]
- Leung, L.S.; Peloquin, P. Cholinergic Modulation Differs between Basal and Apical Dendritic Excitation of Hippocampal CA1 Pyramidal Cells. Cereb. Cortex 2010, 20, 1865–1877. [Google Scholar] [CrossRef]
- Kawakami, R.; Shinohara, Y.; Kato, Y.; Sugiyama, H.; Shigemoto, R.; Ito, I. Asymmetrical Allocation of NMDA Receptor epsilon2 Subunits in Hippocampal Circuitry. Science 2003, 300, 990–994. [Google Scholar] [CrossRef]
- Shinohara, Y.; Hirase, H.; Watanabe, M.; Itakura, M.; Takahashi, M.; Shigemoto, R. Left-right asymmetry of the hippocampal synapses with differential subunit allocation of glutamate receptors. Proc. Natl. Acad. Sci. USA 2008, 105, 19498–19503. [Google Scholar] [CrossRef]
- Kawakami, R.; Dobi, A.; Shigemoto, R.; Ito, I. Right Isomerism of the Brain in Inversus Viscerum Mutant Mice. PLoS ONE 2008, 3, e1945. [Google Scholar] [CrossRef]
- Fujimoto, K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J. Cell Sci. 1995, 108, 3443–3449. [Google Scholar]
- Masugi-Tokita, M.; Shigemoto, R. High-resolution quantitative visualization of glutamate and GABA receptors at central synapses. Curr. Opin. Neurobiol. 2007, 17, 387–393. [Google Scholar] [CrossRef]
- Masugi-Tokita, M.; Tarusawa, E.; Watanabe, M.; Molnar, E.; Fujimoto, K.; Shigemoto, R. Number and Density of AMPA Receptors in Individual Synapses in the Rat Cerebellum as Revealed by SDS-Digested Freeze-Fracture Replica Labeling. J. Neurosci. 2007, 27, 2135–2144. [Google Scholar] [CrossRef]
- Luján, R.; Aguado, C.; Ciruela, F.; Cózar, J.; Kleindienst, D.; de la Ossa, L.; Bettler, B.; Wickman, K.; Watanabe, M.; Shigemoto, R.; et al. Differential association of GABAB receptors with their effector ion channels in Purkinje cells. Brain Struct. Funct. 2018, 223, 1565–1587. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Las Condes, Chile, 11–18 December 2015; pp. 1026–1034. [Google Scholar]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Solass, W.; Schmitt, M.; Klode, J.; Schadendorf, D.; Sondermann, W.; Franklin, C.; Bestvater, F.; et al. Deep learning outperformed 11 pathologists in the classification of histopathological melanoma images. Eur. J. Cancer 2019, 118, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Garcia-Rodriguez, J. A Review on Deep Learning Techniques Applied to Semantic Segmentation. arXiv 2017, arXiv:1704.06857. [Google Scholar]
- Işın, A.; Direkoğlu, C.; Şah, M. Review of MRI-based Brain Tumor Image Segmentation Using Deep Learning Methods. Procedia Comput. Sci. 2016, 102, 317–324. [Google Scholar] [CrossRef]
- Yuan, Y. Hierarchical convolutional-deconvolutional neural networks for automatic liver and tumor segmentation. arXiv 2017, arXiv:1710.04540. [Google Scholar]
- Soltanian-Zadeh, S.; Sahingur, K.; Blau, S.; Gong, Y.; Farsiu, S. Fast and robust active neuron segmentation in two-photon calcium imaging using spatiotemporal deep learning. Proc. Natl. Acad. Sci. USA 2019, 116, 8554–8563. [Google Scholar] [CrossRef]
- Oztel, I.; Yolcu, G.; Ersoy, I.; White, T.; Bunyak, F. Mitochondria segmentation in electron microscopy volumes using deep convolutional neural network. In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE, Kansas City, MO, USA, 13–16 November 2017; pp. 1195–1200. [Google Scholar]
