Pre-Operative Evaluation of DNA Methylation Profile in Oral Squamous Cell Carcinoma Can Predict Tumor Aggressive Potential
Abstract
1. Introduction
2. Results
2.1. Pathological Findings and Tumor Stage
2.2. Methylation Profile and Clinical-Pathological Characteristics
2.3. Methylation Profile and Adverse Event (AE)
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Study Setting and Data Collection
4.3. Treatment Modality
4.4. DNA Methylation Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AE | Adverse Event |
| NGS | Next Generation Sequencing |
| OSCC | Oral Squamous Cell Carcinoma |
| PNI | Perineural invasion |
| POI | Pattern of infiltration |
| WPOI | Worst pattern of infiltration |
| DOI | Depth of Invasion |
| HR | Hazard Ratio |
References
- Rivera, C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11884–11894. [Google Scholar] [PubMed]
- Amin, R.W.; Rivera, B. A spatial study of oral & pharynx cancer mortality and incidence in the U.S.A.: 2000–2015. Sci. Total Environ. 2020, 713, 136688. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Eden, A.; Gaudet, F.; Waghmare, A.; Jaenisch, R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003, 300, 455. [Google Scholar] [CrossRef] [PubMed]
- Van Tongelen, A.; Loriot, A.; De Smet, C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett. 2017, 396, 130–137. [Google Scholar] [CrossRef]
- Mascolo, M.; Siano, M.; Ilardi, G.; Russo, D.; Merolla, F.; De Rosa, G.; Staibano, S. Epigenetic disregulation in oral cancer. Int. J. Mol. Sci. 2012, 13, 2331–2353. [Google Scholar] [CrossRef]
- Demokan, S.; Chang, X.; Chuang, A.; Mydlarz, W.K.; Kaur, J.; Huang, P.; Khan, Z.; Khan, T.; Ostrow, K.L.; Brait, M.; et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int. J. Cancer 2010, 127, 2351–2359. [Google Scholar] [CrossRef]
- Langevin, S.M.; Stone, R.A.; Bunker, C.H.; Grandis, J.R.; Sobol, R.W.; Taioli, E. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis 2010, 31, 864–870. [Google Scholar] [CrossRef]
- Pattani, K.M.; Zhang, Z.; Demokan, S.; Glazer, C.; Loyo, M.; Goodman, S.; Sidransky, D.; Bermudez, F.; Jean-Charles, G.; McCaffrey, T.; et al. Endothelin receptor type B gene promoter hypermethylation in salivary rinses is independently associated with risk of oral cavity cancer and premalignancy. Cancer Prev. Res. 2010, 3, 1093–1103. [Google Scholar] [CrossRef]
- Schussel, J.; Zhou, X.C.; Zhang, Z.; Pattani, K.; Bermudez, F.; Jean-Charles, G.; McCaffrey, T.; Padhya, T.; Phelan, J.; Spivakovsky, S.; et al. EDNRB and DCC salivary rinse hypermethylation has a similar performance as expert clinical examination in discrimination of oral cancer/dysplasia versus benign lesions. Clin. Cancer Res. 2013, 19, 3268–3275. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Hamada, T.; Yamada, N.; Yokoyama, S.; Kitamoto, S.; Kanmura, Y.; Nomura, M.; Kamikawa, Y.; Yonezawa, S.; Sugihara, K. Aberrant DNA methylation of tumor-related genes in oral rinse: A noninvasive method for detection of oral squamous cell carcinoma. Cancer 2012, 118, 4298–4308. [Google Scholar] [CrossRef]
- Sailer, V.; Holmes, E.E.; Gevensleben, H.; Goltz, D.; Dröge, F.; de Vos, L.; Franzen, A.; Schröck, F.; Bootz, F.; Kristiansen, G.; et al. PITX2 and PANCR DNA methylation predicts overall survival in patients with head and neck squamous cell carcinoma. Oncotarget 2016, 7, 75827–75838. [Google Scholar] [CrossRef] [PubMed]
