Evaluating the Genotoxic and Cytotoxic Effects of Thymidine Analogs, 5-Ethynyl-2′-Deoxyuridine and 5-Bromo-2′-Deoxyurdine to Mammalian Cells
Abstract
1. Introduction
2. Results
2.1. The In Vitro Effects of Nucleotide and Nucleoside Supplemented Medium on CHO Cells
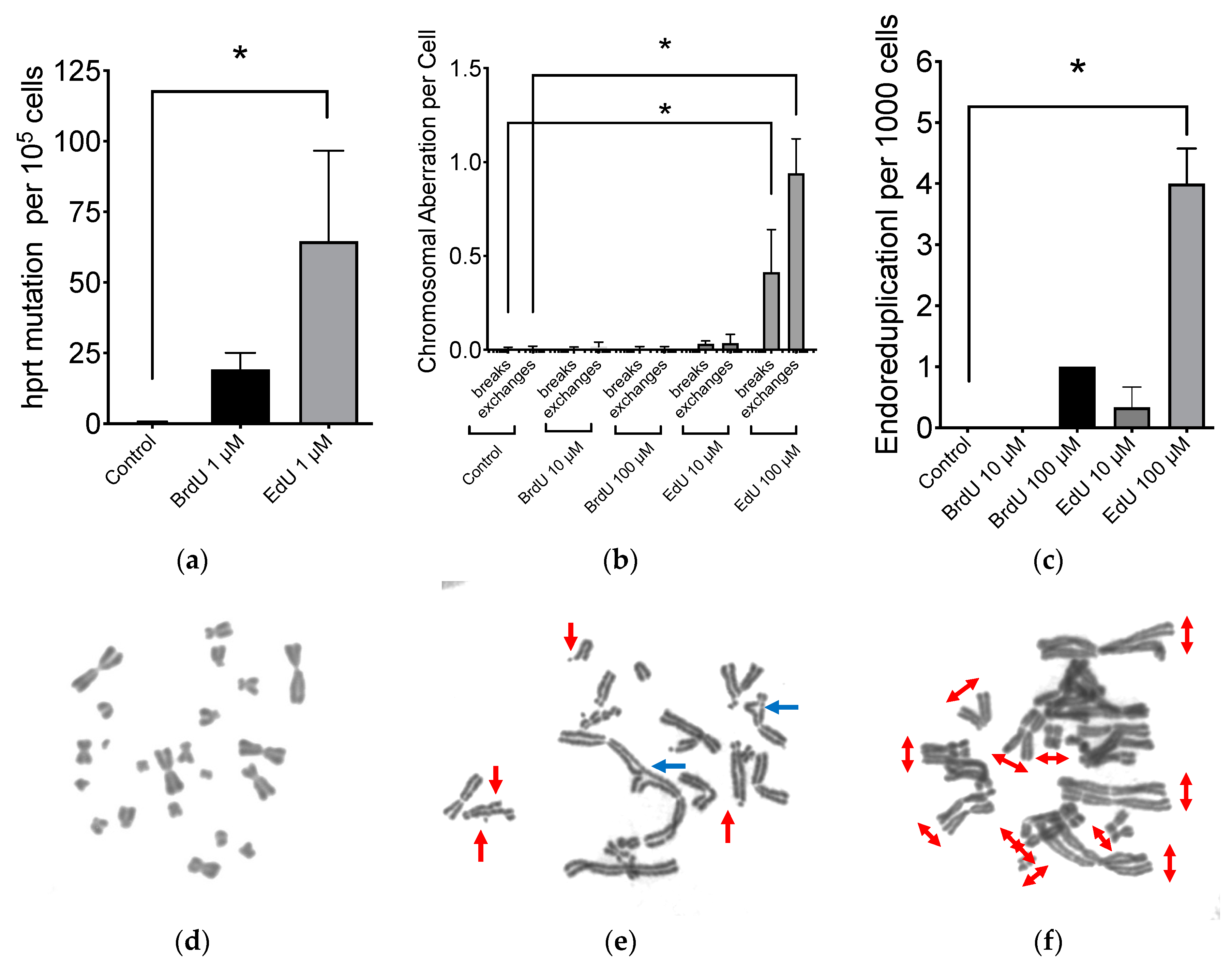
2.2. Mutagenic Properties of EdU
2.3. Effect to DNA Damage Responses
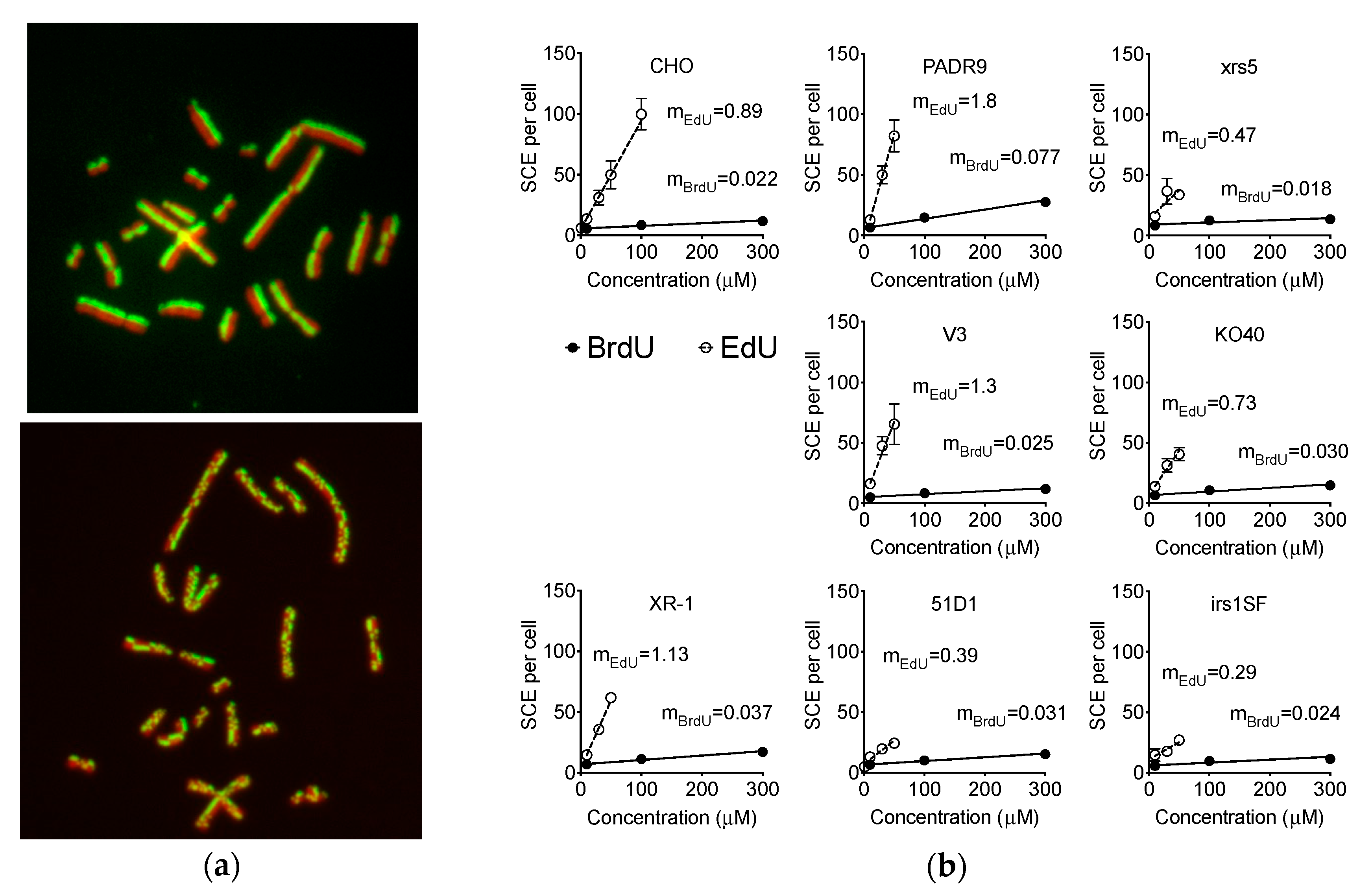
2.4. Effect of EdU on SCE Formation
2.5. Effect of DNA Repair for Cytotoxicity
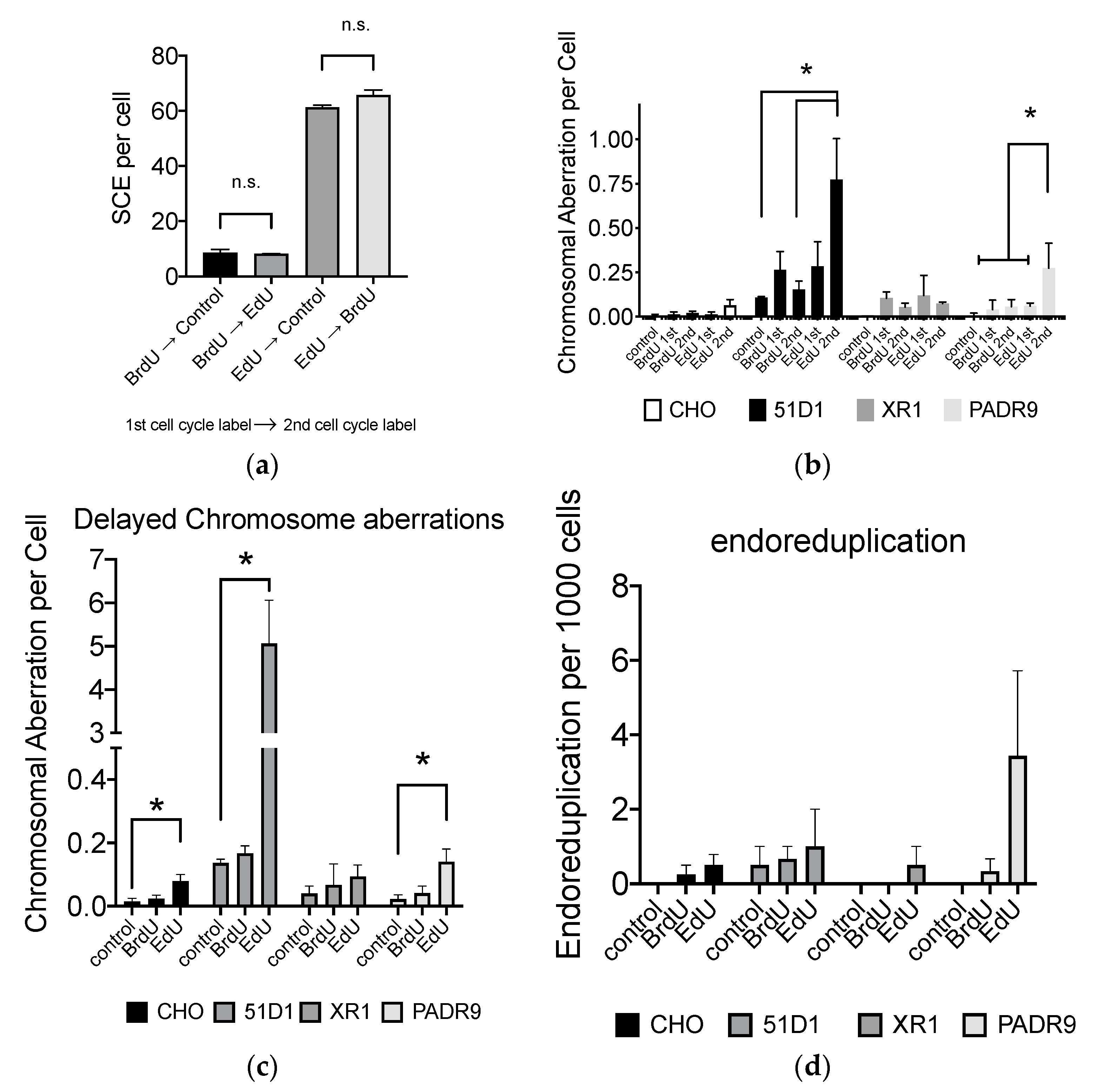
2.6. Effect of EdU Continuous Treatment on Homologous Recombination-Deficient Cells
2.7. Effect of EdU for SCE Formation and Chromosome Aberrations after the Second Cell Cycle
2.8. Loss of BRCA2 Causing Hypersensitivity to EdU
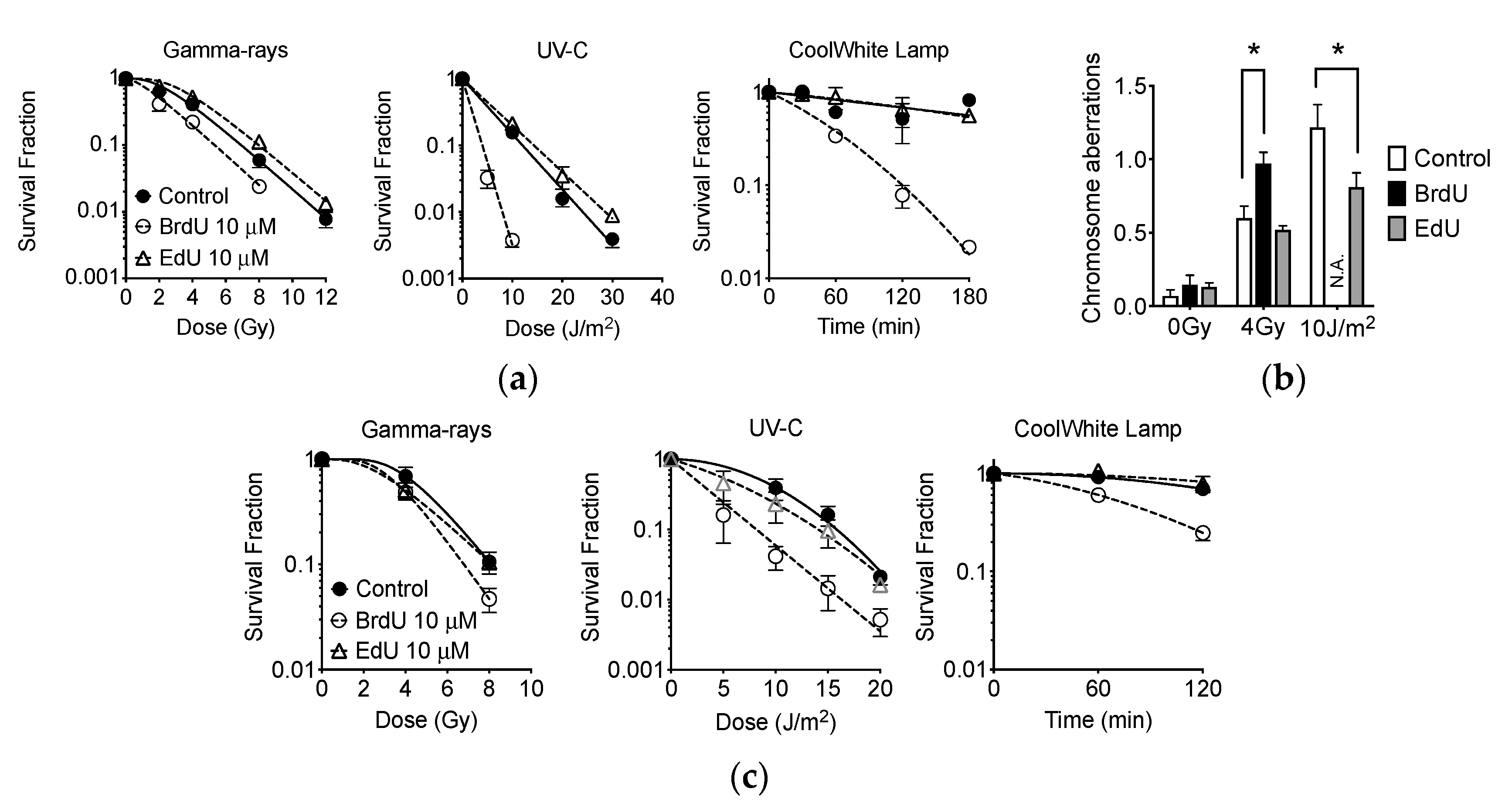
2.9. Effect of EdU Treatment to Gamma-Rays, UVC, and Fluorescent Light
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Cell Doubling Time Analysis
4.3. Colony Formation Assay
4.4. DNA Damage Repair Responses Assay
4.5. Chromosomal Aberration Analysis for Long Term Treatment
4.6. Chromosomal Aberration Analysis for Short Term Treatment
4.7. Hypoxanthine Phosphorybosyl Transferase (HPRT) Mutation Analysis
4.8. Sister Chromatid Exchange
4.9. Sensitization Effects of BrdU and EdU to UV-C, Gamma-Rays, and Light from Fluorescent Lamp
4.10. Statistics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dvorackova, M.; Fajkus, J. Visualization of the Nucleolus Using Ethynyl Uridine. Front. Plant Sci. 2018, 9, 177. [Google Scholar] [CrossRef] [PubMed]
- Salic, A.; Mitchison, T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 2415–2420. [Google Scholar] [CrossRef] [PubMed]