- Heinrich, L.; Funke, J.; Pape, C.; Nunez-Iglesias, J.; Saalfeld, S. Synaptic Cleft Segmentation in Non-isotropic Volume Electron Microscopy of the Complete Drosophila Brain. In Proceedings of Medical Image Computing and Computer Assisted Intervention—MICCAI 2018, Granada, Spain, 16–20 September 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer: Cham, Switzerland, 2018; pp. 317–325. [Google Scholar]
- Quan, T.M.; Hildebrand, D.G.C.; Jeong, W.-K. FusionNet: A deep fully residual convolutional neural network for image segmentation in connectomics. arXiv 2016, arXiv:1612.05360. [Google Scholar]
- Lee, K.; Zung, J.; Li, P.; Jain, V.; Seung, H.S. Superhuman Accuracy on the SNEMI3D Connectomics Challenge. arXiv 2017, arXiv:1706.00120. [Google Scholar]
- Xiao, C.; Liu, J.; Chen, X.; Han, H.; Shu, C.; Xie, Q. Deep contextual residual network for electron microscopy image segmentation in connectomics. In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) IEEE, Washington, DC, USA, 4–7 April 2018; pp. 378–381. [Google Scholar]
- Bhandari, P.; Vandael, D.; Fernández-Fernández, D.; Fritzius, T.; Kleindienst, D.; Montanaro, J.; Gassmann, M.; Jonas, P.; Kulik, A.; Bettler, B.; et al. GABAB receptor auxiliary subunits modulate Cav2.3-mediated release from medial habenula terminals. bioRxiv 2020. [Google Scholar] [CrossRef]
- Eguchi, K.; Velicky, P.; Hollergschwandtner, E.; Itakura, M.; Fukazawa, Y.; Danzl, J.G.; Shigemoto, R. Advantages of Acute Brain Slices Prepared at Physiological Temperature in the Characterization of Synaptic Functions. Front. Cell. Neurosci. 2020, 14, 63. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, D.A.; Trana, R.; Katz, Y.; Kath, W.L.; Spruston, N.; Geinisman, Y. Distance-Dependent Differences in Synapse Number and AMPA Receptor Expression in Hippocampal CA1 Pyramidal Neurons. Neuron 2006, 50, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Reth, M. Matching cellular dimensions with molecular sizes. Nat. Immunol. 2013, 14, 765–767. [Google Scholar] [CrossRef]
- Kharazia, V.N.; Weinberg, R.J. Tangential synaptic distribution of NMDA and AMPA receptors in rat neocortex. Neurosci. Lett. 1997, 238, 41–44. [Google Scholar] [CrossRef]
- Racca, C.; Stephenson, F.A.; Streit, P.; Roberts, J.D.B.; Somogyi, P. NMDA Receptor Content of Synapses in Stratum Radiatum of the Hippocampal CA1 Area. J. Neurosci. 2000, 20, 2512–2522. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Winters, C.; Azzam, R.; Li, X.; Galbraith, J.A.; Leapman, R.D.; Reese, T.S. Organization of the core structure of the postsynaptic density. Proc. Natl. Acad. Sci. USA 2008, 105, 4453–4458. [Google Scholar] [CrossRef]
- Goncalves, J.; Bartol, T.M.; Camus, C.; Levet, F.; Menegolla, A.P.; Sejnowski, T.J.; Sibarita, J.-B.; Vivaudou, M.; Choquet, D.; Hosy, E. Nanoscale co-organization and coactivation of AMPAR, NMDAR, and mGluR at excitatory synapses. Proc. Natl. Acad. Sci. USA 2020, 117, 14503–14511. [Google Scholar] [CrossRef]
- Liao, D.; Hessler, N.A.; Malinow, R. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature 1995, 375, 400–404. [Google Scholar] [CrossRef]
- Groc, L.; Gustafsson, B.; Hanse, E. Spontaneous Unitary Synaptic Activity in CA1 Pyramidal Neurons during Early Postnatal Development: Constant Contribution of AMPA and NMDA Receptors. J. Neurosci. 2002, 22, 5552–5562. [Google Scholar] [CrossRef]