- Morandi, L.; Gissi, D.; Tarsitano, A.; Asioli, S.; Monti, V.; Del Corso, G.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P. DNA methylation analysis by bisulfite next-generation sequencing for early detection of oral squamous cell carcinoma and high-grade squamous intraepithelial lesion from oral brushing. J. Cranio-Maxillofac. Surg. 2015, 43, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- Morandi, L.; Gissi, D.; Tarsitano, A.; Asioli, S.; Gabusi, A.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P. CpG location and methylation level are crucial factors for the early detection of oral squamous cell carcinoma in brushing samples using bisulfite sequencing of a 13-gene panel. Clin. Epigenetics 2017, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Gissi, D.B.; Gabusi, A.; Tarsitano, A.; Asioli, S.; Rossi, R.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P.; Morandi, L. Application of a non-invasive oral brushing procedure based on bisulfite sequencing of a 13-gene panel to study high-risk OSCC patients. Cancer Biomark. 2020. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Preston, R.; Michailidi, C.; Marchionni, L.; Pickering, C.R.; Frederick, M.J.; Myers, J.N.; Yegnasubramanian, S.; Hadar, T.; Noordhuis, M.G.; Zizkova, V.; et al. Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics 2014, 9, 1031–1046. [Google Scholar] [CrossRef]
- Marsit, C.J.; Christensen, B.C.; Houseman, E.A.; Karagas, M.R.; Wrensch, M.R.; Yeh, R.-F.; Nelson, H.H.; Wiemels, J.L.; Zheng, S.; Posner, M.R.; et al. Epigenetic profiling reveals etiologically distinct patterns of DNA methylation in head and neck squamous cell carcinoma. Carcinogenesis 2009, 30, 416–422. [Google Scholar] [CrossRef]
- Kozaki, K.; Imoto, I.; Mogi, S.; Omura, K.; Inazawa, J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008, 68, 2094–2105. [Google Scholar] [CrossRef]
- Li, Y.-F.; Hsiao, Y.-H.; Lai, Y.-H.; Chen, Y.-C.; Chen, Y.-J.; Chou, J.-L.; Chan, M.W.Y.; Lin, Y.-H.; Tsou, Y.-A.; Tsai, M.-H.; et al. DNA methylation profiles and biomarkers of oral squamous cell carcinoma. Epigenetics 2015, 10, 229–236. [Google Scholar] [CrossRef]
- Cheriyan, V.T.; Thomas, C.; Balaram, P. Augmentation of T-cell immune responses and signal transduction proteins in oral cancer patients: Potential for IL-2-mediated immunotherapy. J. Cancer Res. Clin. Oncol. 2011, 137, 1435–1444. [Google Scholar] [CrossRef] [PubMed]
- Shintani, S.; Hamakawa, H.; Nakashiro, K.; Shirota, T.; Hatori, M.; Tanaka, M.; Kuroshita, Y.; Kurokawa, Y. Friend leukaemia insertion (Fli)-1 is a prediction marker candidate for radiotherapy resistant oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2010, 39, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, M.; Fujihara, H.; Fujimori, H.; Kawaguchi, K.; Yamada, H.; Nakayama, R.; Yamamoto, N.; Kishi, Y.; Hamada, Y.; Masutani, M. Synergetic Effects of PARP Inhibitor AZD2281 and Cisplatin in Oral Squamous Cell Carcinoma in Vitro and in Vivo. Int. J. Mol. Sci. 2016, 17, 272. [Google Scholar] [CrossRef] [PubMed]
- Severino, P.; Oliveira, L.S.; Andreghetto, F.M.; Torres, N.; Curioni, O.; Cury, P.M.; Toporcov, T.N.; Paschoal, A.R.; Durham, A.M. Small RNAs in metastatic and non-metastatic oral squamous cell carcinoma. BMC Med. Genom. 2015, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Ghantous, Y.; Bahouth, Z.; Abu El-Naaj, I. Clinical and genetic signatures of local recurrence in oral squamous cell carcinoma. Arch. Oral Biol. 2018, 95, 141–148. [Google Scholar] [CrossRef]
- Boscolo-Rizzo, P.; Rampazzo, E.; Perissinotto, E.; Piano, M.A.; Giunco, S.; Baboci, L.; Spinato, G.; Spinato, R.; Tirelli, G.; Da Mosto, M.C.; et al. Telomere shortening in mucosa surrounding the tumor: Biosensor of field cancerization and prognostic marker of mucosal failure in head and neck squamous cell carcinoma. Oral Oncol. 2015, 51, 500–507. [Google Scholar] [CrossRef]
