The New Paradigm of Network Medicine to Analyze Breast Cancer Phenotypes
Abstract
1. Introduction
2. Results
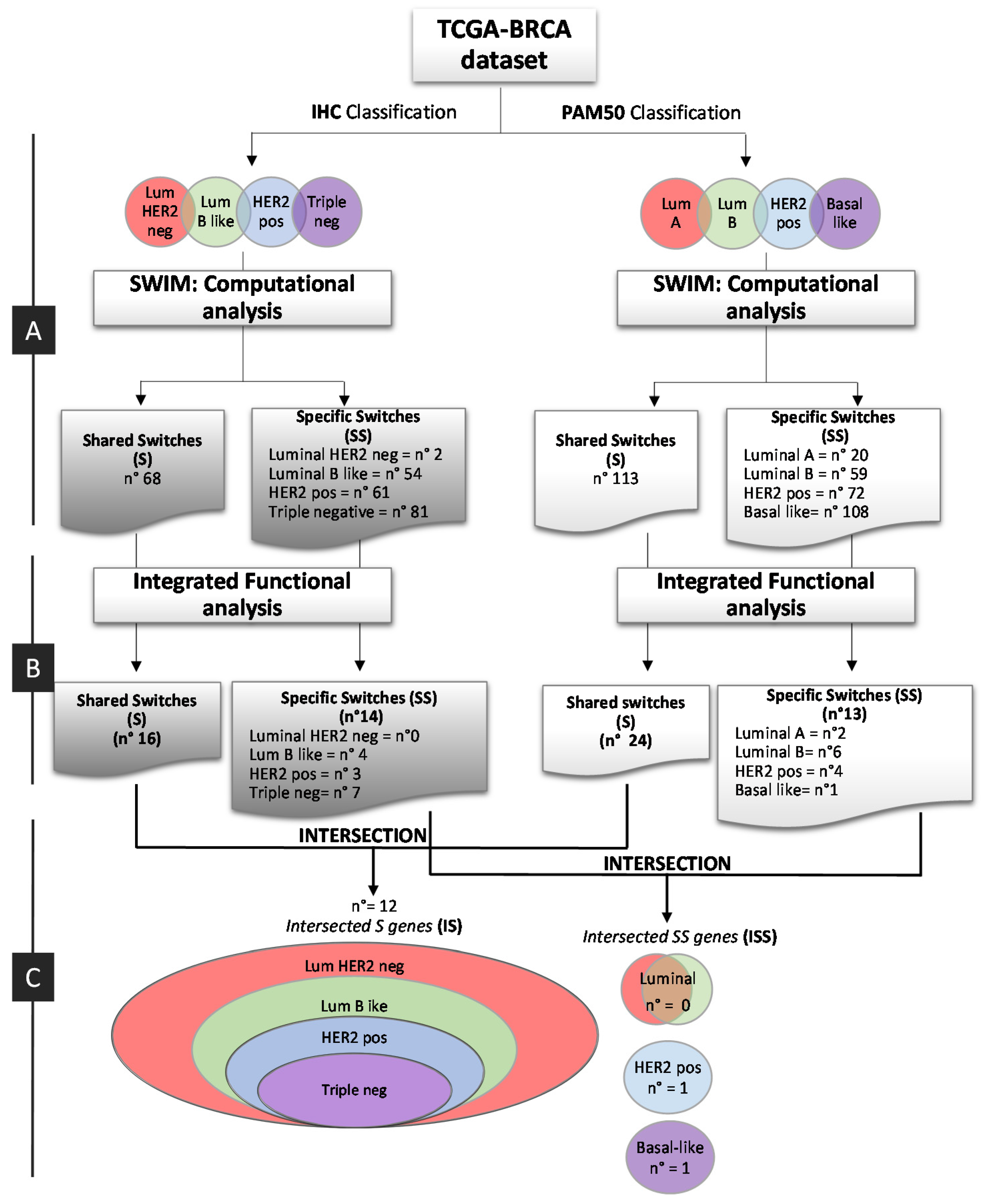
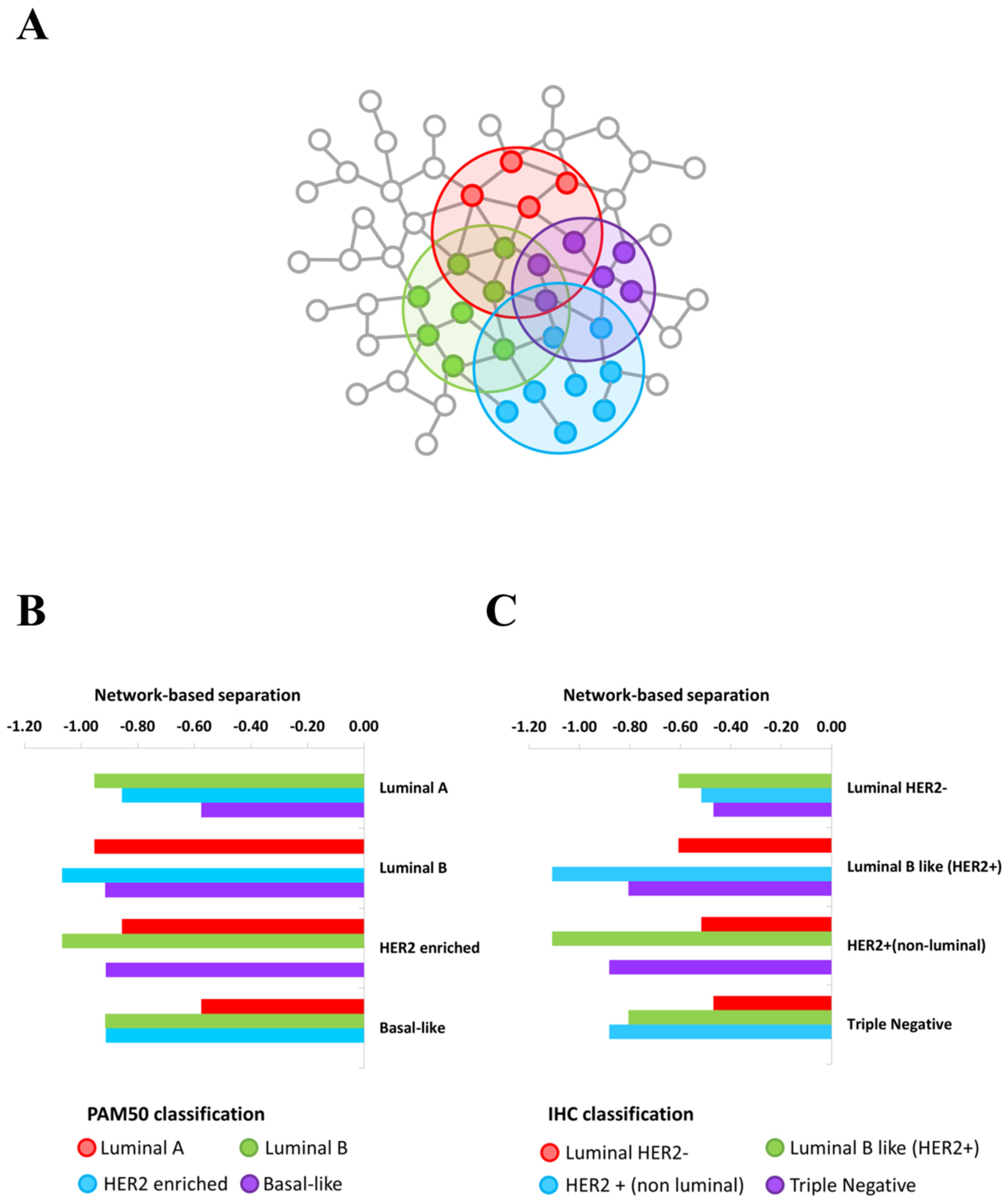
2.1. Overlapping BC Subtype PPI-Based Modules by Switch Genes
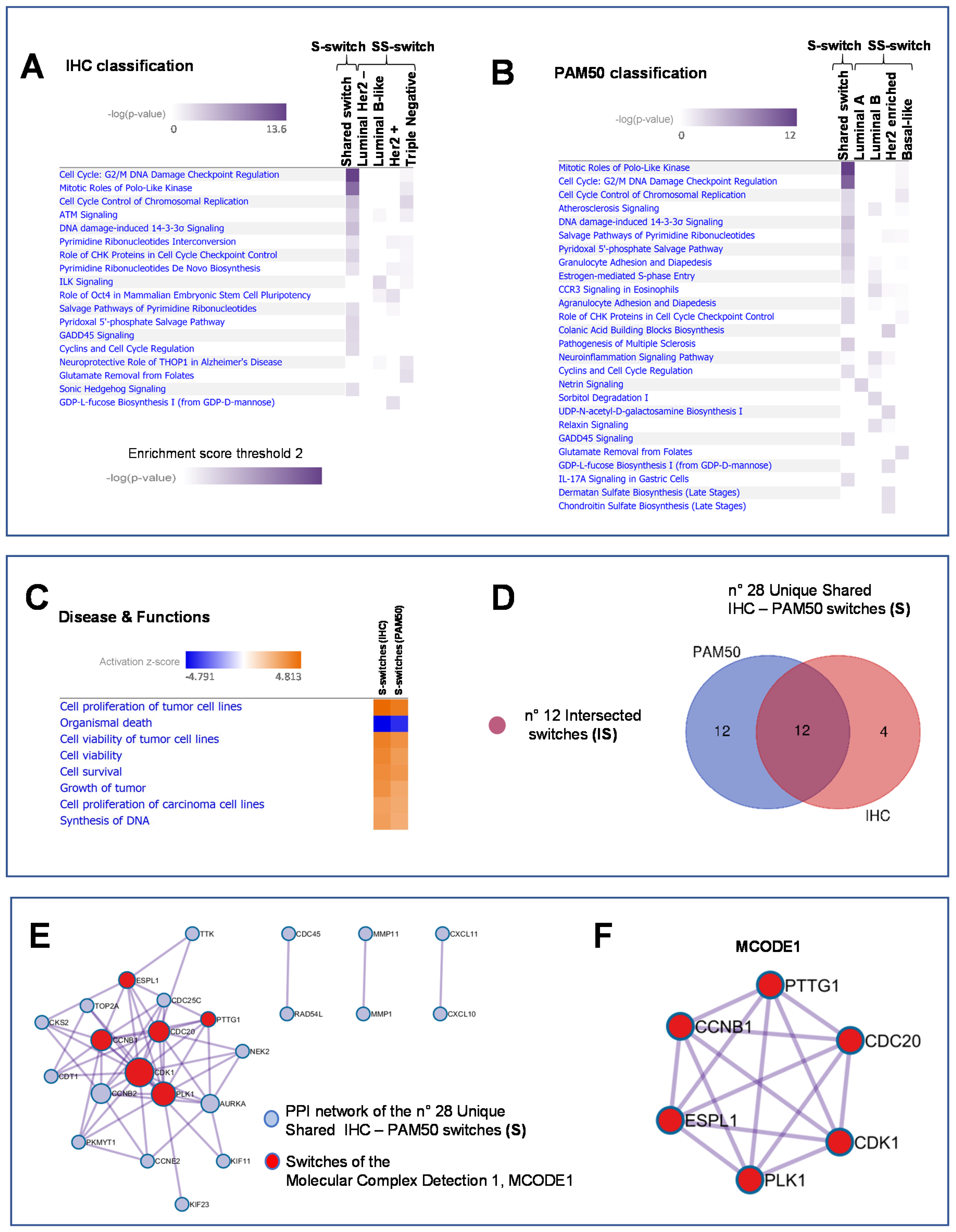
2.2. Integration and Prediction of IHC and PAM50 Key Regulatory Switches
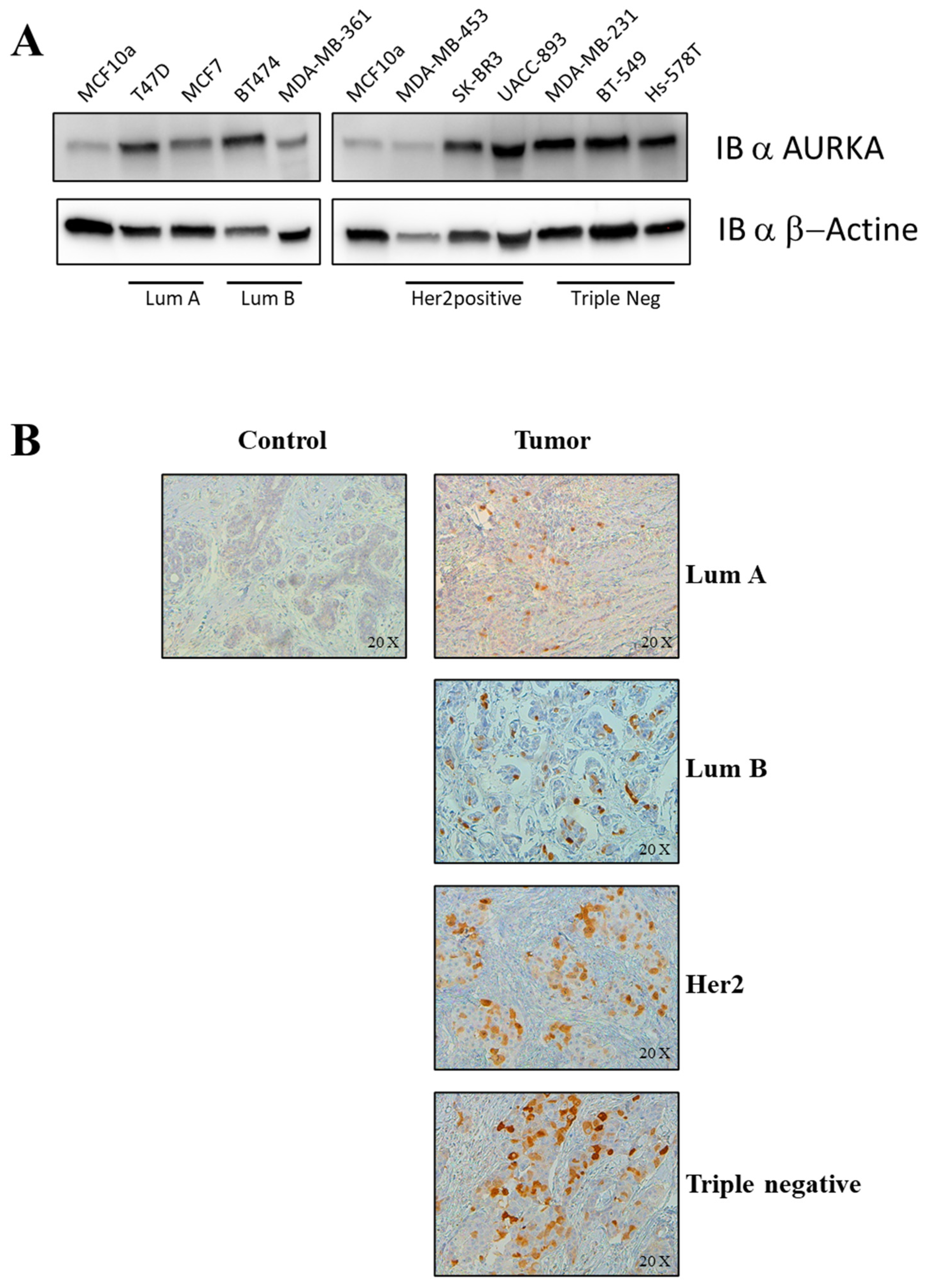
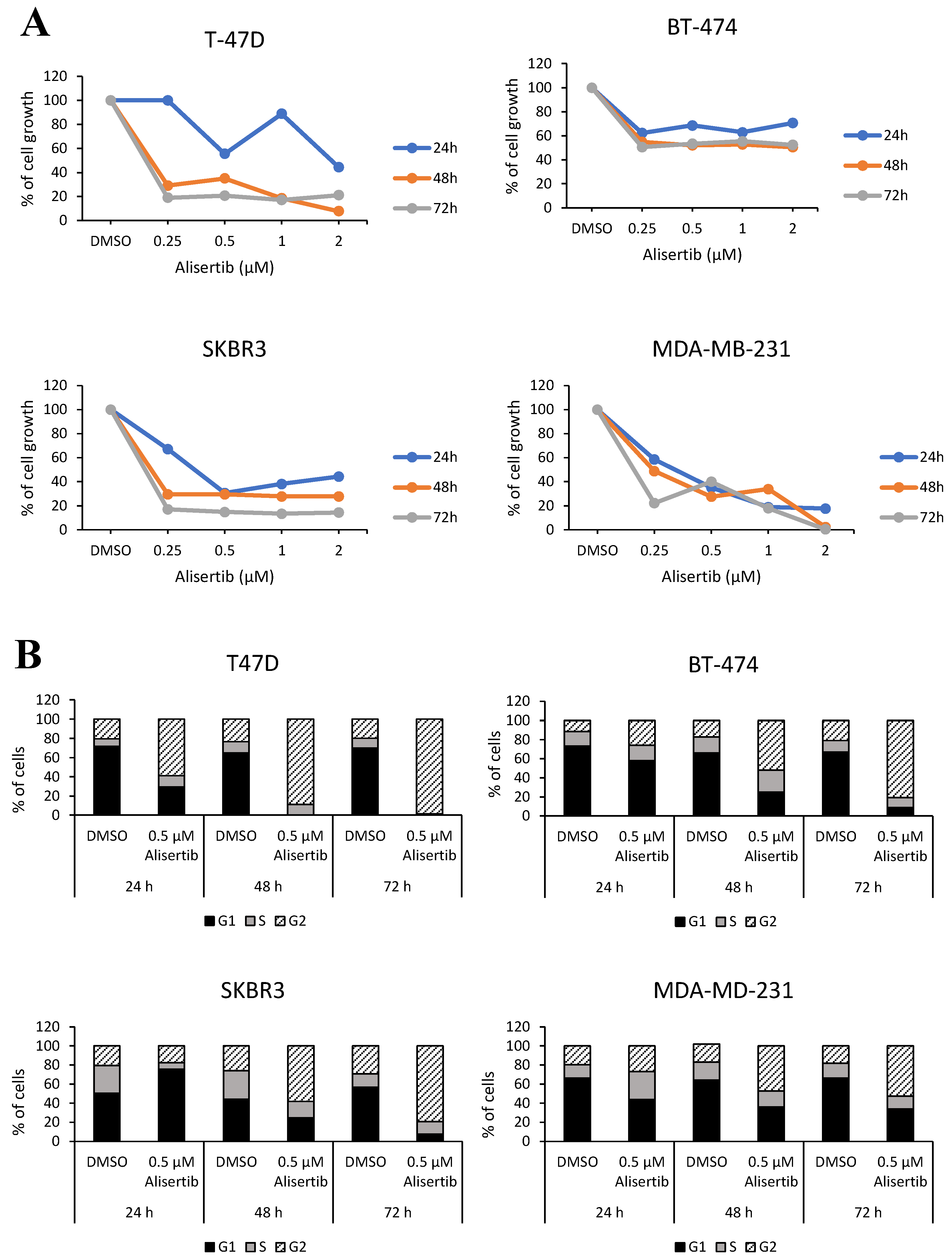
2.3. In Vitro and Ex Vivo Validation of Switch Genes and Functional Studies
3. Discussion
4. Materials and Methods
4.1. Dataset
4.2. Subtype Stratification
4.3. Gene Expression Data Analysis
4.4. Network-Based Separation Measure
4.5. Integrated Analyses for Identifying Key Regulatory Switches
4.6. Cell Culture and Treatment
4.7. RNA Extraction, Real-Time PCR, and Tissues QPCR Array
4.8. Immunohistochemistry
4.9. Protein Isolation and Western Blotting
4.10. Cell Death Assessment by Annexin V Staining and Cell-Cycle Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Ethical Approval
Informed Consent
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thuerlimann, B.; Senn, H.J.; Panel, M. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Vidal, M.; Cusick, M.E.; Barabasi, A.-L. Interactome Networks and Human Disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef]
- Chan, S.Y.; Loscalzo, J. The Emerging Paradigm of Network Medicine in the Study of Human Disease. Circ. Res. 2012, 111, 359–374. [Google Scholar] [CrossRef]
- Paci, P.; Colombo, T.; Fiscon, G.; Gurtner, A.; Pavesi, G.; Farina, L. SWIM: A computational tool to unveiling crucial nodes in complex biological networks. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Licursi, V.; Nasi, S.; Paci, P. Computational identification of specific genes for glioblastoma stem-like cells identity. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Falcone, R.; Conte, F.; Fiscon, G.; Pecce, V.; Sponziello, M.; Durante, C.; Farina, L.; Filetti, S.; Paci, P.; Verrienti, A. BRAF(V600E)-mutant cancers display a variety of networks by SWIM analysis: Prediction of vemurafenib clinical response. Endocrine 2019, 64, 406–413. [Google Scholar] [CrossRef]