- Kuriya, K.; Higashiyama, E.; Avsar-Ban, E.; Okochi, N.; Hattori, K.; Ogata, S.; Takebayashi, S.I.; Ogata, M.; Tamaru, Y.; Okumura, K. Direct visualization of replication dynamics in early zebrafish embryos. Biosci. Biotechnol. Biochem. 2016, 80, 945–948. [Google Scholar] [CrossRef] [PubMed]
- Diermeier-Daucher, S.; Clarke, S.T.; Hill, D.; Vollmann-Zwerenz, A.; Bradford, J.A.; Brockhoff, G. Cell type specific applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytom. A 2009, 75, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Yamakoshi, H.; Dodo, K.; Palonpon, A.; Ando, J.; Fujita, K.; Kawata, S.; Sodeoka, M. Alkyne-tag Raman imaging for visualization of mobile small molecules in live cells. J. Am. Chem. Soc. 2012, 134, 20681–20689. [Google Scholar] [CrossRef]
- Yamakoshi, H.; Dodo, K.; Okada, M.; Ando, J.; Palonpon, A.; Fujita, K.; Kawata, S.; Sodeoka, M. Imaging of EdU, an alkyne-tagged cell proliferation probe, by Raman microscopy. J. Am. Chem. Soc. 2011, 133, 6102–6105. [Google Scholar] [CrossRef]
- Davis, W.B.; Oakes, J.E.; Taylor, J.A. Effect of treatment with 5-ethyl-2′-deoxyuridine on herpes simplex virus encephalitis in normal and immunosuppressed mice. Antimicrob. Agents Chemother. 1978, 14, 743–748. [Google Scholar] [CrossRef]
- Frum, R.A.; Deb, S.; Deb, S.P. Use of the DNA fiber spreading technique to detect the effects of mutant p53 on DNA replication. Methods Mol. Biol. 2013, 962, 147–155. [Google Scholar] [CrossRef]
- Cartwright, I.M.; Genet, M.D.; Fujimori, A.; Kato, T.A. Role of LET and chromatin structure on chromosomal inversion in CHO10B2 cells. Genome Integr. 2014, 5, 1. [Google Scholar] [CrossRef]
- Heartlein, M.W.; O’Neill, J.P.; Pal, B.C.; Preston, R.J. The induction of specific-locus mutations and sister-chromatid exchanges by 5-bromo- and 5-chloro-deoxyuridine. Mutat. Res. 1982, 92, 411–416. [Google Scholar] [CrossRef]
