MiR-581/SMAD7 Axis Contributes to Colorectal Cancer Metastasis: A Bioinformatic and Experimental Validation-Based Study
Abstract
1. Introduction
2. Results
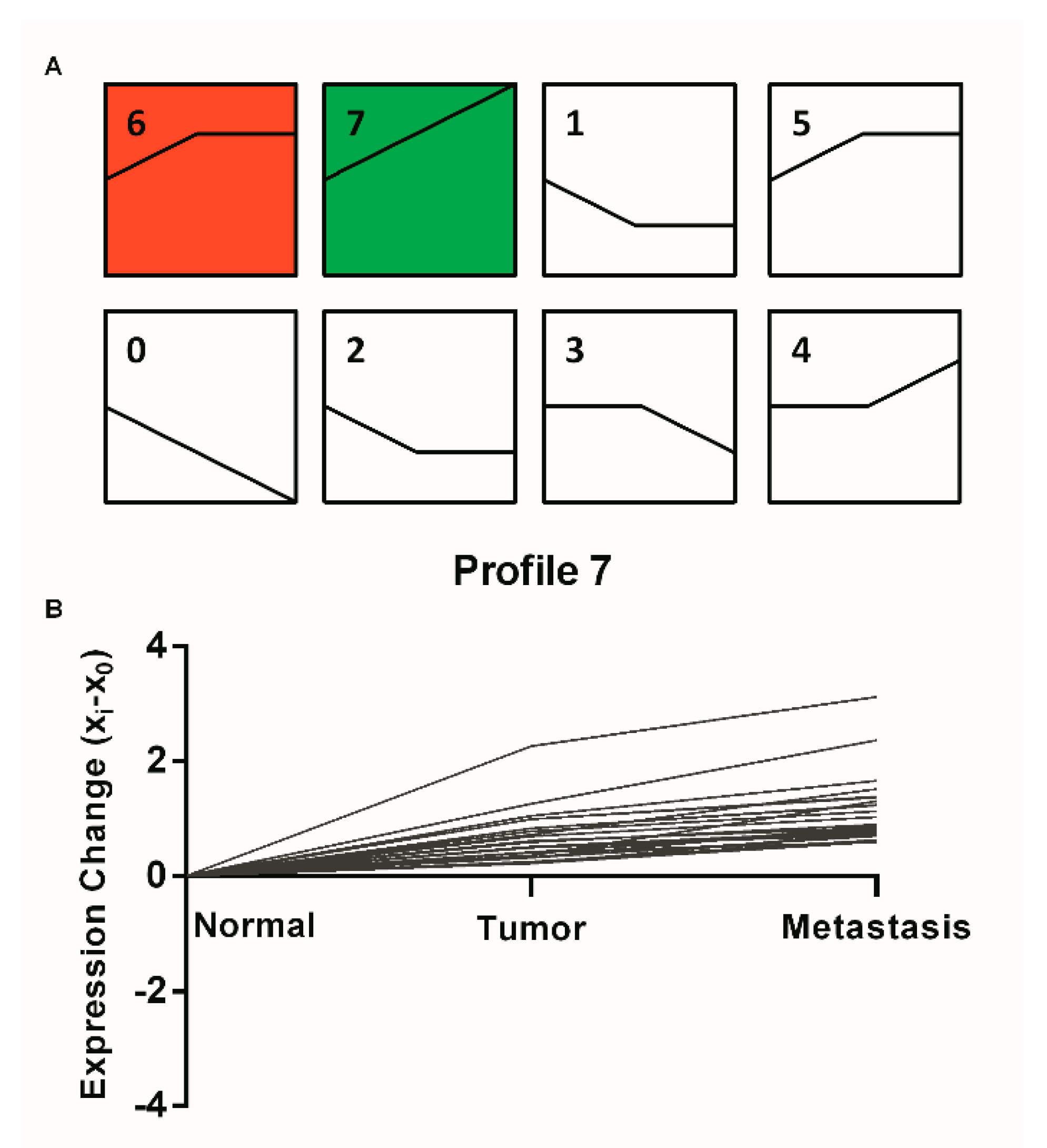
2.1. STEM Clustering on MiRNA Expression
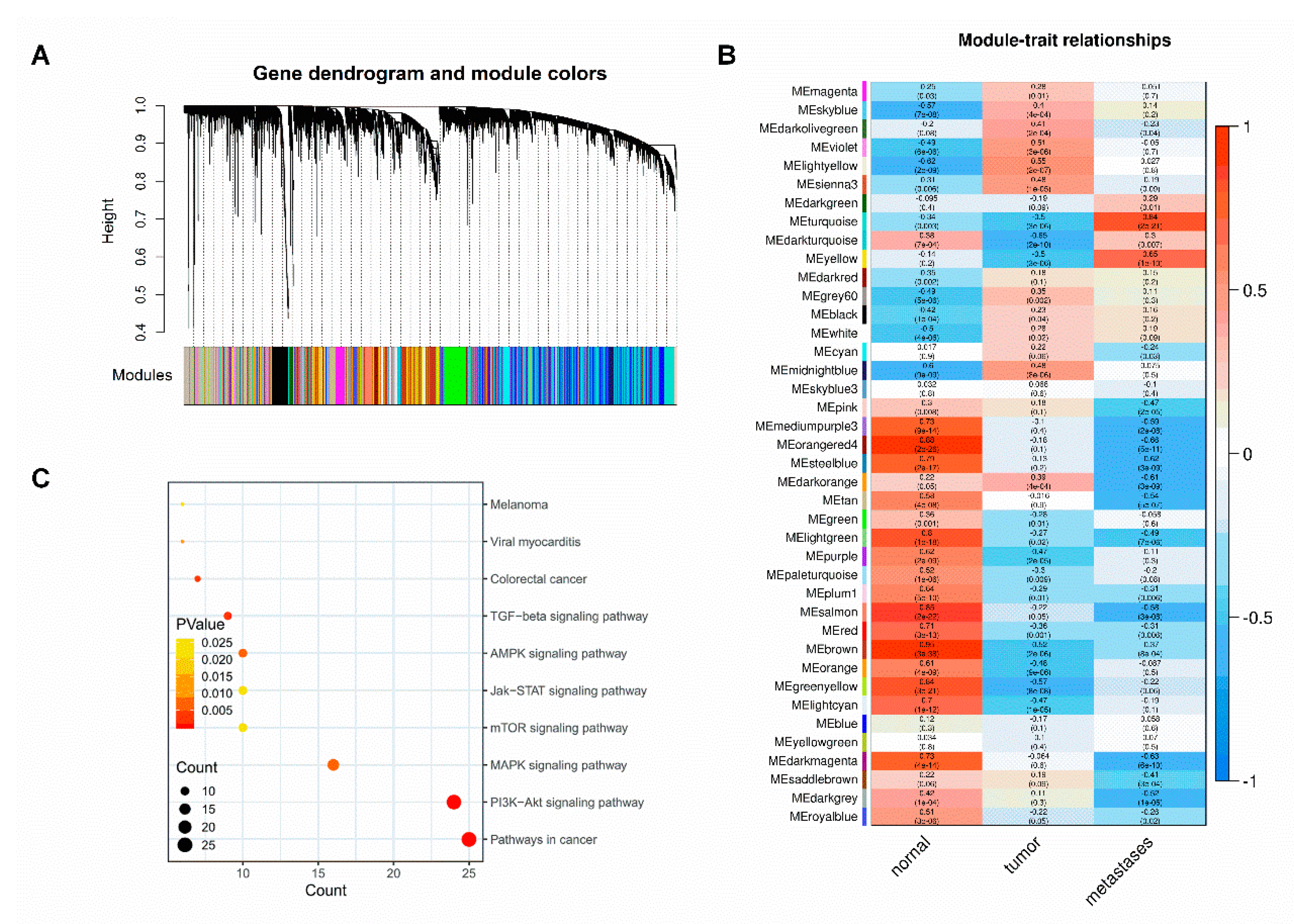
2.2. Identification of CRC Metastasis-Related Gene Modules and Pathways
2.3. Conjoint Analysis of MiRNA and mRNA Expression Profile in CRC.
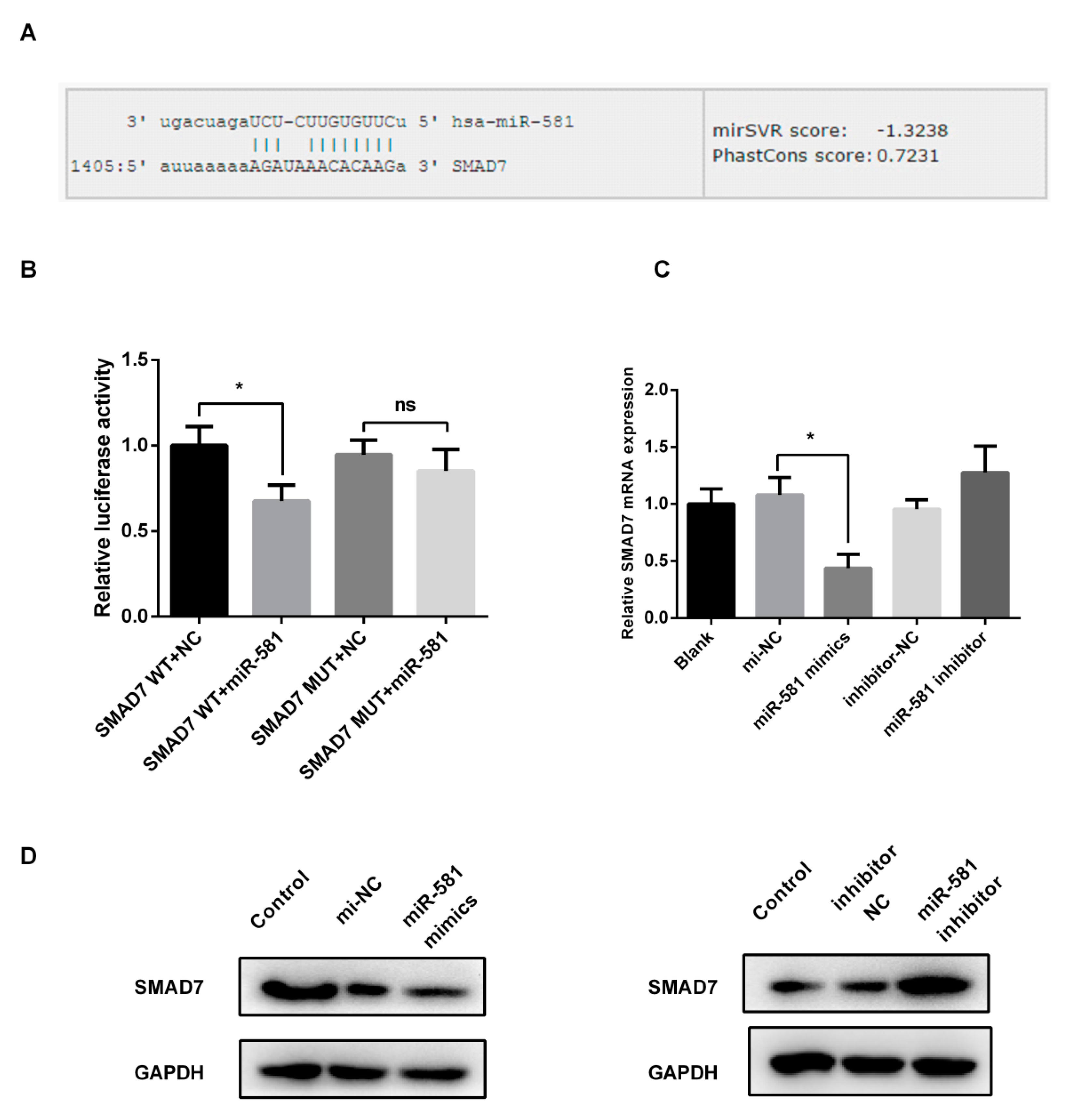
2.4. MiR-581 Tightly Binds to 3′-UTR of SMAD7
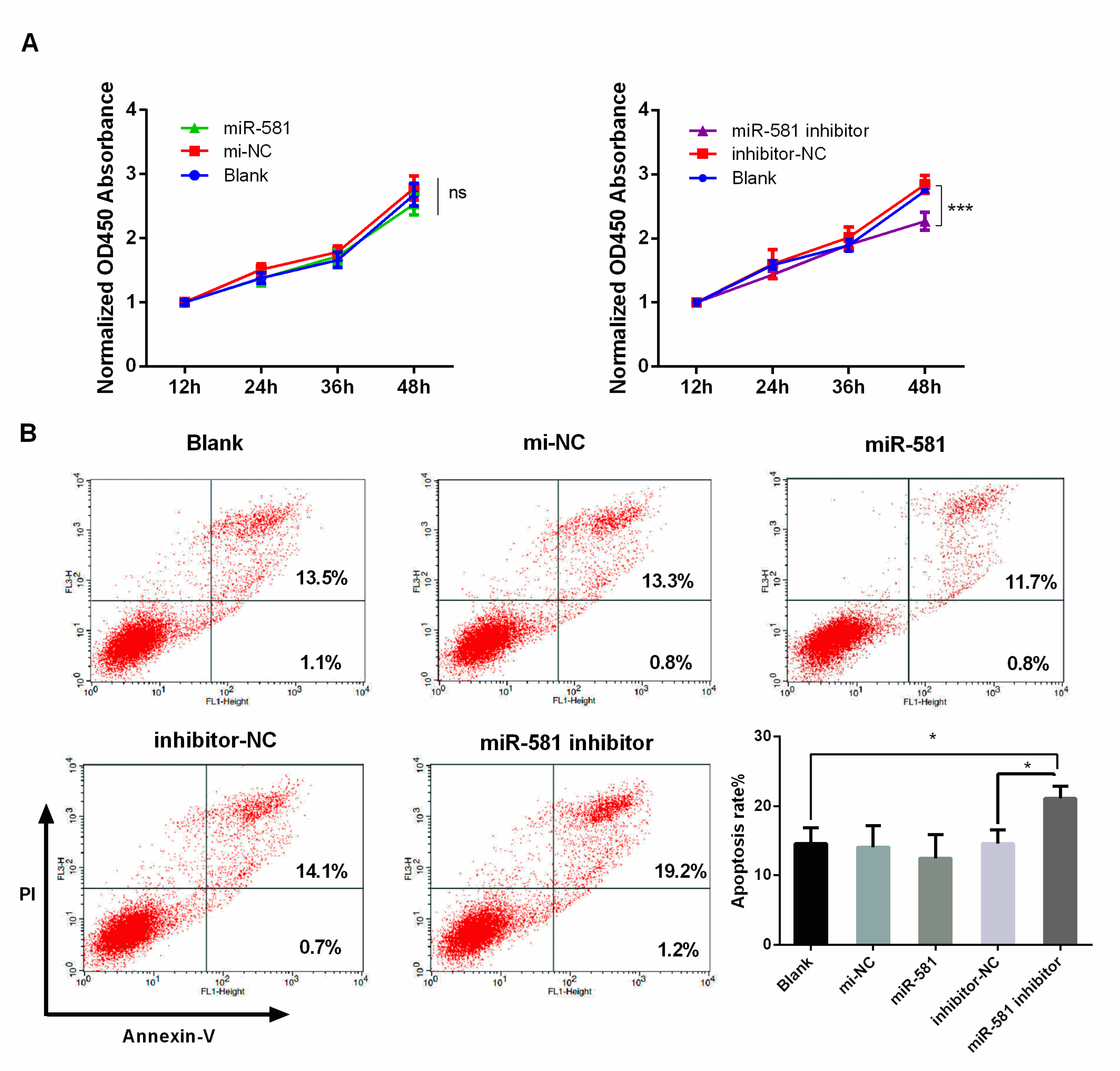
2.5. Decreased Level of MiR-581 Inhibited Cell Proliferation and Induced Apoptosis
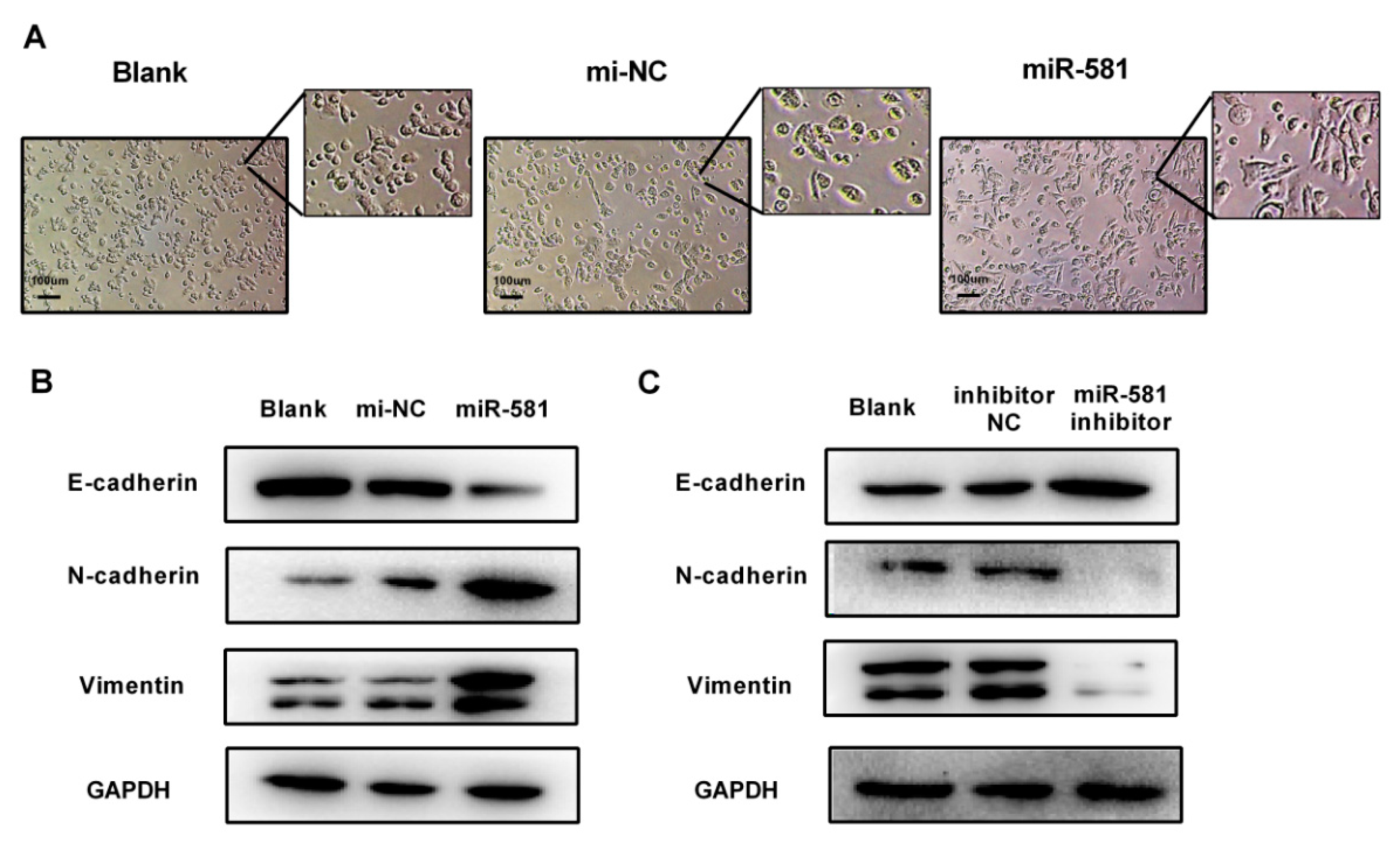
2.6. MiR-581 Promotes Cell Invasion Capacity and Modulates Epithelial to Mesenchymal Process
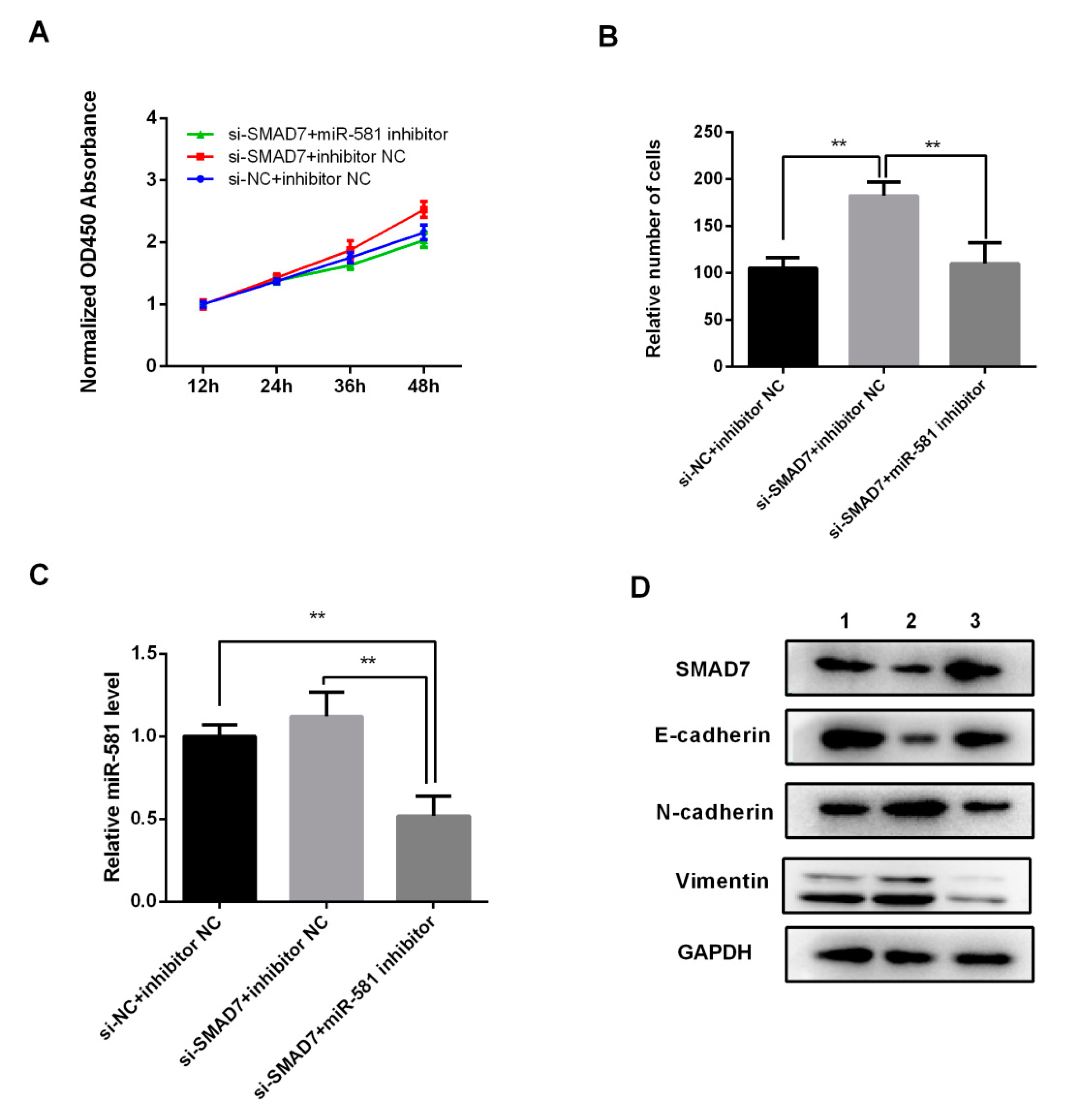
2.7. MiR-581 Induced Malignant Phenotype of CRC via SMAD7/TGFβ Pathway
3. Discussion
4. Materials and Methods
4.1. Affymetrix Microarray Data and Pre-Processing
4.2. STEM Analysis of MiRNA Expression Profile
4.3. Weighted Gene Coexpression Network Analysis
4.4. Cell Line and Culture
4.5. Transfection and Reagents
4.6. Dual Luciferase Reporter Assay
4.7. Western Blot
4.8. Cell Proliferation Assay
4.9. Cell Invasion Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, E.; Li, Q.; Wang, H.; Yang, F.; Min, L.; Yang, J. MiR-92a promotes tumorigenesis of colorectal cancer, a transcriptomic and functional based study. Biomed. Pharmacother. 2018, 106, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, S.D.; Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009, 361, 2449–2460. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Yang, F.; He, H.; Li, Q.; Zhang, W.; Xing, J.; Zhu, Z.; Jiang, J.; Wang, H.; Zhao, X.; et al. Alteration of tumor suppressor BMP5 in sporadic colorectal cancer: A genomic and transcriptomic profiling based study. Mol. Cancer 2018, 17, 176. [Google Scholar] [CrossRef]
- Ventura, A.; Jacks, T. MicroRNAs and cancer: Short RNAs go a long way. Cell 2009, 136, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Nicoloso, M.S.; Spizzo, R.; Shimizu, M.; Rossi, S.; Calin, G.A. MicroRNAs--The micro steering wheel of tumour metastases. Nat. Rev. Cancer 2009, 9, 293–302. [Google Scholar] [CrossRef]
- Wang, H.; Yan, B.; Zhang, P.; Liu, S.; Li, Q.; Yang, J.; Yang, F.; Chen, E. MiR-496 promotes migration and epithelial-mesenchymal transition by targeting RASSF6 in colorectal cancer. J. Cell. Physiol. 2020, 235, 1469–1479. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Chaudhry, M.A.; Omaruddin, R.A.; Brumbaugh, C.D.; Tariq, M.A.; Pourmand, N. Identification of radiation-induced microRNA transcriptome by next-generation massively parallel sequencing. J. Radiat. Res. 2013, 54, 808–822. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Zhao, S.; Sun, H.; Jiang, W.; Mi, Y.; Zhang, D.; Wen, Y.; Cheng, D.; Tang, H.; Wu, S.; Yu, Y.; et al. miR-4775 promotes colorectal cancer invasion and metastasis via the Smad7/TGFbeta-mediated epithelial to mesenchymal transition. Mol. Cancer 2017, 16, 12. [Google Scholar] [CrossRef]