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. (Eds.) AJCC Cancer Staging Manual, 8th ed.; Springer International Publishing: Chicago, IL, USA, 2017; ISBN 978-3-319-40617-6. [Google Scholar]
- Chang, Y.-C.; Nieh, S.; Chen, S.-F.; Jao, S.-W.; Lin, Y.-L.; Fu, E. Invasive pattern grading score designed as an independent prognostic indicator in oral squamous cell carcinoma. Histopathology 2010, 57, 295–303. [Google Scholar] [CrossRef]
- Carsuzaa, F.; Thariat, J.; Gorphe, P.; Righini, C.; Cosmidis, A.; Thureau, S.; Roge, M.; De Mones, E.; Servagi-Vernat, S.; Tonnerre, D.; et al. Surgery or Radiotherapy of the Primary Tumor in T1-2 Head and Neck Squamous Cell Carcinoma with Resectable N3 Nodes: A Multicenter GETTEC Study. Ann. Surg. Oncol. 2019, 26, 3673–3680. [Google Scholar] [CrossRef]
- Herman, J.G.; Baylin, S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003, 349, 2042–2054. [Google Scholar] [CrossRef]
- Bock, C.; Reither, S.; Mikeska, T.; Paulsen, M.; Walter, J.; Lengauer, T. BiQ Analyzer: Visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 2005, 21, 4067–4068. [Google Scholar] [CrossRef]
- Mercer, T.R.; Qureshi, I.A.; Gokhan, S.; Dinger, M.E.; Li, G.; Mattick, J.S.; Mehler, M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Shan, K.; Jiang, Q.; Wang, X.-Q.; Wang, Y.-N.-Z.; Yang, H.; Yao, M.-D.; Liu, C.; Li, X.-M.; Yao, J.; Liu, B.; et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016, 7, e2248. [Google Scholar] [CrossRef]
- Bell, A.; Bell, D.; Weber, R.S.; El-Naggar, A.K. CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer 2011, 117, 2898–2909. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Dang, S.; Hou, P. Gene methylation in gastric cancer. Clin. Chim. Acta 2013, 424, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.; Pan, K.; Linnekamp, J.F.; Medema, J.P.; Kandimalla, R. DNA methylation based biomarkers in colorectal cancer: A systematic review. Biochim. Biophys. Acta 2016, 1866, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Wang, G.; Shi, Q.; Zhang, R.; Zhao, Y.; Wei, Y.; Chen, F.; Christiani, D.C. Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma. Clin. Epigenet. 2017, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Tian, G.; Gao, J. Construction of prognostic risk prediction model of oral squamous cell carcinoma based on co-methylated genes. Int. J. Mol. Med. 2019, 44, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Song, Y.; Cheng, L.; Xu, H.; Liu, J. Analysis of methylation-driven genes for predicting the prognosis of patients with head and neck squamous cell carcinoma. J. Cell. Biochem. 2019, 120, 19482–19495. [Google Scholar] [CrossRef]
- The International Agency for Research on Cancer; Eveson, J.W.; Reichart, P.; Sidransky, D.; World Health Organization Classification of Tumours. Pathology and Genetics of Head and Neck Tumours, 1st ed.; IARC Press: Lyon, France, 2017. [Google Scholar]
- Kademani, D.; Bell, R.B.; Bagheri, S.; Holmgren, E.; Dierks, E.; Potter, B.; Homer, L. Prognostic factors in intraoral squamous cell carcinoma: The influence of histologic grade. J. Oral Maxillofac. Surg. 2005, 63, 1599–1605. [Google Scholar] [CrossRef]
- Adelstein, D.; Gillison, M.L.; Pfister, D.G.; Spencer, S.; Adkins, D.; Brizel, D.M.; Burtness, B.; Busse, P.M.; Caudell, J.J.; Cmelak, A.J.; et al. NCCN Guidelines Insights: Head and Neck Cancers, Version 2.2017. J. Natl. Compr. Cancer Netw. 2017, 15, 761–770. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Ting, A.H.; Li, J. BSPAT: A fast online tool for DNA methylation co-occurrence pattern analysis based on high-throughput bisulfite sequencing data. BMC Bioinform. 2015, 16, 220. [Google Scholar] [CrossRef] [PubMed]