- Paci, P.; Fiscon, G.; Conte, F.; Licursi, V.; Morrow, J.; Hersh, C.; Cho, M.; Castaldi, P.; Glass, K.; Silverman, E.K.; et al. Integrated transcriptomic correlation network analysis identifies COPD molecular determinants. Sci. Rep. 2020, 10, 3361. [Google Scholar] [CrossRef] [PubMed]
- Fiscon, G.; Conte, F.; Paci, P. SWIM tool application to expression data of glioblastoma stem-like cell lines, corresponding primary tumors and conventional glioma cell lines. BMC Bioinform. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, M.C.; Zenoni, S.; Fasoli, M.; Massonnet, M.; Farina, L.; Castiglione, F.; Pezzotti, M.; Paci, P. Integrated Network Analysis Identifies Fight-Club Nodes as a Class of Hubs Encompassing Key Putative Switch Genes That Induce Major Transcriptome Reprogramming during Grapevine Development. Plant Cell 2014, 26, 4617–4635. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M.; Canc Genome Atlas Res, N. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Barabasi, A.-L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Lever, J.; Zhao, E.Y.; Grewal, J.; Jones, M.R.; Jones, S.J.M. CancerMine: A literature-mined resource for drivers, oncogenes and tumor suppressors in cancer. Nat. Methods 2019, 16, 505–507. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Wang, C.; He, B.; Yang, M.; Tong, M.; Long, Z.; Liu, B.; Peng, F.; Xu, L.; Zhang, Y.; et al. Aurora-A Kinase: A Potent Oncogene and Target for Cancer Therapy. Med. Res. Rev. 2016, 36, 1036–1079. [Google Scholar] [CrossRef]
- Damodaran, A.P.; Vaufrey, L.; Gavard, O.; Prigent, C. Aurora A Kinase Is a Priority Pharmaceutical Target for the Treatment of Cancers. Trends Pharmacol. Sci. 2017, 38, 687–700. [Google Scholar] [CrossRef]