- Tong, L.; Shen, S.; Huang, Q.; Fu, J.; Wang, T.; Pan, L.; Zhang, P.; Chen, G.; Huang, T.; Li, K.; et al. Proteasome-dependent degradation of Smad7 is critical for lung cancer metastasis. Cell Death Differ. 2019, 27, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, D.; Ren, M.; Zhao, Y.; Wang, X.; Chen, Y.; Liu, Y.; Lu, G.; He, S. Long non-coding RNA SNAI3-AS1 promotes the proliferation and metastasis of hepatocellular carcinoma by regulating the UPF1/Smad7 signalling pathway. J. Cell. Mol. Med. 2019, 23, 6271–6282. [Google Scholar] [CrossRef] [PubMed]
- Stolfi, C.; Marafini, I.; De Simone, V.; Pallone, F.; Monteleone, G. The dual role of Smad7 in the control of cancer growth and metastasis. Int. J. Mol. Sci. 2013, 14, 23774–23790. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Winter, E.; Stotz, M.; Eberhard, K.; Samonigg, H.; Lax, S.; Hoefler, G. Down-regulation of KRAS-interacting miRNA-143 predicts poor prognosis but not response to EGFR-targeted agents in colorectal cancer. Br. J. Cancer 2012, 106, 1826–1832. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Chen, M.; Peng, L.; Wang, X.; Ma, Q.; Ma, F.; Jiang, B. MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Mol. Cancer 2012, 11, 23. [Google Scholar] [CrossRef]
- Shirafkan, N.; Mansoori, B.; Mohammadi, A.; Shomali, N.; Ghasbi, M.; Baradaran, B. MicroRNAs as novel biomarkers for colorectal cancer: New outlooks. Biomed. Pharmacother. 2018, 97, 1319–1330. [Google Scholar] [CrossRef]
