Databases and Bioinformatic Tools for Glycobiology and Glycoproteomics
Abstract
1. Introduction
2. Glycan Structure Databases
2.1. CFG Glycan Structure Database
2.2. Glycan Mass Spectral DataBase
2.3. UniCarbKB
2.4. KEGG Glycan
2.5. GLYCOSCIENCE.de
2.6. UniCarb-DB
2.7. GlyTouCan
2.8. GlycoStore
2.9. CSDB
3. Glycoprotein Databases
3.1. GlycoProtDB (GPDB)
3.2. UniPep and N-GlycositeAtlas
3.3. O-GalNAc Protein Databases
3.4. O-GlcNAc Protein Database
4. Glycogene Databases
4.1. CAZy
4.2. GGDB
4.3. CFG Glycosyltransferases Database
4.4. CSDB_GT Subdatabase
4.5. GlyMAP
5. Glycan-Protein Interaction Databases
5.1. LfDB
5.2. UniLectin
5.3. PACDB
5.4. SugarBindDB
5.5. GLAD: Glycan Array Dashboard
5.6. MCAW-DB
5.7. GlyMDB
5.8. MatrixDB
6. Software Tools for Glycan and Intact Glycopeptide Analysis
6.1. Software Tools for Glycan Analysis
6.2. Software Tools for Intact N-Glycopeptide Analysis
6.3. Software Tools for Intact O-Glycopeptide Analysis
7. The Latest Integrated Glycoscience Portal
7.1. Glycomics@ExPASy
7.2. Glygen
7.3. GlyCosmos
8. Discussion and Conclusions
Funding
Conflicts of Interest
References
- Apweiler, R.; Hermjakob, H.; Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the swiss-prot database. Biochim. Biophys. Acta 1999, 1473, 4–8. [Google Scholar] [CrossRef]
- Mereiter, S.; Balmana, M.; Campos, D.; Gomes, J.; Reis, C.A. Glycosylation in the era of cancer-targeted therapy: Where are we heading? Cancer Cell 2019, 36, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Dalziel, M.; Crispin, M.; Scanlan, C.N.; Zitzmann, N.; Dwek, R.A. Emerging principles for the therapeutic exploitation of glycosylation. Science 2014, 343, 1235681. [Google Scholar] [CrossRef] [PubMed]
- Strum, J.S.; Nwosu, C.C.; Hua, S.; Kronewitter, S.R.; Seipert, R.R.; Bachelor, R.J.; An, H.J.; Lebrilla, C.B. Automated assignments of n- and o-site specific glycosylation with extensive glycan heterogeneity of glycoprotein mixtures. Anal. Chem. 2013, 85, 5666–5675. [Google Scholar] [CrossRef] [PubMed]
- Kailemia, M.J.; Xu, G.; Wong, M.; Li, Q.; Goonatilleke, E.; Leon, F.; Lebrilla, C.B. Recent advances in the mass spectrometry methods for glycomics and cancer. Anal. Chem. 2018, 90, 208–224. [Google Scholar] [CrossRef]
- Xiao, K.; Han, Y.; Yang, H.; Lu, H.; Tian, Z. Mass spectrometry-based qualitative and quantitative n-glycomics: An update of 2017-2018. Anal. Chim. Acta 2019, 1091, 1–22. [Google Scholar] [CrossRef]
- Hirabayashi, J.; Kuno, A.; Tateno, H. Development and applications of the lectin microarray. Top. Curr. Chem. 2015, 367, 105–124. [Google Scholar]
- Hyun, J.Y.; Pai, J.; Shin, I. The glycan microarray story from construction to applications. Acc. Chem. Res. 2017, 50, 1069–1078. [Google Scholar] [CrossRef]
- Saravanan, C.; Cao, Z.; Head, S.R.; Panjwani, N. Analysis of differential expression of glycosyltransferases in healing corneas by glycogene microarrays. Glycobiology 2019, 29, 188–189. [Google Scholar] [CrossRef]
- Doubet, S.; Bock, K.; Smith, D.; Darvill, A.; Albersheim, P. The complex carbohydrate structure database. Trends Biochem. Sci. 1989, 14, 475–477. [Google Scholar] [CrossRef]
- Doubet, S.; Albersheim, P. Carbbank. Glycobiology 1992, 2, 505. [Google Scholar] [CrossRef] [PubMed]
- Lutteke, T.; Bohne-Lang, A.; Loss, A.; Goetz, T.; Frank, M.; von der Lieth, C.W. Glycosciences. De: An internet portal to support glycomics and glycobiology research. Glycobiology 2006, 16, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Goto, S.; Kawano, S.; Aoki-Kinoshita, K.F.; Ueda, N.; Hamajima, M.; Kawasaki, T.; Kanehisa, M. Kegg as a glycome informatics resource. Glycobiology 2006, 16, 63–70. [Google Scholar] [CrossRef]
- Raman, R.; Venkataraman, M.; Ramakrishnan, S.; Lang, W.; Raguram, S.; Sasisekharan, R. Advancing glycomics: Implementation strategies at the consortium for functional glycomics. Glycobiology 2006, 16, 82–90. [Google Scholar] [CrossRef]
- Narimatsu, H. Construction of a human glycogene library and comprehensive functional analysis. Glycoconj. J. 2004, 21, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Kameyama, A.; Kikuchi, N.; Nakaya, S.; Ito, H.; Sato, T.; Shikanai, T.; Takahashi, Y.; Takahashi, K.; Narimatsu, H. A strategy for identification of oligosaccharide structures using observational multistage mass spectral library. Anal. Chem. 2005, 77, 4719–4725. [Google Scholar] [CrossRef]
- Kaji, H.; Saito, H.; Yamauchi, Y.; Shinkawa, T.; Taoka, M.; Hirabayashi, J.; Kasai, K.; Takahashi, N.; Isobe, T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify n-linked glycoproteins. Nat. Biotechnol. 2003, 21, 667–672. [Google Scholar] [CrossRef]
- York, W.S.; Mazumder, R.; Ranzinger, R.; Edwards, N.; Kahsay, R.; Aoki-Kinoshita, K.F.; Campbell, M.P.; Cummings, R.D.; Feizi, T.; Martin, M.; et al. Glygen: Computational and informatics resources for glycoscience. Glycobiology 2020, 30, 72–73. [Google Scholar] [CrossRef] [PubMed]
- Mariethoz, J.; Alocci, D.; Gastaldello, A.; Horlacher, O.; Gasteiger, E.; Rojas-Macias, M.; Karlsson, N.G.; Packer, N.H.; Lisacek, F. Glycomics@expasy: Bridging the gap. Mol. Cell. Proteom. 2018, 17, 2164–2176. [Google Scholar] [CrossRef] [PubMed]