- Borisa, A.C.; Bhatt, H.G. A comprehensive review on Aurora kinase: Small molecule inhibitors and clinical trial studies. Eur. J. Med. Chem. 2017, 140, 1–19. [Google Scholar] [CrossRef]
- Tang, A.; Gao, K.; Chu, L.; Zhang, R.; Yang, J.; Zheng, J. Aurora kinases: Novel therapy targets in cancers. Oncotarget 2017, 8, 23937–23954. [Google Scholar] [CrossRef] [PubMed]
- Durlacher, C.T.; Li, Z.L.; Chen, X.W.; He, Z.X.; Zhou, S.F. An update on the pharmacokinetics and pharmacodynamics of alisertib, a selective Aurora kinase A inhibitor. Clin. Exp. Pharmacol. Physiol. 2016, 43, 585–601. [Google Scholar] [CrossRef] [PubMed]
- Aradottir, M.; Reynisdottir, S.T.; Stefansson, O.A.; Jonasson, J.G.; Sverrisdottir, A.; Tryggvadottir, L.; Eyfjord, J.E.; Bodvarsdottir, S.K. Aurora A is a prognostic marker for breast cancer arising in BRCA2 mutation carriers. J. Pathol. Clin. Res. 2015, 1, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.M.; Jang, B.G.; Hyun, C.L.; Kim, Y.S.; Hyun, J.W.; Chang, W.Y.; Maeng, Y.H. Aurora Kinase A Is a Prognostic Marker in Colorectal Adenocarcinoma. J. Pathol. Transl. Med. 2017, 51, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-G.; Yi, W.; Tao, Y.-L.; Chan, H.C.; Zeng, M.-S.; Xia, Y.-F. Aurora-A is an efficient marker for predicting poor prognosis in human nasopharyngeal carcinoma with aggressive local invasion: 208 cases with a 10-year follow-up from a single institution. Oncol. Lett. 2012, 3, 1237–1244. [Google Scholar] [CrossRef]
- Cammareri, P.; Scopelliti, A.; Todaro, M.; Eterno, V.; Francescangeli, F.; Moyer, M.P.; Agrusa, A.; Dieli, F.; Zeuner, A.; Stassi, G. Aurora-A Is Essential for the Tumorigenic Capacity and Chemoresistance of Colorectal Cancer Stem Cells. Cancer Res. 2010, 70, 4655–4665. [Google Scholar] [CrossRef]