- Heartlein, M.W.; O’Neill, J.P.; Preston, R.J. SCE induction is proportional to substitution in DNA for thymidine by CldU and BrdU. Mutat. Res. 1983, 107, 103–109. [Google Scholar] [CrossRef]
- Jeney, A.; Barrie, S.E.; Taylor, G.A.; Newell, D.R.; Harrap, K.R.; Szabolcs, A.; Lapis, K.; Otvos, L. 5-Ethyl-2’-deoxyuridine: An explanation for its lack of cytotoxic action in vivo. Eur. J. Cancer Clin. Oncol. 1986, 22, 557–562. [Google Scholar] [CrossRef]
- Ning, H.; Albersen, M.; Lin, G.; Lue, T.F.; Lin, C.S. Effects of EdU labeling on mesenchymal stem cells. Cytotherapy 2013, 15, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Tuominen, H.; Bergstrom, D.; Vilpo, J.A. 5-Ethyl-2’-deoxyuridine. Cytotoxicity and DNA incorporation demonstrated with human leukemic cells and PHA-stimulated lymphocytes in vitro. Acta Chem. Scand. B 1985, 39, 735–743. [Google Scholar] [CrossRef]
- Gupta, S.K.; Agarwal, R.; Srivastava, S. Textbook on Clinical Ocular Pharmacology and Therapeutics; Jaypee Brothers Medical: New Delhi, India, 2014; Volome 1. [Google Scholar]
- Rhind, N. Incorporation of thymidine analogs for studying replication kinetics in fission yeast. Methods Mol. Biol. 2009, 521, 509–515. [Google Scholar] [CrossRef]
- Anda, S.; Boye, E.; Grallert, B. Cell-cycle analyses using thymidine analogues in fission yeast. PLoS ONE 2014, 9, e88629. [Google Scholar] [CrossRef]
- Gutierrez, C.; Gonzalez-Gil, G.; Hernandez, P. Analysis of baseline and BrdU-dependent SCEs at different BrdU concentrations. Exp. Cell Res. 1983, 149, 461–469. [Google Scholar] [CrossRef]
- O’Neill, J.P.; Heartlein, M.W.; Preston, R.J. Sister-chromatid exchanges and gene mutations are induced by the replication of 5-bromo- and 5-chloro-deoxyuridine substituted DNA. Mutat. Res. 1983, 109, 259–270. [Google Scholar] [CrossRef]