- Schepeler, T.; Holm, A.; Halvey, P.; Nordentoft, I.; Lamy, P.; Riising, E.M.; Christensen, L.L.; Thorsen, K.; Liebler, D.C.; Helin, K.; et al. Attenuation of the beta-catenin/TCF4 complex in colorectal cancer cells induces several growth-suppressive microRNAs that target cancer promoting genes. Oncogene 2012, 31, 2750–2760. [Google Scholar] [CrossRef]
- Guo, C.; Sah, J.F.; Beard, L.; Willson, J.K.; Markowitz, S.D.; Guda, K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 2008, 47, 939–946. [Google Scholar] [CrossRef]
- Wang, F.; Chang, Y.; Li, J.; Wang, H.; Zhou, R.; Qi, J.; Liu, J.; Zhao, Q. Strong correlation between ASPM gene expression and HCV cirrhosis progression identified by co-expression analysis. Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2017, 49, 70–76. [Google Scholar] [CrossRef]
- Yuan, L.; Qian, G.; Chen, L.; Wu, C.L.; Dan, H.C.; Xiao, Y.; Wang, X. Co-expression Network Analysis of Biomarkers for Adrenocortical Carcinoma. Front. Genet. 2018, 9, 328. [Google Scholar] [CrossRef]
- Nabatchian, F.; Rahimi Naiini, M.; Moradi, A.; Tabatabaeian, H.; Hoghoughi, N.; Azadeh, M.; Ghaedi, K. miR-581-Related Single Nucleotide Polymorphism, rs2641726, Located in MUC4 Gene, is Associated with Gastric Cancer Incidence. Indian J. Clin. Biochem. 2019, 34, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Katayama, Y.; Maeda, M.; Miyaguchi, K.; Nemoto, S.; Yasen, M.; Tanaka, S.; Mizushima, H.; Fukuoka, Y.; Arii, S.; Tanaka, H. Identification of pathogenesis-related microRNAs in hepatocellular carcinoma by expression profiling. Oncol. Lett. 2012, 4, 817–823. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, X.; Zeng, X. Shikonin suppresses progression and epithelial-mesenchymal transition in hepatocellular carcinoma (HCC) cells by modulating miR-106b/SMAD7/TGF-beta signaling pathway. Cell Biol. Int. 2019, 44, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, P.; Yuan, M.; Li, X. Overexpression of miRNA-21 Promotes the Proliferation and Invasion in Hepatocellular Carcinoma Cells via Suppressing SMAD7. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhang, N.; He, S. Transcription factor KLF4 modulates microRNA-106a that targets Smad7 in gastric cancer. Pathol. Res. Pract. 2019, 215, 152467. [Google Scholar] [CrossRef]
- Sim, W.J.; Iyengar, P.V. c-Met activation leads to the establishment of a TGFbeta-receptor regulatory network in bladder cancer progression. Nat. Commun. 2019, 10, 4349. [Google Scholar] [CrossRef]
- Zhou, T.; Yu, L.; Huang, J.; Zhao, X.; Li, Y.; Hu, Y.; Lei, Y. GDF10 inhibits proliferation and epithelial-mesenchymal transition in triple-negative breast cancer via upregulation of Smad7. Aging 2019, 11, 3298–3314. [Google Scholar] [CrossRef]
- Fu, Y.; Tang, Y.; Wang, J.; Guo, Z. MicroRNA-181c Suppresses the Biological Progression of Osteosarcoma via Targeting SMAD7 and Regulating Transforming Growth Factor-beta (TGF-beta) Signaling Pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 4801–4810. [Google Scholar]