- Isaac, J. Postsynaptic silent synapses: Evidence and mechanisms. Neuropharmacology 2003, 45, 450–460. [Google Scholar] [CrossRef]
- Voronin, L.L.; Cherubini, E. ‘Deaf, mute and whispering’ silent synapses: Their role in synaptic plasticity: Silent synapses and LTP. J. Physiol. 2004, 557, 3–12. [Google Scholar] [CrossRef]
- Luebke, J.I.; Dunlap, K.; Turner, T.J. Multiple calcium channel types control glutamatergic synaptic transmission in the hippocampus. Neuron 1993, 11, 895–902. [Google Scholar] [CrossRef]
- Wheeler, D.; Randall, A.; Tsien, R. Roles of N-type and Q-type Ca2+ channels in supporting hippocampal synaptic transmission. Science 1994, 264, 107–111. [Google Scholar] [CrossRef]
- Scheuber, A. Presynaptic Cav2.1 and Cav2.2 Differentially Influence Release Dynamics at Hippocampal Excitatory Synapses. J. Neurosci. 2004, 24, 10402–10409. [Google Scholar] [CrossRef]
- Rebola, N.; Reva, M.; Kirizs, T.; Szoboszlay, M.; Lőrincz, A.; Moneron, G.; Nusser, Z.; DiGregorio, D.A. Distinct Nanoscale Calcium Channel and Synaptic Vesicle Topographies Contribute to the Diversity of Synaptic Function. Neuron 2019, 104, 693–710.e9. [Google Scholar] [CrossRef]
- Nakamura, Y.; Harada, H.; Kamasawa, N.; Matsui, K.; Rothman, J.S.; Shigemoto, R.; Silver, R.A.; DiGregorio, D.A.; Takahashi, T. Nanoscale Distribution of Presynaptic Ca2+ Channels and Its Impact on Vesicular Release during Development. Neuron 2015, 85, 145–158. [Google Scholar] [CrossRef]
- Miki, T.; Kaufmann, W.A.; Malagon, G.; Gomez, L.; Tabuchi, K.; Watanabe, M.; Shigemoto, R.; Marty, A. Numbers of presynaptic Ca 2+ channel clusters match those of functionally defined vesicular docking sites in single central synapses. Proc. Natl. Acad. Sci. USA 2017, 114, E5246–E5255. [Google Scholar]
- Schikorski, T.; Stevens, C.F. Quantitative Ultrastructural Analysis of Hippocampal Excitatory Synapses. J. Neurosci. 1997, 17, 5858–5867. [Google Scholar] [CrossRef]
- Fortin, D.A.; Davare, M.A.; Srivastava, T.; Brady, J.D.; Nygaard, S.; Derkach, V.A.; Soderling, T.R. Long-Term Potentiation-Dependent Spine Enlargement Requires Synaptic Ca2+-Permeable AMPA Receptors Recruited by CaM-Kinase I. J. Neurosci. 2010, 30, 11565–11575. [Google Scholar] [CrossRef]
- Hunt, D.L.; Castillo, P.E. Synaptic plasticity of NMDA receptors: Mechanisms and functional implications. Curr. Opin. Neurobiol. 2012, 22, 496–508. [Google Scholar] [CrossRef]
- Frank, R.A.W.; Komiyama, N.H.; Ryan, T.J.; Zhu, F.; O’Dell, T.J.; Grant, S.G.N. NMDA receptors are selectively partitioned into complexes and supercomplexes during synapse maturation. Nat. Commun. 2016, 7, 11264. [Google Scholar] [CrossRef] [PubMed]
- Frank, R.A.; Grant, S.G. Supramolecular organization of NMDA receptors and the postsynaptic density. Curr. Opin. Neurobiol. 2017, 45, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Nadkarni, S.; Bartol, T.M.; Stevens, C.F.; Sejnowski, T.J.; Levine, H. Short-term plasticity constrains spatial organization of a hippocampal presynaptic terminal. Proc. Natl. Acad. Sci. USA 2012, 109, 14657–14662. [Google Scholar] [CrossRef]