- Pivazyan, A.D.; Birks, E.M.; Wood, T.G.; Lin, T.S.; Prusoff, W.H. Inhibition of poly(ADP-ribose)polymerase activity by nucleoside analogs of thymidine. Biochem. Pharmacol. 1992, 44, 947–953. [Google Scholar] [CrossRef]
- Attardi, G.; Keeley, B.; Tu, C. Photosensitivity and heat resistance conferred by BrdU incorporation upon a thymidine kinase-deficient mouse cell line with persistent mitochondrial enzyme activity. J. Cell Sci. 1975, 19, 55–68. [Google Scholar]
- Huang, X.; King, M.A.; Halicka, H.D.; Traganos, F.; Okafuji, M.; Darzynkiewicz, Z. Histone H2AX phosphorylation induced by selective photolysis of BrdU-labeled DNA with UV light: Relation to cell cycle phase. Cytom. A 2004, 62, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Genet, M.D.; Roybal, E.J.; Kubota, N.; Okayasu, R.; Miyagawa, K.; Fujimori, A.; Kato, T.A. Comparison of the bromodeoxyuridine-mediated sensitization effects between low-LET and high-LET ionizing radiation on DNA double-strand breaks. Oncol. Rep. 2013, 29, 2133–2139. [Google Scholar] [CrossRef] [PubMed]
- Erikson, R.L.; Szybalski, W. Molecular radiobiology of human cell lines. I. Comparative sensitivity to x-rays and ultraviolet light of cells containing halogen-substituted DNA. Biochem. Biophys. Res. Commun. 1961, 4, 258–261. [Google Scholar] [PubMed]
- Sabatinos, S.A.; Mastro, T.L.; Green, M.D.; Forsburg, S.L. A mammalian-like DNA damage response of fission yeast to nucleoside analogs. Genetics 2013, 193, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Ross, H.H.; Rahman, M.; Levkoff, L.H.; Millette, S.; Martin-Carreras, T.; Dunbar, E.M.; Reynolds, B.A.; Laywell, E.D. Ethynyldeoxyuridine (EdU) suppresses in vitro population expansion and in vivo tumor progression of human glioblastoma cells. J. Neurooncol. 