- Krainer, J.; Weinhäusel, A.; Hanak, K.; Pulverer, W.; Özen, S.; Vierlinger, K.; Pabinger, S. EPIC-TABSAT: Analysis tool for targeted bisulfite sequencing experiments and array-based methylation studies. Nucleic Acids Res. 2019, 47, W166–W170. [Google Scholar] [CrossRef] [PubMed]
- Gruntman, E.; Qi, Y.; Slotkin, R.K.; Roeder, T.; Martienssen, R.A.; Sachidanandam, R. Kismeth: Analyzer of plant methylation states through bisulfite sequencing. BMC Bioinform. 2008, 9, 371. [Google Scholar] [CrossRef] [PubMed]
- Mallona, I.; Díez-Villanueva, A.; Peinado, M.A. Methylation plotter: A web tool for dynamic visualization of DNA methylation data. Source Code Biol. Med. 2014, 9, 11. [Google Scholar] [CrossRef]
- Tutz, G.; Binder, H. Generalized additive modeling with implicit variable selection by likelihood-based boosting. Biometrics 2006, 62, 961–971. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.; Schumacher, M. Allowing for mandatory covariates in boosting estimation of sparse high-dimensional survival models. BMC Bioinform. 2008, 9, 14. [Google Scholar] [CrossRef]
- Mayr, A.; Binder, H.; Gefeller, O.; Schmid, M. The evolution of boosting algorithms. From machine learning to statistical modelling. Methods Inf. Med. 2014, 53, 419–427. [Google Scholar] [CrossRef]
- Sauerbrei, W.; Perperoglou, A.; Schmid, M.; Abrahamowicz, M.; Becher, H.; Binder, H.; Dunkler, D.; Harrell, F.E.; Royston, P.; Heinze, G.; et al. State of the art in selection of variables and functional forms in multivariable analysis-outstanding issues. Diagn. Progn. Res. 2020, 4, 3. [Google Scholar] [CrossRef]
- De Bin, R. Boosting in Cox regression: A Comparison between the Likelihood-Based and the Model-Based Approaches with Focus on the R-Packages CoxBoost and Mboost. Available online: https://epub.ub.uni-muenchen.de/24466/ (accessed on 3 July 2020).
- Seibold, H.; Bernau, C.; Boulesteix, A.-L.; De Bin, R. On the choice and influence of the number of boosting steps for high-dimensional linear Cox-models. Comput. Stat. 2018. [Google Scholar] [CrossRef]
- Binder, H.; Porzelius, C.; Schumacher, M. Rank-Based p-Values for Sparse High-Dimensional Risk Prediction Models Fitted by Componentwise Boosting; FDM-Preprint Nr. 101; University of Freiburg: Breisgau, Germany, 2009. [Google Scholar]
- Binder, H.; Schumacher, M. Incorporating pathway information into boosting estimation of high-dimensional risk prediction models. BMC Bioinform. 2009, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Gerds, T.A.; Schumacher, M. Consistent estimation of the expected Brier score in general survival models with right-censored event times. Biom. J. 2006, 48, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology 2010, 21, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Uno, H.; Cai, T.; Pencina, M.J.; D’Agostino, R.B.; Wei, L.J. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med. 2011, 30, 1105–1117. [Google Scholar] [CrossRef]
- Uno, H.; Cai, T.; Tian, L.; Wei, L.J. Evaluating Prediction Rules for t-Year Survivors with Censored Regression Models. J. Am. Stat. Assoc. 2007, 102, 527–537. [Google Scholar] [CrossRef]
- Blanche, P.; Kattan, M.W.; Gerds, T.A. The c-index is not proper for the evaluation of $t$-year predicted risks. Biostatistics 2019, 20, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zhang, X.; Huang, C.-C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 3 July 2020).