- Scimemi, A.; Diamond, J.S. The Number and Organization of Ca2+ Channels in the Active Zone Shapes Neurotransmitter Release from Schaffer Collateral Synapses. J. Neurosci. 2012, 32, 18157–18176. [Google Scholar] [CrossRef] [PubMed]
- Bornschein, G.; Schmidt, H. Synaptotagmin Ca2+ Sensors and Their Spatial Coupling to Presynaptic Cav Channels in Central Cortical Synapses. Front. Mol. Neurosci. 2019, 11, 494. [Google Scholar] [CrossRef]
- Seif, G. Semantic Segmentation Suite. Available online: https://github.com/GeorgeSeif/Semantic-Segmentation-Suite (accessed on 27 October 2018).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. arXiv 2015, arXiv:1511.00561. [Google Scholar] [CrossRef]
- Yang, M.; Yu, K.; Zhang, C.; Li, Z.; Yang, K. DenseASPP for Semantic Segmentation in Street Scenes. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 3684–3692. [Google Scholar]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Salt Lake City, UT, USA, 18–23 June 2018; pp. 801–818. [Google Scholar]
- Jegou, S.; Drozdzal, M.; Vazquez, D.; Romero, A.; Bengio, Y. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 11–19. [Google Scholar]

| Antibody | Reference | Concentration or Dilution |
|---|---|---|
| Rb-anti-GluA1-3 | [45] | 4.3 µg/mL |
| Ms-anti-GluN1 | Millipore, Burlington, MA, USA, Cat.No. MAB363 | 5.63 µg/mL |
| Gp-anti-Cav2.1 | Synaptic Systems, Göttingen, Germany, Cat.No. 152205 | 2.5 µg/mL |
| Gt-anti-ms-6nm | Jackson ImmunoResearch, West Grove, PA, USA, Cat.No. 115-195-146 | 1:30 |
| Gt-anti-ms-12nm | Jackson ImmunoResearch, Cat.No. 115-205-146 | 1:30 |
| Gt-anti-rb-6nm | Jackson ImmunoResearch, Cat.No. 111-195-144 | 1:30 |
| Gt-anti-rb-12nm | Jackson ImmunoResearch, Cat.No. 111-205-144 | 1:30 |
| Gt-anti-rb-5nm | BBI Solutions, Crumlin, UK, Cat.No. EM.GAR5 | 1:30 |
| Gt-anti-gp-5nm | BBI Solutions, Cat.No. EM.GAG5 | 1:50 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleindienst, D.; Montanaro, J.; Bhandari, P.; Case, M.J.; Fukazawa, Y.; Shigemoto, R. Deep Learning-Assisted High-Throughput Analysis of Freeze-Fracture Replica Images Applied to Glutamate Receptors and Calcium Channels at Hippocampal Synapses. Int. J. Mol. Sci. 2020, 21, 6737. https://doi.org/10.3390/ijms21186737
Kleindienst D, Montanaro J, Bhandari P, Case MJ, Fukazawa Y, Shigemoto R. Deep Learning-Assisted High-Throughput Analysis of Freeze-Fracture Replica Images Applied to Glutamate Receptors and Calcium Channels at Hippocampal Synapses. International Journal of Molecular Sciences. 2020; 21(18):6737. https://doi.org/10.3390/ijms21186737
Chicago/Turabian StyleKleindienst, David, Jacqueline Montanaro, Pradeep Bhandari, Matthew J. Case, Yugo Fukazawa, and Ryuichi Shigemoto. 2020. "Deep Learning-Assisted High-Throughput Analysis of Freeze-Fracture Replica Images Applied to Glutamate Receptors and Calcium Channels at Hippocampal Synapses" International Journal of Molecular Sciences 21, no. 18: 6737. https://doi.org/10.3390/ijms21186737
APA StyleKleindienst, D., Montanaro, J., Bhandari, P., Case, M. J., Fukazawa, Y., & Shigemoto, R. (2020). Deep Learning-Assisted High-Throughput Analysis of Freeze-Fracture Replica Images Applied to Glutamate Receptors and Calcium Channels at Hippocampal Synapses. International Journal of Molecular Sciences, 21(18), 6737. https://doi.org/10.3390/ijms21186737