2011, 105, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Halicka, H.D.; Li, J.; Biela, E.; Berniak, K.; Dobrucki, J.; Darzynkiewicz, Z. DNA damage signaling, impairment of cell cycle progression, and apoptosis triggered by 5-ethynyl-2’-deoxyuridine incorporated into DNA. Cytom. A 2013, 83, 979–988. [Google Scholar] [CrossRef]
- Shealy, Y.F.; O’Dell, C.A.; Arnett, G.; Shannon, W.M. Synthesis and antiviral activity of the carbocyclic analogues of 5-ethyl-2’-deoxyuridine and of 5-ethynyl-2′-deoxyuridine. J. Med. Chem. 1986, 29, 79–84. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. Click Nucleic Acid Ligation. Nucleic Acid Ther. 2011, 21, A6. [Google Scholar]
- Cortes, F.; Pastor, N.; Mateos, S.; Dominguez, I. The nature of DNA plays a role in chromosome segregation: Endoreduplication in halogen-substituted chromosomes. DNA Repair 2003, 2, 719–726. [Google Scholar] [CrossRef]
- Schuermann, D.; Fritsch, O.; Lucht, J.M.; Hohn, B. Replication stress leads to genome instabilities in Arabidopsis DNA polymerase delta mutants. Plant Cell 2009, 21, 2700–2714. [Google Scholar] [CrossRef][Green Version]
- Stamato, T.D.; Weinstein, R.; Giaccia, A.; Mackenzie, L. Isolation of cell cycle-dependent gamma ray-sensitive Chinese hamster ovary cell. Somatic.Cell Genet. 1983, 9, 165–173. [Google Scholar] [CrossRef]
- Witmer, M.V.; Aboul-Ela, N.; Jacobson, M.K.; Stamato, T.D. Increased sensitivity to DNA-alkylating agents in CHO mutants with decreased poly(ADP-ribose) polymerase activity. Mutat. Res. 1994, 314, 249–260. [Google Scholar] [CrossRef]
- Batsanov, S.S. Van der Waals radii of elements. Inorg. Mater. 2001, 37, 871–885. [Google Scholar] [CrossRef]
- Saha, B.K.; Nangia, A. Ethynyl group as a supramolecular halogen and C equivalent to C-H---C equivalent to C trimer synthon in 2,4,6-tris(4-ethynylphenoxy)-1,3,5-triazine. Cryst. Growth Des. 2007, 7, 393–401. [Google Scholar] [CrossRef]