- Xie, Y.; Zhu, S.; Zhong, M.; Yang, M.; Sun, X.; Liu, J.; Kroemer, G.; Lotze, M.; Zeh, H.J., III; Kang, R.; et al. Inhibition of Aurora Kinase A Induces Necroptosis in Pancreatic Carcinoma. Gastroenterology 2017, 153, 1429–1443. [Google Scholar] [CrossRef]
- Kulbe, H.; Otto, R.; Darb-Esfahani, S.; Lammert, H.; Abobaker, S.; Welsch, G.; Chekerov, R.; Schaefer, R.; Dragun, D.; Hummel, M.; et al. Discovery and Validation of Novel Biomarkers for Detection of Epithelial Ovarian Cancer. Cells 2019, 8, 713. [Google Scholar] [CrossRef]
- Zou, L.; Mitchell, J.; Stillman, B. CDC45, a novel yeast gene that functions with the origin recognition complex and MCM proteins in initiation of DNA replication. Mol. Cell. Biol. 1997, 17, 553–563. [Google Scholar] [CrossRef]
- Pacek, M.; Tutter, A.V.; Kubota, Y.; Takisawa, H.; Walter, J.C. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 2006, 21, 581–587. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, L.; Li, Z.; Wan, Z.; Shao, M.; Wu, S.; Wang, G. Potential Prognostic and Diagnostic Values of CDC6, CDC45, ORC6 and SNHG7 in Colorectal Cancer. Oncotargets Ther. 2019, 12, 11609–11621. [Google Scholar] [CrossRef]
- Huang, J.; Li, Y.; Lu, Z.; Che, Y.; Sun, S.; Mao, S.; Lei, Y.; Zang, R.; Li, N.; Zheng, S.; et al. Analysis of functional hub genes identifies CDC45 as an oncogene in non-small cell lung cancer–A short report. Cell. Oncol. 2019, 42, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Shi, R.; Zhao, S.; Li, X.; Lu, S.; Bu, H.; Ma, X. Cell division cycle 45 promotes papillary thyroid cancer progression via regulating cell cycle. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Pollok, S.; Bauerschmidt, C.; Saenger, J.; Nasheuer, H.P.; Grosse, F. Human Cdc45 is a proliferation-associated antigen. Febs J. 2007, 274, 3669–3684. [Google Scholar] [CrossRef]
- Mukherjee, M.; Ge, G.; Zhang, N.; Edwards, D.G.; Sumazin, P.; Sharan, S.K.; Rao, P.H.; Medina, D.; Pati, D. MMTV-Espl1 transgenic mice develop aneuploid, estrogen receptor alpha (ER alpha)-positive mammary adenocarcinomas. Oncogene 2014, 33, 5511–5522. [Google Scholar] [CrossRef] [PubMed]