- Binder, H. CoxBoost: Cox Models by Likelihood Based Boosting for a Single Survival Endpoint or Competing Risks. 2013. Available online: https://CRAN.R-project.org/package=CoxBoost (accessed on 3 July 2020).


| Clinical-Pathological Variables | ||||
|---|---|---|---|---|
| Patients | Relapses Observed | p-Value | ||
| Sex | Male | 17 (47%) | 6 (35%) | 0.82 |
| Female | 19 (53%) | 6 (31%) | ||
| Age | <65 | 15 (42%) | 8 (53%) | 0.35 |
| >65 | 21 (58%) | 4 (19%) | ||
| Smoke | Yes | 7 (19%) | 3 (43%) | 0.26 |
| No | 29 (81%) | 9 (31%) | ||
| Site | Tongue and floor of mouth | 13 (36%) | 4 (31%) | 0.96 |
| Buccal and labial mucosa | 7 (19%) | 3 (43%) | ||
| Gingiva, Hard Palate, Retromolar region | 16 (45%) | 5 (31%) | ||
| T stage | T1-T2 | 27 (75%) | 8 (29%) | 0.33 |
| T3-T4 | 9 (25%) | 4 (44%) | ||
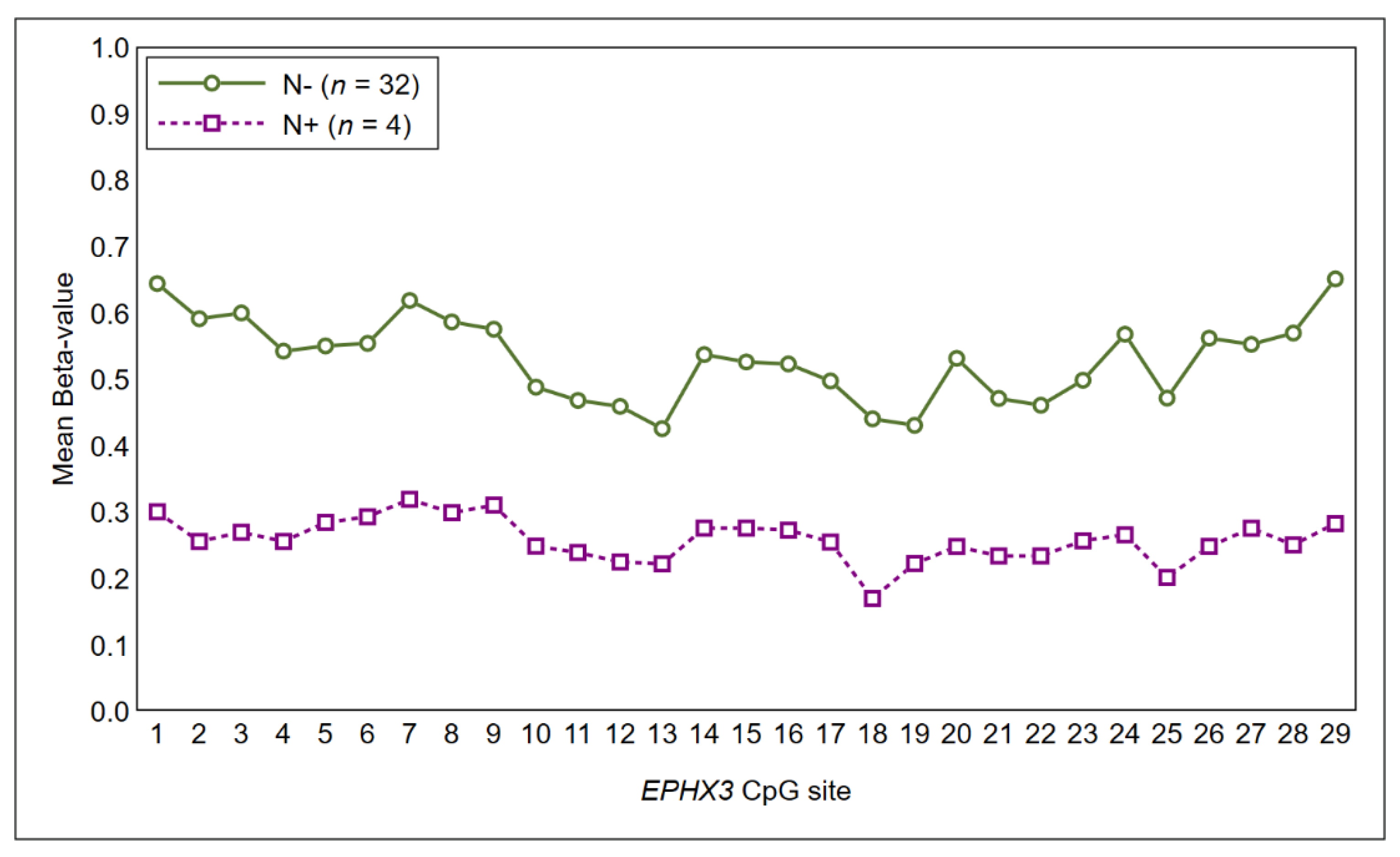
| N stage | N− | 32 (89%) | 9 (28%) | 0.06 |
| N+ | 4 (11%) | 3 (75%) | ||
| Grading | G1 | 20 (56%) | 5 (25%) | 0.18 |
| G2 | 14 (39%) | 6 (43%) | ||
| G3 | 2 (5%) | 1(50%) | ||
| Surgical margins | Free | 29 (81%) | 9 (31%) | 0.83 |
| Close | 4 (11%) | 1 (25%) | ||
| Displasia | 3 (8%) | 2 (66%) | ||
| Involved | 0 (0%) | |||
| Presence of associated OPMD | None | 26 (72%) | 11 (42%) | 0.07 |