- Whitmore, G.F.; Varghese, A.J.; Gulyas, S. Cell cycle responses of two X-ray sensitive mutants defective in DNA repair. Int. J. Radiat. Biol. 1989, 56, 657–665. [Google Scholar]
- Jeggo, P.A.; Kemp, L.M. X-ray-sensitive mutants of Chinese hamster ovary cell line. Isolation and cross-sensitivity to other DNA-damaging agents. Mutat. Res. 1983, 112, 313–327. [Google Scholar] [PubMed]
- Verhaegh, G.W.; Jongmans, W.; Morolli, B.; Jaspers, N.G.; van der Schans, G.P.; Lohman, P.H.; Zdzienicka, M.Z. A novel type of X-ray-sensitive Chinese hamster cell mutant with radioresistant DNA synthesis and hampered DNA double-strand break repair. Mutat. Res. 1995, 337, 119–129. [Google Scholar] [CrossRef]
- Hinz, J.M.; Tebbs, R.S.; Wilson, P.F.; Nham, P.B.; Salazar, E.P.; Nagasawa, H.; Urbin, S.S.; Bedford, J.S.; Thompson, L.H. Repression of mutagenesis by Rad51D-mediated homologous recombination. Nucleic Acids Res. 2006, 34, 1358–1368. [Google Scholar] [CrossRef]
- Tebbs, R.S.; Hinz, J.M.; Yamada, N.A.; Wilson, J.B.; Salazar, E.P.; Thomas, C.B.; Jones, I.M.; Jones, N.J.; Thompson, L.H. New insights into the Fanconi anemia pathway from an isogenic FancG hamster CHO mutant. DNA Repair 2005, 4, 11–22. [Google Scholar] [CrossRef]
- Fuller, L.F.; Painter, R.B. A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res. 1988, 193, 109–121. [Google Scholar] [CrossRef]
- Gerelchuluun, A.; Maeda, J.; Manabe, E.; Brents, C.A.; Sakae, T.; Fujimori, A.; Chen, D.J.; Tsuboi, K.; Kato, T.A. Histone Deacetylase Inhibitor Induced Radiation Sensitization Effects on Human Cancer Cells after Photon and Hadron Radiation Exposure. Int. J. Mol. Sci. 2018, 19, 496. [Google Scholar] [CrossRef]