- Finetti, P.; Guille, A.; Adelaide, J.; Birnbaum, D.; Chaffanet, M.; Bertucci, F. ESPL1 is a candidate oncogene of luminal B breast cancers. Breast Cancer Res. Treat. 2014, 147, 51–59. [Google Scholar] [CrossRef]
- Liu, J.; Wan, Y.; Li, S.; Qiu, H.; Jiang, Y.; Ma, X.; Zhou, S.; Cheng, W. Identification of aberrantly methylated differentially expressed genes and associated pathways in endometrial cancer using integrated bioinformatic analysis. Cancer Med. 2020, 9, 3522–3536. [Google Scholar] [CrossRef]
- Yan, X.; Liu, X.-P.; Guo, Z.-X.; Tong-Zu, L.; Li, S. Identification of Hub Genes Associated With Progression and Prognosis in Patients With Bladder Cancer. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Li, D.H.; Liu, H.; Jiao, L.; Chang, D.Z.; Beinart, G.; Wolff, R.A.; Evans, D.B.; Hassan, M.M.; Abbruzzese, J.L. Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival. Cancer Res. 2006, 66, 3323–3330. [Google Scholar] [CrossRef]
- Gee, H.E.; Buffa, F.M.; Harris, A.L.; Toohey, J.M.; Carroll, S.L.; Cooper, C.L.; Beith, J.; McNeil, C.; Carmalt, H.; Mak, C.; et al. MicroRNA-Related DNA Repair/Cell-Cycle Genes Independently Associated With Relapse After Radiation Therapy for Early Breast Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2015, 93, 1104–1114. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwinska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. (Pozn. Pol.) 2015, 19, A68–A77. [Google Scholar] [CrossRef] [PubMed]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabasi, A.-L. Uncovering disease-disease relationships through the incomplete interactome. Science 2015, 347. [Google Scholar] [CrossRef]