- Yamada, I.; Shiota, M.; Shinmachi, D.; Ono, T.; Tsuchiya, S.; Hosoda, M.; Fujita, A.; Aoki, N.P.; Watanabe, Y.; Fujita, N.; et al. The glycosmos portal: A unified and comprehensive web resource for the glycosciences. Nat. Methods 2020, 17, 649–650. [Google Scholar] [CrossRef]
- Akune, Y.; Lin, C.H.; Abrahams, J.L.; Zhang, J.; Packer, N.H.; Aoki-Kinoshita, K.F.; Campbell, M.P. Comprehensive analysis of the n-glycan biosynthetic pathway using bioinformatics to generate unicorn: A theoretical n-glycan structure database. Carbohydr. Res. 2016, 431, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Hu, Y.; Ao, M.; Shah, P.; Chen, J.; Yang, W.; Jia, X.; Tian, Y.; Thomas, S.; Zhang, H. N-glycositeatlas: A database resource for mass spectrometry-based human n-linked glycoprotein and glycosylation site mapping. Clin. Proteom. 2019, 16, 35. [Google Scholar] [CrossRef] [PubMed]
- Egorova, K.S.; Toukach, P.V. Glycoinformatics: Bridging isolated islands in the sea of data. Angew. Chem. Int. Ed. Engl. 2018, 57, 14986–14990. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, J.L.; Taherzadeh, G.; Jarvas, G.; Guttman, A.; Zhou, Y.; Campbell, M.P. Recent advances in glycoinformatic platforms for glycomics and glycoproteomics. Curr. Opin. Struct. Biol. 2020, 62, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.P.; Hayes, C.A.; Struwe, W.B.; Wilkins, M.R.; Aoki-Kinoshita, K.F.; Harvey, D.J.; Rudd, P.M.; Kolarich, D.; Lisacek, F.; Karlsson, N.G.; et al. Unicarbkb: Putting the pieces together for glycomics research. Proteomics 2011, 11, 4117–4121. [Google Scholar] [CrossRef]
- Campbell, M.P.; Packer, N.H. Unicarbkb: New database features for integrating glycan structure abundance, compositional glycoproteomics data, and disease associations. Biochim. Biophys. Acta 2016, 1860, 1669–1675. [Google Scholar] [CrossRef]
- Campbell, M.P.; Peterson, R.; Mariethoz, J.; Gasteiger, E.; Akune, Y.; Aoki-Kinoshita, K.F.; Lisacek, F.; Packer, N.H. Unicarbkb: Building a knowledge platform for glycoproteomics. Nucleic Acids Res. 2014, 42, D215–D221. [Google Scholar] [CrossRef]
- Cooper, C.A.; Harrison, M.J.; Wilkins, M.R.; Packer, N.H. Glycosuitedb: A new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. 2001, 29, 332–335. [Google Scholar] [CrossRef]
- Cooper, C.A.; Joshi, H.J.; Harrison, M.J.; Wilkins, M.R.; Packer, N.H. Glycosuitedb: A curated relational database of glycoprotein glycan structures and their biological sources. 2003 update. Nucleic Acids Res. 2003, 31, 511–513. [Google Scholar] [CrossRef]
- Royle, L.; Radcliffe, C.M.; Dwek, R.A.; Rudd, P.M. Detailed structural analysis of n-glycans released from glycoproteins in sds-page gel bands using hplc combined with exoglycosidase array digestions. Methods Mol. Biol. 2006, 347, 125–143. [Google Scholar]
- von der Lieth, C.W.; Freire, A.A.; Blank, D.; Campbell, M.P.; Ceroni, A.; Damerell, D.R.; Dell, A.; Dwek, R.A.; Ernst, B.; Fogh, R.; et al. Eurocarbdb: An open-access platform for glycoinformatics. Glycobiology 2011, 21, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Gotz, L.; Abrahams, J.L.; Mariethoz, J.; Rudd, P.M.; Karlsson, N.G.; Packer, N.H.; Campbell, M.P.; Lisacek, F. Glycodigest: A tool for the targeted use of exoglycosidase digestions in glycan structure determination. Bioinformatics 2014, 30, 3131–3133. [Google Scholar] [CrossRef] [PubMed]
- Lutteke, T.; von der Lieth, C.W. Data mining the pdb for glyco-related data. Methods Mol. Biol. 2009, 534, 293–310. [Google Scholar] [PubMed]
- Bohne, A.; Lang, E.; von der Lieth, C.W. Sweet-www-based rapid 3d construction of oligo- and polysaccharides. Bioinformatics 1999, 15, 767–768. [Google Scholar] [CrossRef] [PubMed]
- Bohm, M.; Bohne-Lang, A.; Frank, M.; Loss, A.; Rojas-Macias, M.A.; Lutteke, T. Glycosciences.Db: An annotated data collection linking glycomics and proteomics data (2018 update). Nucleic Acids Res. 2019, 47, D1195–D1201. [Google Scholar] [CrossRef]
- Campbell, M.P.; Nguyen-Khuong, T.; Hayes, C.A.; Flowers, S.A.; Alagesan, K.; Kolarich, D.; Packer, N.H.; Karlsson, N.G. Validation of the curation pipeline of unicarb-db: Building a global glycan reference ms/ms repository. Biochim. Biophys. Acta 2014, 1844, 108–116. [Google Scholar] [CrossRef]
- Aoki-Kinoshita, K.; Agravat, S.; Aoki, N.P.; Arpinar, S.; Cummings, R.D.; Fujita, A.; Fujita, N.; Hart, G.M.; Haslam, S.M.; Kawasaki, T.; et al. Glytoucan 1.0—The international glycan structure repository. Nucleic Acids Res. 2016, 44, D1237–D1242. [Google Scholar] [CrossRef]
- Ranzinger, R.; Herget, S.; Wetter, T.; von der Lieth, C.W. Glycomedb-integration of open-access carbohydrate structure databases. BMC Bioinform. 2008, 9, 384. [Google Scholar] [CrossRef]
- Ranzinger, R.; Herget, S.; von der Lieth, C.W.; Frank, M. Glycomedb—A unified database for carbohydrate structures. Nucleic Acids Res. 2011, 39, D373–D376. [Google Scholar] [CrossRef]