- Fedorova, M.S.; Snezhkina, A.V.; Pudova, E.A.; Abramov, I.S.; Lipatova, A.V.; Kharitonov, S.L.; Sadritdinova, A.F.; Nyushko, K.M.; Klimina, K.M.; Belyakov, M.M.; et al. Upregulation of NETO2 gene in colorectal cancer. BMC Genet. 2017, 18 (Suppl. 1), 117. [Google Scholar] [CrossRef]
- Kamiza, A.B.; Wang, W.C.; You, J.F.; Tang, R.; Wang, Y.T.; Chien, H.T.; Lai, C.H.; Chiu, L.L.; Lo, T.P.; Hung, K.Y.; et al. EGFR, SMAD7, and TGFBR2 Polymorphisms Are Associated with Colorectal Cancer in Patients with Lynch Syndrome. Anticancer Res. 2018, 38, 5983–5990. [Google Scholar] [CrossRef]
- Zhai, W.; Li, S.; Zhang, J.; Chen, Y.; Ma, J.; Kong, W.; Gong, D.; Zheng, J.; Xue, W.; Xu, Y. Sunitinib-suppressed miR-452-5p facilitates renal cancer cell invasion and metastasis through modulating SMAD4/SMAD7 signals. Mol. Cancer 2018, 17, 157. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jiang, D. Hypoxia-responsive miRNA-21-5p inhibits Runx2 suppression by targeting SMAD7 in MC3T3-E1 cells. J. Cell. Biochem. 2019, 120, 16867–16875. [Google Scholar] [CrossRef] [PubMed]
- Pande, H.O.; Tesfaye, D.; Hoelker, M.; Gebremedhn, S.; Held, E.; Neuhoff, C.; Tholen, E.; Schellander, K.; Wondim, D.S. MicroRNA-424/503 cluster members regulate bovine granulosa cell proliferation and cell cycle progression by targeting SMAD7 gene through activin signalling pathway. J. Ovarian Res. 2018, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Li, W.; Wang, X.; Qi, S. MicroRNA-17-5p aggravates lipopolysaccharide-induced injury in nasal epithelial cells by targeting Smad7. BMC Cell Biol. 2018, 19, 1. [Google Scholar] [CrossRef]