- Cheng, F.; Desai, R.J.; Handy, D.E.; Wang, R.; Schneeweiss, S.; Barabasi, A.-L.; Loscalzo, J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Tilli, T.M.; Castro, C.d.S.; Tuszynski, J.A.; Carels, N. A strategy to identify housekeeping genes suitable for analysis in breast cancer diseases. BMC Genom. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, P.; Incoronato, M.; Coppola, L.; Infante, T.; Parente, C.A.; Nicolai, E.; Soricelli, A.; Salvatore, M. SDN biobank: Bioresource of human samples associated with functional and/or morphological bioimaging results for the study of oncological, cardiological, neurological, and metabolic diseases. Open J. Bioresour. 2017, 4, 2. [Google Scholar] [CrossRef]


| IHC Classification | PAM50 Classification | |||||||
|---|---|---|---|---|---|---|---|---|
| Lum HER2− | Lum B (HER2+) | HER2+ (Non-Luminal) | Triple Negative | Luminal A | Luminal B | HER2 Enriched | Basal-Like | |
| Samples | ||||||||
| # normal | 111 | 111 | 111 | 111 | 111 | 111 | 111 | 111 |
| # tumor | 574 | 123 | 37 | 153 | 229 | 120 | 58 | 98 |
| Thresholds | ||||||||
| FC threshold | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| FDR threshold | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 |
| PC threshold | 0.6 | 0.72 | 0.75 | 0.69 | 0.64 | 0.63 | 0.68 | 0.66 |
| DEGs | ||||||||
| # total | 1365 | 1603 | 1862 | 1714 | 1338 | 2062 | 1954 | 1829 |
| # up | 363 (27%) | 448 (28%) | 496 (27%) | 515 (30%) | 373 (28%) | 468 (23%) | 540 (28%) | 510 (28%) |
| # down | 1002 (73%) | 1155 (72%) | 1366 (73%) | 1199 (70%) | 965 (72%) | 1594 (77%) | 1414 (72%) | 1319 (72%) |
| Switch | ||||||||
| # total | 84 | 261 | 278 | 251 | 222 | 358 | 363 | 343 |
| # up | 84 (100%) | 261 (100%) | 272 (98%) | 217 (86%) | 221 (99.5%) | 356 (99%) | 363 (100%) | 329 (96%) |
| # down | 0 | 0 | 6 (2%) | 34 (14%) | 1 (0.5%) | 2 (1%) | 0 | 14 (4%) |
| IHC Clinicopathologic Surrogate Definition | TCGA-BRCA Clinicopathologic Surrogate Definition |
|---|---|