- Tiemeyer, M.; Aoki, K.; Paulson, J.; Cummings, R.D.; York, W.S.; Karlsson, N.G.; Lisacek, F.; Packer, N.H.; Campbell, M.P.; Aoki, N.P.; et al. Glytoucan: An accessible glycan structure repository. Glycobiology 2017, 27, 915–919. [Google Scholar] [CrossRef]
- Zhao, S.; Walsh, I.; Abrahams, J.L.; Royle, L.; Nguyen-Khuong, T.; Spencer, D.; Fernandes, D.L.; Packer, N.H.; Rudd, P.M.; Campbell, M.P. Glycostore: A database of retention properties for glycan analysis. Bioinformatics 2018, 34, 3231–3232. [Google Scholar] [CrossRef] [PubMed]
- Toukach, P.V.; Egorova, K.S. Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts. Nucleic Acids Res. 2016, 44, D1229–D1236. [Google Scholar] [CrossRef]
- Toukach, P.V.; Egorova, K.S. Bacterial, plant, and fungal carbohydrate structure databases: Daily usage. Methods Mol. Biol. 2015, 1273, 55–85. [Google Scholar] [PubMed]
- Toukach, P.; Egorova, K. Carbohydrate structure database (csdb): Examples of usage. In A Practical Guide to Using Glycomics Databases; Aoki-Kinoshita, K.F., Ed.; Springer: Tokyo, Japan, 2017; pp. 75–113. [Google Scholar]
- Bennett, E.P.; Mandel, U.; Clausen, H.; Gerken, T.A.; Fritz, T.A.; Tabak, L.A. Control of mucin-type o-glycosylation: A classification of the polypeptide galnac-transferase gene family. Glycobiology 2012, 22, 736–756. [Google Scholar] [CrossRef] [PubMed]
- Holt, G.D.; Hart, G.W. The subcellular distribution of terminal n-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, o-linked glcnac. J. Biol. Chem. 1986, 261, 8049–8057. [Google Scholar]
- Walsh, I.; Zhao, S.; Campbell, M.; Taron, C.H.; Rudd, P.M. Quantitative profiling of glycans and glycopeptides: An informatics’ perspective. Curr. Opin. Struct. Biol. 2016, 40, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Lisacek, F.; Mariethoz, J.; Alocci, D.; Rudd, P.M.; Abrahams, J.L.; Campbell, M.P.; Packer, N.H.; Stahle, J.; Widmalm, G.; Mullen, E.; et al. Databases and associated tools for glycomics and glycoproteomics. Methods Mol. Biol. 2017, 1503, 235–264. [Google Scholar]
- Cao, W.; Liu, M.; Kong, S.; Wu, M.; Zhang, Y.; Yang, P. Recent advances in software tools for more generic and precise intact glycopeptide analysis. Mol. Cell Proteom. 2020. [Google Scholar] [CrossRef]
- Klein, J.; Carvalho, L.; Zaia, J. Application of network smoothing to glycan lc-ms profiling. Bioinformatics 2018, 34, 3511–3518. [Google Scholar] [CrossRef]
- Sun, S.; Shah, P.; Eshghi, S.T.; Yang, W.; Trikannad, N.; Yang, S.; Chen, L.; Aiyetan, P.; Hoti, N.; Zhang, Z.; et al. Comprehensive analysis of protein glycosylation by solid-phase extraction of n-linked glycans and glycosite-containing peptides. Nat. Biotechnol. 2016, 34, 84–88. [Google Scholar] [CrossRef]
- Mao, J.; You, X.; Qin, H.; Wang, C.; Wang, L.; Ye, M. A new searching strategy for the identification of o-linked glycopeptides. Anal. Chem. 2019, 91, 3852–3859. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Mao, Y.; Clausen, H.; Vakhrushev, S.Y. Glyco-dia: A method for quantitative o-glycoproteomics with in silico-boosted glycopeptide libraries. Nat. Methods 2019, 16, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Ao, M.; Hu, Y.; Li, Q.K.; Zhang, H. Mapping the o-glycoproteome using site-specific extraction of o-linked glycopeptides (exoo). Mol. Syst. Biol. 2018, 14, e8486. [Google Scholar] [CrossRef]
- Kaji, H.; Yamauchi, Y.; Takahashi, N.; Isobe, T. Mass spectrometric identification of n-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat. Protoc. 2006, 1, 3019–3027. [Google Scholar] [CrossRef]
- Kaji, H.; Shikanai, T.; Sasaki-Sawa, A.; Wen, H.; Fujita, M.; Suzuki, Y.; Sugahara, D.; Sawaki, H.; Yamauchi, Y.; Shinkawa, T.; et al. Large-scale identification of n-glycosylated proteins of mouse tissues and construction of a glycoprotein database, glycoprotdb. J. Proteome Res. 2012, 11, 4553–4566. [Google Scholar] [CrossRef]
- Sugahara, D.; Kaji, H.; Sugihara, K.; Asano, M.; Narimatsu, H. Large-scale identification of target proteins of a glycosyltransferase isozyme by lectin-igot-lc/ms, an lc/ms-based glycoproteomic approach. Sci. Rep. 2012, 2, 680. [Google Scholar] [CrossRef]
- Kaji, H.; Ocho, M.; Togayachi, A.; Kuno, A.; Sogabe, M.; Ohkura, T.; Nozaki, H.; Angata, T.; Chiba, Y.; Ozaki, H.; et al. Glycoproteomic discovery of serological biomarker candidates for hcv/hbv infection-associated liver fibrosis and hepatocellular carcinoma. J. Proteome Res. 2013, 12, 2630–2640. [Google Scholar] [CrossRef]
- Zhang, H.; Loriaux, P.; Eng, J.; Campbell, D.; Keller, A.; Moss, P.; Bonneau, R.; Zhang, N.; Zhou, Y.; Wollscheid, B.; et al. Unipep—A database for human n-linked glycosites: A resource for biomarker discovery. Genome Biol. 2006, 7, R73. [Google Scholar] [CrossRef]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 310–322. [Google Scholar]