| Lichen | 6 (17%) | 0 (0%) | ||
| Leucoplakia | 4 (11%) | 1 (25%) | ||
| Depth of invasion (DOI) | <4 mm | 21 (58%) | 5 (24%) | 0.07 |
| >4 mm | 15 (42%) | 7 (47%) | ||
| Pattern of invasion | P1-P2 | 22 (61%) | 4 (18%) | 0.01 * |
| P3-P4 | 14 (39%) | 8 (57%) | ||
| Radiotherapy | Yes | 8 (22%) | 4 (50%) | 0.2 |
| No | 28 (78%) | 8 (29%) | ||
| CpG Site | Ln HR | HR | p-Value |
|---|---|---|---|
| EPHX3-24 (Chr19:15232040) | −0.0234 | 0.9769 | 0.0157 |
| EPHX3-26 (Chr19:15232034) | −0.0226 | 0.9777 | 0.0172 |
| ITGA4-3 (Chr2:181458175) | 0.0163 | 1.0165 | 0.0078 |
| ITGA4-4 (Chr2:181458181) | 0.0306 | 1.0310 | 0.0027 |
| MiR193-3 (Chr17:31559856) | 0.0089 | 1.0090 | 0.0099 |
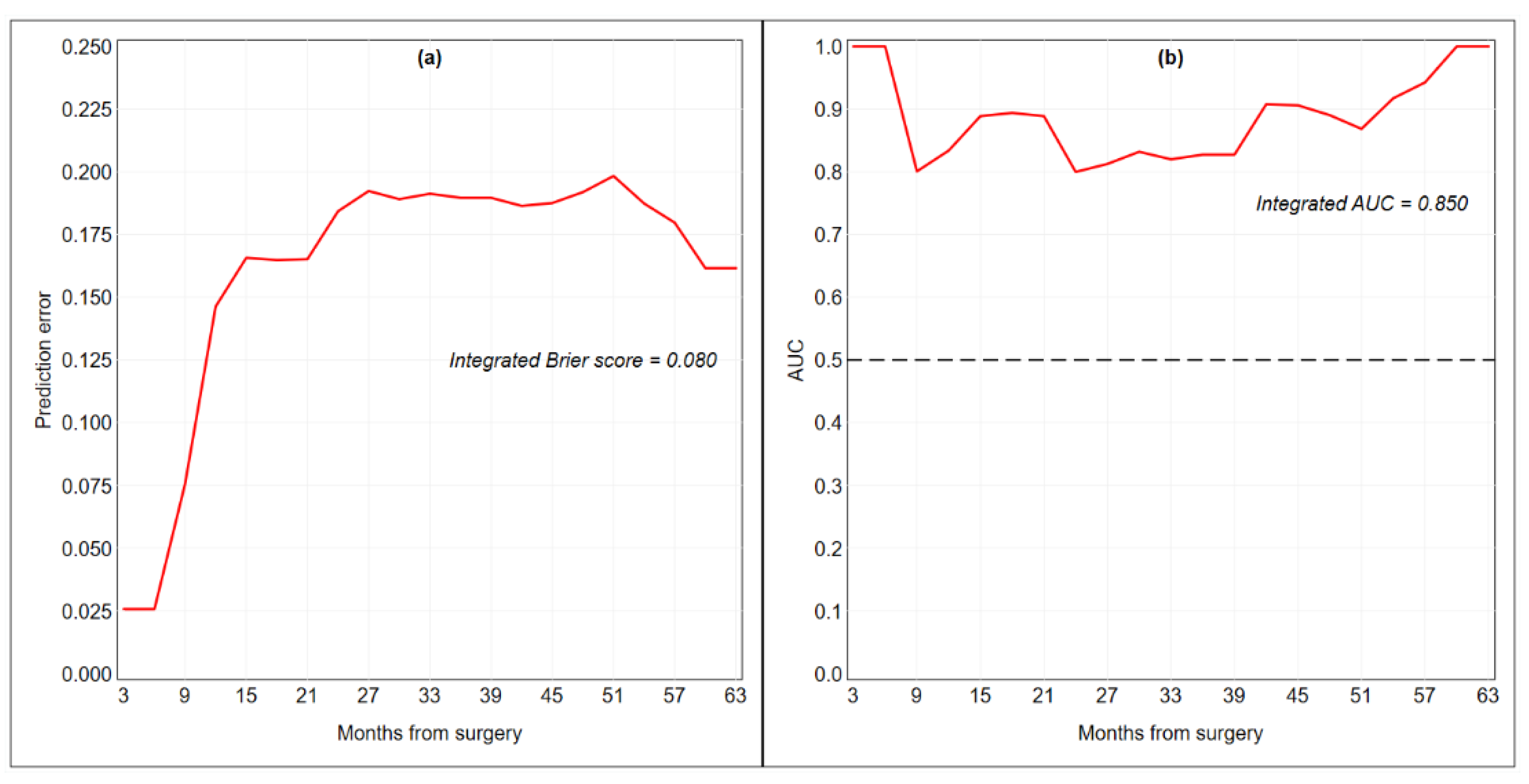
| Integrated Brier score | 0.080 | ||
| C-index | 0.802 | ||
| Integrated AUC | 0.850 | ||
| CpG Site | Ln HR | HR | p-Value |
|---|---|---|---|
| ITGA4-3(Chr2:181458175) | 0.0616 | 1.0636 | 0.0001 |
| ITGA4-4(Chr2:181458181) | 0.0467 | 1.0478 | 0.0003 |
| ITGA4-7(Chr2: 181458229) | 0.0176 | 1.0177 | 0.0046 |
| ITGA4-12(Chr2: 181458289) | 0.0152 | 1.0153 | 0.0052 |
| Integrated Brier score | 0.059 | ||
| C-index | 0.892 | ||
| Integrated AUC | 0.903 | ||
| Gene | Map | Position | Amplicon Length | Position Respect to TSS | Number of Interrogated CpG | hg38 Coordinates |