| ‘Luminal A-like’ all: ER and PgR positive, HER2 negative, Ki-67 ‘low’. | ‘Luminal HER2 negative’ all: ER and PgR positive, HER2 negative. |
| ‘Luminal B-like (HER2 negative)’ ER positive HER2 negative and at least one of: Ki-67 ‘high’, PgR ‘negative or low’. | |
| ‘Luminal B-like (HER2 positive)’ ER positive, HER2 over-expressed or amplified, Any Ki-67 and Any PgR. | ‘Luminal B-like (HER2 positive)’ ER positive, HER2 over-expressed or amplified, Any PgR. |
| ‘HER2 positive (non-luminal)’ HER2 over-expressed or amplified, ER and PgR absent. | ‘HER2 positive (non-luminal)’ HER2 over-expressed or amplified, ER and PgR absent. |
| ‘Triple negative’ ER and PgR absent, HER2 negative. | ‘Triple negative’ ER and PgR absent, HER2 negative. |
| Gene Name | Gene Description | Location | Type(s) | Gene Stable ID | Drug(s) |
|---|---|---|---|---|---|
| AURKA | aurora kinase A | Nucleus | kinase | ENSG00000087586 | ilorasertib, MK 5108, SNS 314, AT-9283, alisertib, MLN8054, TTP607, CYC 116, TAS-119, tozasertib, AMG 900, danusertib |
| CCNB1 | cyclin B1 | Cytoplasm | kinase | ENSG00000134057 | bertilimumab |
| CCNB2 | cyclin B2 | Cytoplasm | other | ENSG00000157456 | - |
| CDC20 | cell division cycle 20 | Nucleus | other | ENSG00000117399 | - |
| CDC45 | cell division cycle 45 | Nucleus | other | ENSG00000093009 | - |
| CDK1 | cyclin dependent kinase 1 | Nucleus | kinase | ENSG00000170312 | AZD5438, SB-1317, alvocidib, AG 024322, milciclib, riviciclib, roniciclib, dinaciclib |
| ESPL1 | extra spindle pole bodies like 1, separase | Nucleus | peptidase | ENSG00000135476 | - |
| NEK2 | NIMA related kinase 2 | Cytoplasm | kinase | ENSG00000117650 | - |
| PLK1 | polo like kinase 1 | Nucleus | kinase | ENSG00000166851 | lipid encapsulated anti-PLK1 siRNA TKM-080301, onvansertib, GSK461364, TAK-960, MK1496, rigosertib, volasertib, BI 2536 |
| PTTG1 | PTTG1 regulator of sister chromatid separation, securin | Nucleus | transcription regulator | ENSG00000164611 | - |