- Blom, N.; Sicheritz-Ponten, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The carbohydrate-active enzymes database (cazy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Consortium, C.A. Ten years of cazypedia: A living encyclopedia of carbohydrate-active enzymes. Glycobiology 2018, 28, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Egorova, K.S.; Knirel, Y.A.; Toukach, P.V. Expanding csdb_gt glycosyltransferase database with escherichia coli. Glycobiology 2019, 29, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.; Lind-Thomsen, A.; Joshi, H.J.; Pedersen, N.B.; Have, C.T.; Kong, Y.; Wang, S.; Sparso, T.; Grarup, N.; Vester-Christensen, M.B.; et al. A glycogene mutation map for discovery of diseases of glycosylation. Glycobiology 2015, 25, 211–224. [Google Scholar] [CrossRef]
- Narimatsu, H.; Sawaki, H.; Kuno, A.; Kaji, H.; Ito, H.; Ikehara, Y. A strategy for discovery of cancer glyco-biomarkers in serum using newly developed technologies for glycoproteomics. FEBS J. 2010, 277, 95–105. [Google Scholar] [CrossRef]
- Hirabayashi, J.; Tateno, H.; Shikanai, T.; Aoki-Kinoshita, K.F.; Narimatsu, H. The lectin frontier database (lfdb), and data generation based on frontal affinity chromatography. Molecules 2015, 20, 951–973. [Google Scholar] [CrossRef]
- Bonnardel, F.; Perez, S.; Lisacek, F.; Imberty, A. Structural database for lectins and the unilectin web platform. Methods Mol. Biol. 2020, 2132, 1–14. [Google Scholar]
- Maeda, M.; Fujita, N.; Suzuki, Y.; Sawaki, H.; Shikanai, T.; Narimatsu, H. Jcggdb: Japan consortium for glycobiology and glycotechnology database. Methods Mol. Biol. 2015, 1273, 161–179. [Google Scholar]
- Mariethoz, J.; Khatib, K.; Alocci, D.; Campbell, M.P.; Karlsson, N.G.; Packer, N.H.; Mullen, E.H.; Lisacek, F. Sugarbinddb, a resource of glycan-mediated host-pathogen interactions. Nucleic Acids Res. 2016, 44, D1243–D1250. [Google Scholar] [CrossRef]
- Mehta, A.Y.; Cummings, R.D. Glad: Glycan array dashboard, a visual analytics tool for glycan microarrays. Bioinformatics 2019, 35, 3536–3537. [Google Scholar] [CrossRef]
- Hosoda, M.; Takahashi, Y.; Shiota, M.; Shinmachi, D.; Inomoto, R.; Higashimoto, S.; Aoki-Kinoshita, K.F. Mcaw-db: A glycan profile database capturing the ambiguity of glycan recognition patterns. Carbohydr. Res. 2018, 464, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Park, S.J.; Mehta, A.Y.; Cummings, R.D.; Im, W. Glymdb: Glycan microarray database and analysis toolset. Bioinformatics 2020, 36, 2438–2442. [Google Scholar] [CrossRef]
- Chautard, E.; Ballut, L.; Thierry-Mieg, N.; Ricard-Blum, S. Matrixdb, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinformatics 2009, 25, 690–691. [Google Scholar] [CrossRef]
- Chautard, E.; Fatoux-Ardore, M.; Ballut, L.; Thierry-Mieg, N.; Ricard-Blum, S. Matrixdb, the extracellular matrix interaction database. Nucleic Acids Res. 2011, 39, D235–D240. [Google Scholar] [CrossRef]
- Launay, G.; Salza, R.; Multedo, D.; Thierry-Mieg, N.; Ricard-Blum, S. Matrixdb, the extracellular matrix interaction database: Updated content, a new navigator and expanded functionalities. Nucleic Acids Res. 2015, 43, D321–D327. [Google Scholar] [CrossRef]
- Clerc, O.; Deniaud, M.; Vallet, S.D.; Naba, A.; Rivet, A.; Perez, S.; Thierry-Mieg, N.; Ricard-Blum, S. Matrixdb: Integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 2019, 47, D376–D381. [Google Scholar] [CrossRef]
- Damerell, D.; Ceroni, A.; Maass, K.; Ranzinger, R.; Dell, A.; Haslam, S.M. The glycanbuilder and glycoworkbench glycoinformatics tools: Updates and new developments. Biol. Chem. 2012, 393, 1357–1362. [Google Scholar] [CrossRef]
- Maxwell, E.; Tan, Y.; Tan, Y.; Hu, H.; Benson, G.; Aizikov, K.; Conley, S.; Staples, G.O.; Slysz, G.W.; Smith, R.D.; et al. Glycresoft: A software package for automated recognition of glycans from lc/ms data. PLoS ONE 2012, 7, e45474. [Google Scholar] [CrossRef] [PubMed]
- Toghi Eshghi, S.; Shah, P.; Yang, W.; Li, X.; Zhang, H. Gpquest: A spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact n-glycopeptides. Anal. Chem. 2015, 87, 5181–5188. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Hu, Y.; Sun, S.; Ouyang, C.; Yang, W.; Wang, Q.; Betenbaugh, M.; Zhang, H. Comprehensive glycoproteomic analysis of chinese hamster ovary cells. Anal. Chem. 2018, 90, 14294–14302. [Google Scholar] [CrossRef]
- Zeng, W.F.; Liu, M.Q.; Zhang, Y.; Wu, J.Q.; Fang, P.; Peng, C.; Nie, A.; Yan, G.; Cao, W.; Liu, C.; et al. Pglyco: A pipeline for the identification of intact n-glycopeptides by using hcd- and cid-ms/ms and ms3. Sci. Rep. 2016, 6, 25102. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Q.; Zeng, W.F.; Fang, P.; Cao, W.Q.; Liu, C.; Yan, G.Q.; Zhang, Y.; Peng, C.; Wu, J.Q.; Zhang, X.J.; et al. Pglyco 2.0 enables precision n-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 2017, 8, 438. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Jiang, B.; Zhao, H.; Wu, M.; Kong, S.; Liu, M.; Yang, P.; Cao, W. Development of a computational tool for automated interpretation of intact o-glycopeptide tandem mass spectra from single proteins. Anal. Chem. 2020, 92, 6777–6784. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Liu, Y.; Lajoie, G.A.; Ma, B.; Zhang, K. An improved approach for n-linked glycan structure identification from hcd ms/ms spectra. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Alocci, D.; Mariethoz, J.; Gastaldello, A.; Gasteiger, E.; Karlsson, N.G.; Kolarich, D.; Packer, N.H.; Lisacek, F. Glyconnect: Glycoproteomics goes visual, interactive, and analytical. J. Proteome Res. 2019, 18, 664–677. [Google Scholar] [CrossRef]