- Halder, S.K.; Beauchamp, R.D.; Datta, P.K. Smad7 induces tumorigenicity by blocking TGF-beta-induced growth inhibition and apoptosis. Exp. Cell Res. 2005, 307, 231–246. [Google Scholar] [CrossRef]
- Huang, B.; Yu, H.; Li, Y.; Zhang, W.; Liu, X. Upregulation of long noncoding TNFSF10 contributes to osteoarthritis progression through the miR-376-3p/FGFR1 axis. J. Cell. Biochem. 2019, 120, 19610–19620. [Google Scholar] [CrossRef]
- Qu, Y.; Liao, Z.; Wang, X.; Zhang, J.; Liu, C. EFLDO sensitizes liver cancer cells to TNFSF10-induced apoptosis in a p53-dependent manner. Mol. Med. Rep. 2019, 19, 3799–3806. [Google Scholar] [CrossRef]
- Shi, Z.; Wu, D.; Tang, R.; Li, X.; Chen, R.; Xue, S.; Zhang, C.; Sun, X. Silencing of HMGA2 promotes apoptosis and inhibits migration and invasion of prostate cancer cells. J. Biosci. 2016, 41, 229–236. [Google Scholar] [CrossRef]
- Nittka, S.; Böhm, C.; Zentgraf, H.; Neumaier, M. The CEACAM1-mediated apoptosis pathway is activated by CEA and triggers dual cleavage of CEACAM1. Oncogene 2008, 27, 3721–3728. [Google Scholar] [CrossRef]
- Xu, J.; Liu, B.; Ma, S.; Zhang, J.; Ji, Y.; Xu, L.; Zhu, M.; Chen, S.; Wu, X.; Wu, D. Characterizing the Tumor Suppressor Role of CEACAM1 in Multiple Myeloma. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 45, 1631–1640. [Google Scholar] [CrossRef]
- Feng, F.B.; Qiu, H.Y. Effects of Artesunate on chondrocyte proliferation, apoptosis and autophagy through the PI3K/AKT/mTOR signaling pathway in rat models with rheumatoid arthritis. Biomed. Pharmacother. 2018, 102, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef]