| RAD54L | RAD54 like | Nucleus | enzyme | ENSG00000085999 | - |
| N° Healthy Control | 4 |
| Age 40.75 (39–55 years) | |
| N° Breast Cancer Patients | 23 |
| Age 63.39 (41–82 years) | |
| Histological Type | |
| Invasive Ductal carcinoma | 23 |
| Hyperplasia | 2 |
| Gynecomastia | 2 |
| Subtype | |
| Luminal A | 6 |
| Luminal B | 5 |
| Her2+ | 8 |
| Triple negative | 4 |
| Ki67 | |
| Low (0–29%) | 15 |
| High (30–100%) | 8 |
| Grade | |
| G1 | 3 |
| G2 | 12 |
| G3 | 8 |
| Tumor Size (cm) | |
| 0.1–2 | 15 |
| 2–5 | 6 |
| >5 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grimaldi, A.M.; Conte, F.; Pane, K.; Fiscon, G.; Mirabelli, P.; Baselice, S.; Giannatiempo, R.; Messina, F.; Franzese, M.; Salvatore, M.; et al. The New Paradigm of Network Medicine to Analyze Breast Cancer Phenotypes. Int. J. Mol. Sci. 2020, 21, 6690. https://doi.org/10.3390/ijms21186690
Grimaldi AM, Conte F, Pane K, Fiscon G, Mirabelli P, Baselice S, Giannatiempo R, Messina F, Franzese M, Salvatore M, et al. The New Paradigm of Network Medicine to Analyze Breast Cancer Phenotypes. International Journal of Molecular Sciences. 2020; 21(18):6690. https://doi.org/10.3390/ijms21186690
Chicago/Turabian StyleGrimaldi, Anna Maria, Federica Conte, Katia Pane, Giulia Fiscon, Peppino Mirabelli, Simona Baselice, Rosa Giannatiempo, Francesco Messina, Monica Franzese, Marco Salvatore, and et al. 2020. "The New Paradigm of Network Medicine to Analyze Breast Cancer Phenotypes" International Journal of Molecular Sciences 21, no. 18: 6690. https://doi.org/10.3390/ijms21186690
APA StyleGrimaldi, A. M., Conte, F., Pane, K., Fiscon, G., Mirabelli, P., Baselice, S., Giannatiempo, R., Messina, F., Franzese, M., Salvatore, M., Paci, P., & Incoronato, M. (2020). The New Paradigm of Network Medicine to Analyze Breast Cancer Phenotypes. International Journal of Molecular Sciences, 21(18), 6690. https://doi.org/10.3390/ijms21186690