- Genet, S.C.; Fujii, Y.; Maeda, J.; Kaneko, M.; Genet, M.D.; Miyagawa, K.; Kato, T.A. Hyperthermia inhibits homologous recombination repair and sensitizes cells to ionizing radiation in a time and temperature dependent manner. J. Cell. Physiol. 2012, 228, 1473–1481. [Google Scholar] [CrossRef]
- Kato, T.A. Human Lymphocyte Metaphase Chromosome Preparation for Radiation-Induced Chromosome Aberration Analysis. Methods Mol. Biol. 2019, 1984, 1–6. [Google Scholar] [CrossRef]
- Wall, A.C.; Gius, J.P.; Buglewicz, D.J.; Banks, A.B.; Kato, T.A. Oxidative stress and endoreduplication induced by blue light exposure to CHO cells. Mutat. Res. 2019, 841, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Buglewicz, D.J.; Mussallem, J.T.; Haskins, A.H.; Su, C.; Maeda, J.; Kato, T.A. Cytotoxicity and Mutagenicity of Narrowband UVB to Mammalian Cells. Genes 2020, 11, 646. [Google Scholar] [CrossRef] [PubMed]
- Little, J.B.; Nagasawa, H.; Pfenning, T.; Vetrovs, H. Radiation-induced genomic instability: Delayed mutagenic and cytogenetic effects of X rays and alpha particles. Radiat. Res. 1997, 148, 299–307. [Google Scholar] [CrossRef]
- Terasima, T.; Tolmach, L.J. Changes in X-ray sensitivity of HeLa cells during the division cycle. Nature 1961, 190, 1210–1211. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Allum, A.J.; Aizawa, Y.; Kato, T.A. Novel glyceryl glucoside is a low toxic alternative for cryopreservation agent. Biochem. Biophys. Res. Commun. 2016, 476, 359–364. [Google Scholar] [CrossRef]
- Maeda, J.; Allum, A.J.; Mussallem, J.T.; Froning, C.E.; Haskins, A.H.; Buckner, M.A.; Miller, C.D.; Kato, T.A. Ascorbic Acid 2-Glucoside Pretreatment Protects Cells from Ionizing Radiation, UVC, and Short Wavelength of UVB. Genes 2020, 11, 238. [Google Scholar] [CrossRef]





| Cell Line | BrdU (μM) | EdU (μM) | Ratio BrdU/EdU | Affected Pathways |