- Chen, E.; Yang, F.; He, H.; Lei, L.; Liu, R.; Du, L.; Dong, J.; Wang, M.; Yang, J. Decreased level of RASSF6 in sporadic colorectal cancer and its anti-tumor effects both in vitro and in vivo. Oncotarget 2016, 7, 19813–19823. [Google Scholar] [CrossRef][Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Liu, S.; Yan, B.; Yang, J.; Chen, E. MiR-581/SMAD7 Axis Contributes to Colorectal Cancer Metastasis: A Bioinformatic and Experimental Validation-Based Study. Int. J. Mol. Sci. 2020, 21, 6499. https://doi.org/10.3390/ijms21186499
Zhao X, Liu S, Yan B, Yang J, Chen E. MiR-581/SMAD7 Axis Contributes to Colorectal Cancer Metastasis: A Bioinformatic and Experimental Validation-Based Study. International Journal of Molecular Sciences. 2020; 21(18):6499. https://doi.org/10.3390/ijms21186499
Chicago/Turabian StyleZhao, Xiaojuan, Shuzhen Liu, Bianbian Yan, Jin Yang, and Erfei Chen. 2020. "MiR-581/SMAD7 Axis Contributes to Colorectal Cancer Metastasis: A Bioinformatic and Experimental Validation-Based Study" International Journal of Molecular Sciences 21, no. 18: 6499. https://doi.org/10.3390/ijms21186499
APA StyleZhao, X., Liu, S., Yan, B., Yang, J., & Chen, E. (2020). MiR-581/SMAD7 Axis Contributes to Colorectal Cancer Metastasis: A Bioinformatic and Experimental Validation-Based Study. International Journal of Molecular Sciences, 21(18), 6499. https://doi.org/10.3390/ijms21186499