- Kahsay, R.; Vora, J.; Navelkar, R.; Mousavi, R.; Fochtman, B.C.; Holmes, X.; Pattabiraman, N.; Ranzinger, R.; Mahadik, R.; Williamson, T.; et al. Glygen data model and processing workflow. Bioinformatics 2020, 36, 3941–3943. [Google Scholar] [CrossRef]
- Watanabe, Y.; Bowden, T.A.; Wilson, I.A.; Crispin, M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1480–1497. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Kuno, A.; Ikehara, Y.; Tanaka, Y.; Ito, K.; Matsuda, A.; Sekiya, S.; Hige, S.; Sakamoto, M.; Kage, M.; Mizokami, M.; et al. A serum “sweet-doughnut” protein facilitates fibrosis evaluation and therapy assessment in patients with viral hepatitis. Sci. Rep. 2013, 3, 1065. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Zhu, M.Y.; Yu, D.M.; Li, W.; Zhang, D.H.; Lu, F.J.; Gong, Q.M.; Liu, F.; Jiang, J.H.; Zheng, M.H.; et al. Serum wfa(+) -m2bp levels for evaluation of early stages of liver fibrosis in patients with chronic hepatitis b virus infection. Liver Int. 2017, 37, 35–44. [Google Scholar] [CrossRef]
- Matsuda, A.; Kuno, A.; Matsuzaki, H.; Kawamoto, T.; Shikanai, T.; Nakanuma, Y.; Yamamoto, M.; Ohkohchi, N.; Ikehara, Y.; Shoda, J.; et al. Glycoproteomics-based cancer marker discovery adopting dual enrichment with wisteria floribunda agglutinin for high specific glyco-diagnosis of cholangiocarcinoma. J. Proteom. 2013, 85, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Yao, F.; Yang, F.; Zhang, F.; Xu, Z.; Shi, J.; Kuno, A.; Zhao, H.; Zhang, Y. Glycomic signatures of plasma igg improve preoperative prediction of the invasiveness of small lung nodules. Molecules 2019, 25, 28. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Macias, M.A.; Mariethoz, J.; Andersson, P.; Jin, C.; Venkatakrishnan, V.; Aoki, N.P.; Shinmachi, D.; Ashwood, C.; Madunic, K.; Zhang, T.; et al. Towards a standardized bioinformatics infrastructure for n- and o-glycomics. Nat. Commun. 2019, 10, 3275. [Google Scholar] [CrossRef] [PubMed]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human o-galnac glycoproteome through simplecell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef]
- Egorova, K.S.; Toukach, P.V. Csdb_gt: A new curated database on glycosyltransferases. Glycobiology 2017, 27, 285–290. [Google Scholar] [CrossRef]
- Bern, M.; Kil, Y.J.; Becker, C. Byonic: Advanced peptide and protein identification software. Curr. Protoc. Bioinform. 2012, 40, 13–20. [Google Scholar] [CrossRef]
- Medzihradszky, K.F.; Kaasik, K.; Chalkley, R.J. Tissue-specific glycosylation at the glycopeptide level. Mol. Cell Proteom. 2015, 14, 2103–2110. [Google Scholar] [CrossRef]
- Lynn, K.S.; Chen, C.C.; Lih, T.M.; Cheng, C.W.; Su, W.C.; Chang, C.H.; Cheng, C.Y.; Hsu, W.L.; Chen, Y.J.; Sung, T.Y. Magic: An automated n-linked glycoprotein identification tool using a y1-ion pattern matching algorithm and in silico ms(2) approach. Anal. Chem. 2015, 87, 2466–2473. [Google Scholar] [CrossRef]
- He, L.; Xin, L.; Shan, B.; Lajoie, G.A.; Ma, B. Glycomaster db: Software to assist the automated identification of n-linked glycopeptides by tandem mass spectrometry. J. Proteome Res. 2014, 13, 3881–3895. [Google Scholar] [CrossRef]
- Cao, L.; Tolic, N.; Qu, Y.; Meng, D.; Zhao, R.; Zhang, Q.; Moore, R.J.; Zink, E.M.; Lipton, M.S.; Pasa-Tolic, L.; et al. Characterization of intact n- and o-linked glycopeptides using higher energy collisional dissociation. Anal. Biochem. 2014, 452, 96–102. [Google Scholar] [CrossRef]
- Wu, S.W.; Liang, S.Y.; Pu, T.H.; Chang, F.Y.; Khoo, K.H. Sweet-heart-an integrated suite of enabling computational tools for automated ms2/ms3 sequencing and identification of glycopeptides. J. Proteom. 2013, 84, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.W.; Pu, T.H.; Viner, R.; Khoo, K.H. Novel lc-ms(2) product dependent parallel data acquisition function and data analysis workflow for sequencing and identification of intact glycopeptides. Anal. Chem. 2014, 86, 5478–5486. [Google Scholar] [CrossRef] [PubMed]
- Mayampurath, A.; Yu, C.Y.; Song, E.; Balan, J.; Mechref, Y.; Tang, H. Computational framework for identification of intact glycopeptides in complex samples. Anal. Chem. 2014, 86, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Pagan, J.D.; Kitaoka, M.; Anthony, R.M. Engineered sialylation of pathogenic antibodies in vivo attenuates autoimmune disease. Cell 2018, 172, 564–577e513. [Google Scholar] [CrossRef]
- Pompach, P.; Chandler, K.B.; Lan, R.; Edwards, N.; Goldman, R. Semi-automated identification of n-glycopeptides by hydrophilic interaction chromatography, nano-reverse-phase lc-ms/ms, and glycan database search. J. Proteome Res. 2012, 11, 1728–1740. [Google Scholar] [CrossRef]
- Chandler, K.B.; Pompach, P.; Goldman, R.; Edwards, N. Exploring site-specific n-glycosylation microheterogeneity of haptoglobin using glycopeptide cid tandem mass spectra and glycan database search. J. Proteome Res. 2013, 12, 3652–3666. [Google Scholar] [CrossRef]