|---|---|---|---|---|
| CHO | 15 | 0.088 | 170 | n.a. |
| V3 | 0.54 | 0.025 | 21 | NHEJ |
| xrs5 | 0.63 | 0.023 | 27 | NHEJ |
| XR-1 | 0.56 | 0.022 | 25 | NHEJ |
| irs1SF | 0.30 | 0.0027 | 111 | HR |
| 51D1 | 0.54 | 0.0021 | 257 | HR |
| KO40 | 0.44 | 0.011 | 40 | Fanconi Anemia |
| PADR9 | 0.38 | 0.010 | 38 | poly(ADP-ribosylation) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haskins, J.S.; Su, C.; Maeda, J.; Walsh, K.D.; Haskins, A.H.; Allum, A.J.; Froning, C.E.; Kato, T.A. Evaluating the Genotoxic and Cytotoxic Effects of Thymidine Analogs, 5-Ethynyl-2′-Deoxyuridine and 5-Bromo-2′-Deoxyurdine to Mammalian Cells. Int. J. Mol. Sci. 2020, 21, 6631. https://doi.org/10.3390/ijms21186631
Haskins JS, Su C, Maeda J, Walsh KD, Haskins AH, Allum AJ, Froning CE, Kato TA. Evaluating the Genotoxic and Cytotoxic Effects of Thymidine Analogs, 5-Ethynyl-2′-Deoxyuridine and 5-Bromo-2′-Deoxyurdine to Mammalian Cells. International Journal of Molecular Sciences. 2020; 21(18):6631. https://doi.org/10.3390/ijms21186631
Chicago/Turabian StyleHaskins, Jeremy S., Cathy Su, Junko Maeda, Kade D. Walsh, Alexis H. Haskins, Allison J. Allum, Coral E. Froning, and Takamitsu A. Kato. 2020. "Evaluating the Genotoxic and Cytotoxic Effects of Thymidine Analogs, 5-Ethynyl-2′-Deoxyuridine and 5-Bromo-2′-Deoxyurdine to Mammalian Cells" International Journal of Molecular Sciences 21, no. 18: 6631. https://doi.org/10.3390/ijms21186631
APA StyleHaskins, J. S., Su, C., Maeda, J., Walsh, K. D., Haskins, A. H., Allum, A. J., Froning, C. E., & Kato, T. A. (2020). Evaluating the Genotoxic and Cytotoxic Effects of Thymidine Analogs, 5-Ethynyl-2′-Deoxyuridine and 5-Bromo-2′-Deoxyurdine to Mammalian Cells. International Journal of Molecular Sciences, 21(18), 6631. https://doi.org/10.3390/ijms21186631