|---|---|---|---|---|---|---|
| ZAP70 | 2q11.2 | Exon 3 | 180 | 10728 | 20 | Chr2: 97724265-97724445 |
| GP1BB | 22q11.21 | Exon 1 | 192 | 363 | 18 | Chr22: 19723282-19723460 |
| KIF1A | 2q37.3 | Exon 1 | 189 | −1 | 27 | Chr2: 240820168-240820310 |
| PARP15 | 3q21.1 | Exon 1 | 206 | 93 | 19 | Chr3:122577695-122577901 |
| ITGA4 | 2q31.3 | Exon 2 | 214 | 912 | 14 | Chr2:181457647-181457879 |
| NTM | 11q25 | Exon 1 | 190 | 62 | 15 | Chr11:131911126-131911314 |
| MIR193A | 17q11.2 | Promoter | 256 | −178 | 26 | Chr17:31559818-31560073 |
| EPHX3 | 19p13.12 | Exon 1 | 223 | 215 | 29 | Chr19:15231995-15232217 |
| LINC00599 | 8p23.1 | Exon 1 | 199 | 69 | 20 | Chr8:9903205-9903403 |
| FLI1 | 11q24.3 | Exon 1 | 186 | 187 | 12 | Chr11:128694103-128694288 |
| MIR296 | 20q13.32 | Exon 1 | 238 | 180 | 15 | Chr20:58817149-58817363 |
| LRRTM1 | 2p12 | Promoter | 179 | −431 | 24 | Chr2:80304527-80304705 |
| TERT | 5p15.33 | Intron4-5 | 109 | 14976 | 6 | Chr5:1279604-1279759 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gissi, D.B.; Fabbri, V.P.; Gabusi, A.; Lenzi, J.; Morandi, L.; Melotti, S.; Asioli, S.; Tarsitano, A.; Balbi, T.; Marchetti, C.; et al. Pre-Operative Evaluation of DNA Methylation Profile in Oral Squamous Cell Carcinoma Can Predict Tumor Aggressive Potential. Int. J. Mol. Sci. 2020, 21, 6691. https://doi.org/10.3390/ijms21186691