- Cheng, K.; Chen, R.; Seebun, D.; Ye, M.; Figeys, D.; Zou, H. Large-scale characterization of intact n-glycopeptides using an automated glycoproteomic method. J. Proteom. 2014, 110, 145–154. [Google Scholar] [CrossRef]
- Mayampurath, A.M.; Wu, Y.; Segu, Z.M.; Mechref, Y.; Tang, H. Improving confidence in detection and characterization of protein n-glycosylation sites and microheterogeneity. Rapid Commun. Mass Spectrom. 2011, 25, 2007–2019. [Google Scholar] [CrossRef]
- Hansen, J.E.; Lund, O.; Tolstrup, N.; Gooley, A.A.; Williams, K.L.; Brunak, S. Netoglyc: Prediction of mucin type o-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 1998, 15, 115–130. [Google Scholar] [CrossRef]
- Gupta, R.; Jung, E.; Gooley, A.A.; Williams, K.L.; Brunak, S.; Hansen, J. Scanning the available dictyostelium discoideum proteome for o-linked glcnac glycosylation sites using neural networks. Glycobiology 1999, 9, 1009–1022. [Google Scholar] [CrossRef]
- Julenius, K. Netcglyc 1.0: Prediction of mammalian c-mannosylation sites. Glycobiology 2007, 17, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Eisenhaber, B.; Bork, P.; Eisenhaber, F. Sequence properties of gpi-anchored proteins near the omega-site: Constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 1998, 11, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Fankhauser, N.; Maser, P. Identification of gpi anchor attachment signals by a kohonen self-organizing map. Bioinformatics 2005, 21, 1846–1852. [Google Scholar] [CrossRef]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. Predgpi: A gpi-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed]
- Poisson, G.; Chauve, C.; Chen, X.; Bergeron, A. Fraganchor: A large-scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring. Genom. Proteom. Bioinform. 2007, 5, 121–130. [Google Scholar] [CrossRef]
- Campbell, M.P.; Aoki-Kinoshita, K.F.; Lisacek, F.; York, W.S.; Packer, N.H. Glycoinformatics. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
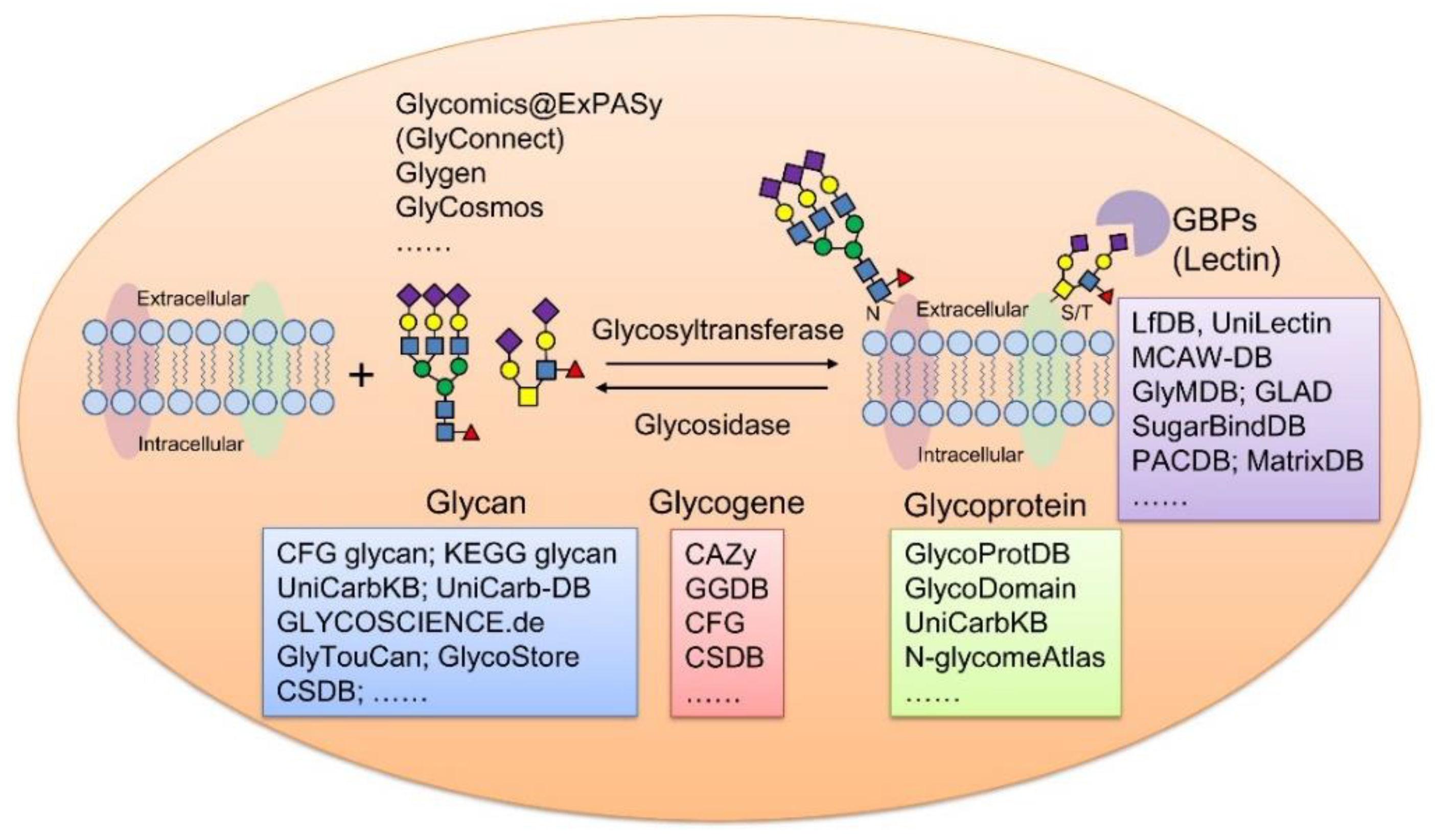
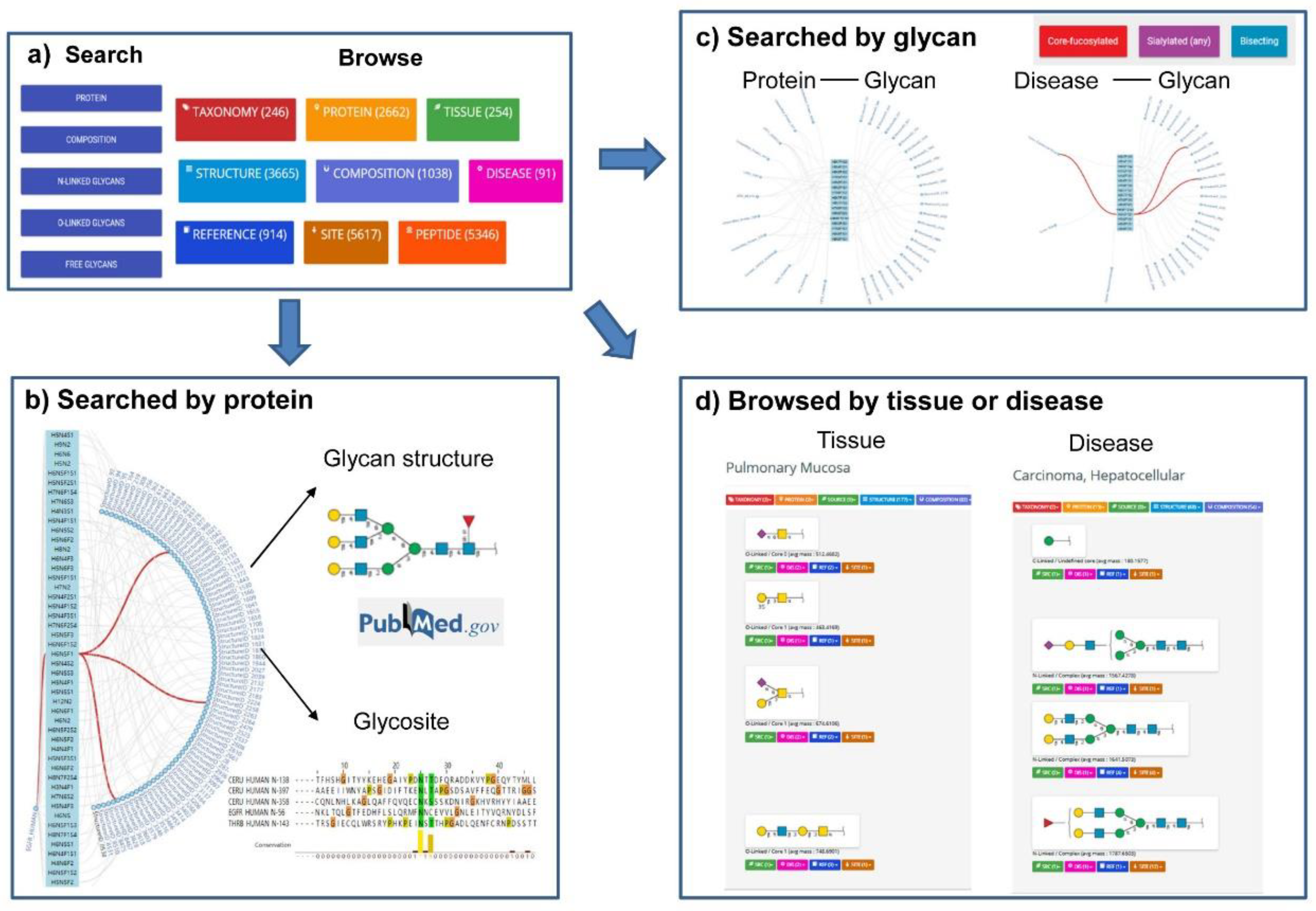
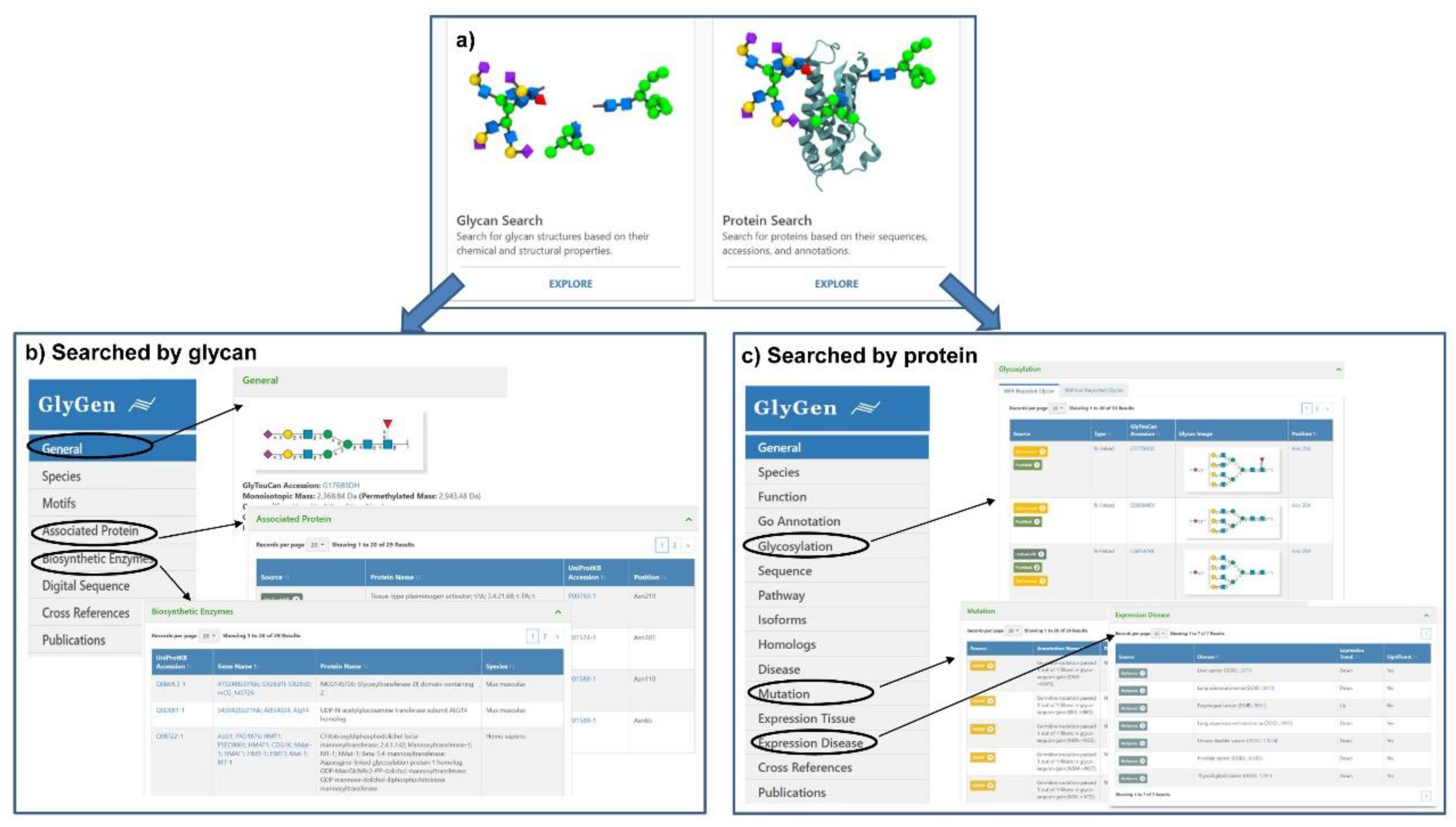

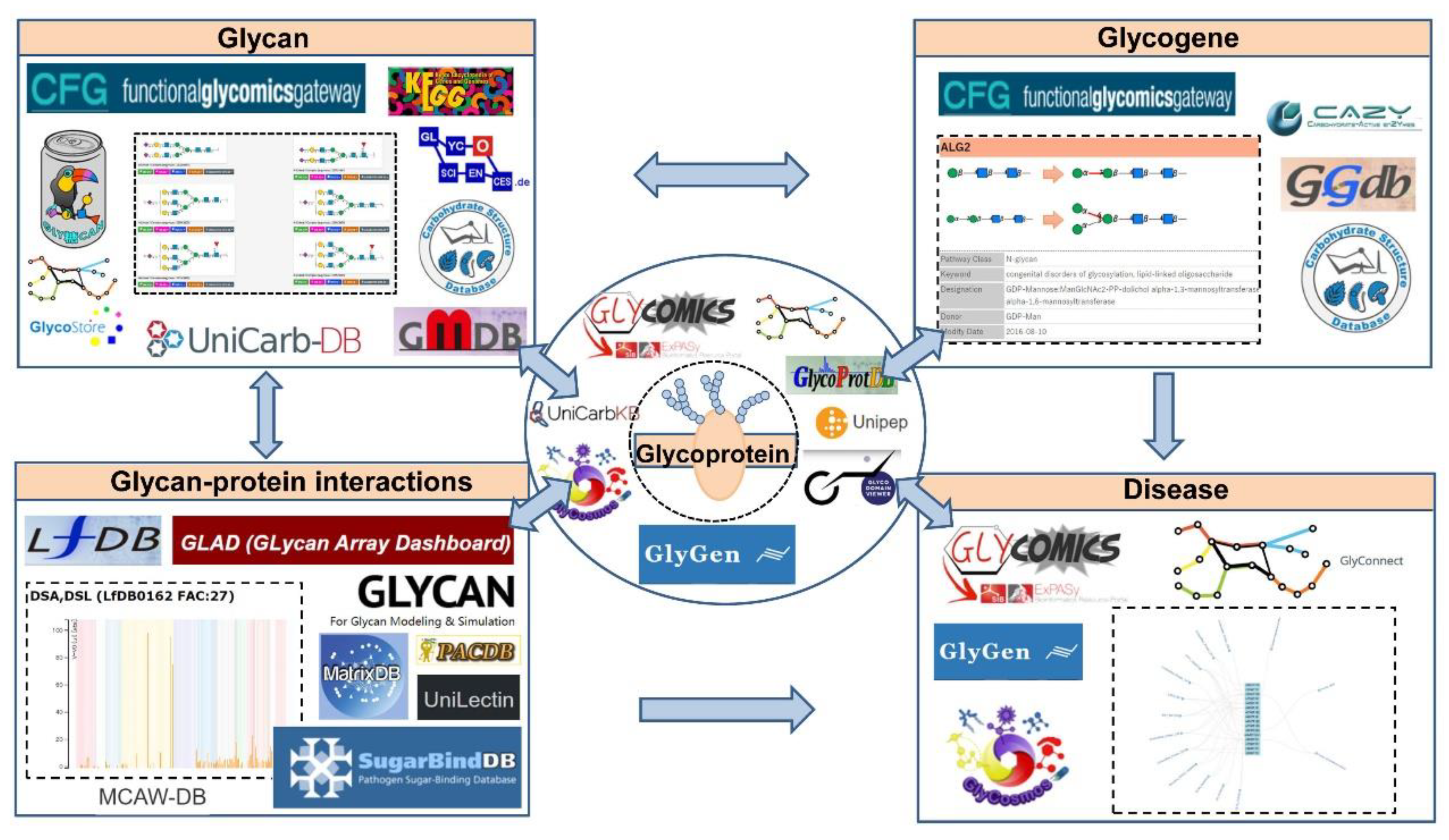
| Name | Description | Retrievable by | URL |
|---|---|---|---|
| Glycan structure databases | |||
| CFG glycan structure database [14] | Database providing structural and chemical information on thousands of glycans, including both synthetic glycans and glycans for mammalian species. | Searched by glycan names, composition, molecular weight, motifs, cell lines or tissue samples. | http://www.functionalglycomics.org/glycomics/molecule/jsp/carbohydrate/carbMoleculeHome.jsp |
| JCGGDB Glycan Mass spectral DataBase [16] | Database containing multi-stage tandem mass spectral of structurally defined N-and O-linked glycans, and glycolipid glycans. | Searched by glycan composition or m/z value of precursor ion. | https://jcggdb.jp/rcmg/glycodb/Ms_ResultSearch |
| UniCarbKB [25,26,27] | A curated database of information on glycan structures of glycoproteins, with descriptions of its biological source, supporting reference and experimental methods. | Searched by monosaccharide composition, attached protein, taxonomy or tissue by using an auto completion feature. | http://unicarbkb.org/ |
| KEGG glycan [13] | Database providing information on experimentally determined glycan structures and their metabolic pathways. | Searched by the G number for each glycan structure. | http://www.genome.jp/kegg/glycan/ |
| GLYCOSCIENCE.de [12,33] | An integrated portal containing databases and tools mainly glycan 3D structure analysis. | Searched by monosaccharide composition, molecular formula, structure classification and motifs, as well as NMR atoms or peaks. | http://www.glycosciences.de/ |
| Glycosciences.DB [35] | The main glycan structure database of GLYCOSCIENCE.de, providing published data on glycan structures, their taxonomy, MS and NMR-experimental data, 3D structure models as well as references to PDB entries. | Searched by glycan (sub-)structure, monosaccharide composition, molecular formula, structure classification and motifs, as well as NMR, MS, PDB query, or bibliography queries. | http://www.glycosciences.de/database/ |
| UniCarb-DB [36] | Database providing LC-MS/MS data of glycan structures. | Searched by taxonomy, tissue, reference, mass, composition or precursor mass. | https://unicarb-db.expasy.org/ |
| GlyTouCan [37] | A international glycan sequence repository with a globally unique accession number assigned to each structure. | Searched by text input, motif, or drawing glycan structures in GlycanBuilder. Registered users can additionally register new glycan structures to obtain unique IDs for each structure. | https://glytoucan.org/ |
| GlycoStore [41] | A curated database of information on glycan retention properties with chromatographic, electrophoretic and mass-spectrometry composition data. | Searched by experimental values (GU, AU or time), monosaccharide composition or metadata labels (taxonomy, sample name and the Oxford linear notation). | https://www.glycostore.org |