Gissi DB, Fabbri VP, Gabusi A, Lenzi J, Morandi L, Melotti S, Asioli S, Tarsitano A, Balbi T, Marchetti C, et al. Pre-Operative Evaluation of DNA Methylation Profile in Oral Squamous Cell Carcinoma Can Predict Tumor Aggressive Potential. International Journal of Molecular Sciences. 2020; 21(18):6691. https://doi.org/10.3390/ijms21186691
Chicago/Turabian StyleGissi, Davide B., Viscardo P. Fabbri, Andrea Gabusi, Jacopo Lenzi, Luca Morandi, Sofia Melotti, Sofia Asioli, Achille Tarsitano, Tiziana Balbi, Claudio Marchetti, and et al. 2020. "Pre-Operative Evaluation of DNA Methylation Profile in Oral Squamous Cell Carcinoma Can Predict Tumor Aggressive Potential" International Journal of Molecular Sciences 21, no. 18: 6691. https://doi.org/10.3390/ijms21186691
APA StyleGissi, D. B., Fabbri, V. P., Gabusi, A., Lenzi, J., Morandi, L., Melotti, S., Asioli, S., Tarsitano, A., Balbi, T., Marchetti, C., & Montebugnoli, L. (2020). Pre-Operative Evaluation of DNA Methylation Profile in Oral Squamous Cell Carcinoma Can Predict Tumor Aggressive Potential. International Journal of Molecular Sciences, 21(18), 6691. https://doi.org/10.3390/ijms21186691