| CSDB [42] | Database on the structures of glycans and glycoconjugates in prokaryotes, plants and fungi. | Searched by CSDB ID, glycan substructure, composition, taxonomy, bibliography, NMR signals, conformation ID or GT name. | http://csdb.glycoscience.ru/database/ |
| Glycoprotein databases | |||
| GlycoProtDB [56] | Database providing information on N-glycoproteins and their glycosylated site(s) identified from C. elegans, mouse tissues and human. | Searched by gene ID, gene name, and its description (protein name). | https://acgg.asia/db/gpdb2/ |
| UniPep [59] | Database providing information on N-glycopeptides identified from human plasma and tissues including bladder, breast, liver, lymphocytes, cerebrospinal fluid, and prostate. | Searched by gene name, gene symbol, Swiss Prot ID, IPI ID, protein sequence or peptide mass. | http://www.unipep.org/ |
| N-GlycositeAtlas [22] | Database providing information on N-glycopeptides identified from over 100 publications and unpublished datasets | Searched by gene/protein name, accession number, glycosylation site location, glycosite containing peptide, tissue/liquid/cell line, or publication | http://nglycositeatlas.biomarkercenter.org |
| GlycoDomain Viewer [95] | Database of O-GalNAc proteinsidentified by SimpleCell technology from human and animal cell lines, associated with the verified and predicted glycosylated sites of N-glycan, O-GalNAc, O-Mannose and O-Xylose mapping on the protein sequence | Searched by the NCBI gene name or the Uniprot ID | https://glycodomain.glycomics.ku.dk/ |
| Glycogene databases | |||
| CAZy [62] | The largest database for display and analysis of genomic, structural and biochemical information on glyco-enzymes | Searched by enzyme family, protein name, organism name, GeneBank or UniProt accession, or EC number. | http://www.cazy.org/ |
| CAZypedia [63] | A comprehensive encyclopedic of detailed structural, and biochemical information on glyco-enzymes, and relevant reference. | Searched by enzyme name or enzyme ID | http://www.cazypedia.org |
| GlycoGene DataBase [15] | Database providing information of glycogenes on gene sequences, substrate specificities, homologous genes, EC numbers, tissue distribution, KO mouse as well as external links to various databases. | Searched by gene symbols or designations or selected from the list of glycogenes | https://acgg.asia/ggdb2/ |
| CFG glycosyltransferases database | Database providing information of glycosyltransferase on enzyme name, EC number, organism, relevant CFG data, and other data from public databases (PubMed, KEGG, CAZy, SwissProt, and others). | Providing a graphical interface of different glycans. By clicking a monosaccharide, users are directed to the information of the glycosyltransferase which forms this structure. | http://www.functionalglycomics.org/glycomics/molecule/jsp/glycoEnzyme/geMolecule.jsp |
| CSDB_GT [96] | A curated database of glycosyltransferases in Arabidopsis thaliana, Escherichia coli and Saccharomyces cerevisiae. | Searched by CSDB ID, glycan structure, composition, taxonomy, bibliography, NMR signals, conformation, or GT activity. | http://csdb.glycoscience.ru/gt.html |
| Glycan-protein interaction database | |||
| Lectin Frontier Database [67] | Database providing quantitative interaction data between various glycan and lectins, as well as basic information such as kingdom, monosaccharide specificity on lectins | Searched by keyword or choose categories among Lectin family, Monosaccharide Specificity, or 3D-fold. | http://acgg.asia/lfdb2/ |
| UniLectin [68] | A interactive database for the classification and curation of lectins (with UniLectin3D module), and the prediction of β-propeller lectins (with PropLec module). | Searched by keywords, kingdom order, historical classification, monosaccharide, associate IUPAC sequence, fold of the binding site, or multiple criteria | https://www.unilectin.eu/ |
| PACDB [69] | Database providing information on the interaction of microbial glycan-binding proteins and glycans with host glycan ligands | Selected from the list of disease, pathogen names, monosaccharides, or glycoepitopes | https://acgg.asia/db/diseases/pacdb |
| SugarBindDB [70] | A curated database providing information on known glycan structure interacted with pathogenic organisms (bacteria, toxins and viruses) in various disease | Searched by pathogenic agents, ligands, recognizing lectins, affected area, references, diseases or multi-criteria. | https://sugarbind.expasy.org/ |
| GLAD [71] | A web-based tool to visualize, analyze, present, and mine glycan array data. | Input data as tab-delimited text files in the correct format | https://glycotoolkit.com/Tools/GLAD/ |
| MCAW-DB [72] | Database providing information on binding affinity of glycan binding proteins to glycan substructures by multiple alignment analysis of glycan array data | Searched by filtering taxa, protein family, investigator and array version. | https://mcawdb.glycoinfo.org/ |
| GlyMDB [73] | Database enabling users to upload their own microarray data, query binder/non-binder classification, discover glycan-binding motif, compare glycan array sample, and cross-link microarray samples to PDB structures | Searched by protein name, protein sequence or PDB ID, or upload microarray spreadsheet file. | http://www.glycanstructure.org/ |
| MatrixDB [74] | A curated database providing information on interactions between extracellular matrix proteins, proteoglycans and polysaccharides. | Searched by a biomolecule, keyword, author, publication or IMEx identifier. | http://matrixdb.univ-lyon1.fr/ |
| Latest integrated glycoscience portal | |||
| Glycomics@ExPASy [19] | The glycomics tab of ExPASy, centralizing web-based glycoinformatics databases and tools resources developed by SIB (such as GlyConnect, SugarBind and UniCarb-DB databases) and other external resources to (such as CAZy, CSDB, GlyTouCan and UniCarbKB) to bridge the glycobiology and protein-oriented bioinformatics resources | Click on the link of interest | https://www.expasy.org/glycomics |
| GlyConnect [86] | The central platform of the Glycomics@ExPASy, providing interactive diagrams that help the user understand relations between glycans, proteins, tissues, diseases, and taxonomy | Either browsed or searched by protein name, ID, or monosaccharide composition, linkage type | https://glyconnect.expasy.org/ |
| Glygen [18] | A web portal data integration, harmonization and dissemination web portal for integrate data and knowledge from diverse disciplines relevant to glycobiology, carbohydrate and glycoconjugate-related data retrieved from multiple international data sources including UniProtKB, GlyTouCan, UniCarbKB and other key resources. | Searched by protein accession, sequences, glycan structure or monosaccharide composition. | https://glygen.org/ |
| GlyCosmos [20] | An integrated web resource including the database of JCGGDB and providing information on glycan-related genes, proteins, lipids, glycomes, pathways and diseases to integrate the glycosciences with the life sciences | Searched by protein name, protein accession, species, or various glycan search tools, such as by mass, composition, graphical glycan structure or monosaccharide composition. | https://glycosmos.org |
| Bioinformatic Tools | Glycan Identification Method | Peptide Identification Method |
|---|---|---|
| GlycoWorkbench [78] | MS, MS/MS | MS |
| GlycReSoft [50,79] | LC-MS | LC-MS/MS |
| Byonic [97] | match by glycan mass | ETD, HCD |
| Protein Prospector [98] | match by glycan mass | ETD |
| pGlyco [82,83] | CID | HCD |
| GlycoNovoDB [85] | HCD | HCD |
| GPQuest [80] | HCD | HCD |
| GlycoPeptide Finder (GPFinder) [4] | QTOF-CID | QTOF-CID |
| MAGIC [99] | QTOF-CID | QTOF-CID |
| GlycoMaster DB [100] | HCD | ETD |
| GlycoFinder [101] | low energy HCD | HCD |
| Sweet-Heart [102] | CID | MS3 |
| Sweet-Heart for HCD [103] | CID | HCD |
| GlycoFragWork [104] | CID | ETD |
| pMatchGlyco [105] | match by MS/MS spectra | HCD |
| GlycoPeptideSearch [106,107] | CID | ETD |
| ArMone [108] | CID | HCD |
| GlypID 2.0 [109] | CID | HCD |
| O-O-Search [52] | HCD | HCD |
| AOGP [84] | HCD | HCD |
| Bioinformatic Tools | Description | URL |
|---|---|---|
| NetNGlyc | N-glycosylation site prediction | http://www.cbs.dtu.dk/services/NetNGlyc/ |
| NetOGlyc [110] | O-GalNAc site prediction | http://www.cbs.dtu.dk/services/NetOGlyc/ |
| YinOYang [60] | O-(beta)-GlcNAc and phosphorylation site prediction | http://www.cbs.dtu.dk/services/YinOYang/ |
| DictyOGlyc [111] | O-(alpha)-GlcNAc site prediction | http://www.cbs.dtu.dk/services/DictyOGlyc/ |
| NetCGlyc [112] | C-mannose site prediction | http://www.cbs.dtu.dk/services/NetCGlyc/ |
| Big-PI Predictor [113] | GPI-anchor prediction | http://mendel.imp.ac.at/sat/gpi/gpi_server.html |
| GPI-SOM [114] | GPI-anchor prediction | http://genomics.unibe.ch/cgi-bin/gpi.cgi |
| PredGPI [115] | GPI-anchor prediction | http://gpcr.biocomp.unibo.it/predgpi/pred.htm |
| FragAnchor [116] | GPI-anchor prediction | http://navet.ics.hawaii.edu/~fraganchor/NNHMM/NNHMM.html. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Xu, Z.; Hong, X.; Zhang, Y.; Zou, X. Databases and Bioinformatic Tools for Glycobiology and Glycoproteomics. Int. J. Mol. Sci. 2020, 21, 6727. https://doi.org/10.3390/ijms21186727
Li X, Xu Z, Hong X, Zhang Y, Zou X. Databases and Bioinformatic Tools for Glycobiology and Glycoproteomics. International Journal of Molecular Sciences. 2020; 21(18):6727. https://doi.org/10.3390/ijms21186727
Chicago/Turabian StyleLi, Xing, Zhijue Xu, Xiaokun Hong, Yan Zhang, and Xia Zou. 2020. "Databases and Bioinformatic Tools for Glycobiology and Glycoproteomics" International Journal of Molecular Sciences 21, no. 18: 6727. https://doi.org/10.3390/ijms21186727
APA StyleLi, X., Xu, Z., Hong, X., Zhang, Y., & Zou, X. (2020). Databases and Bioinformatic Tools for Glycobiology and Glycoproteomics. International Journal of Molecular Sciences, 21(18), 6727. https://doi.org/10.3390/ijms21186727





