Phylogenetic Utility of rRNA ITS2 Sequence-Structure under Functional Constraint
Abstract
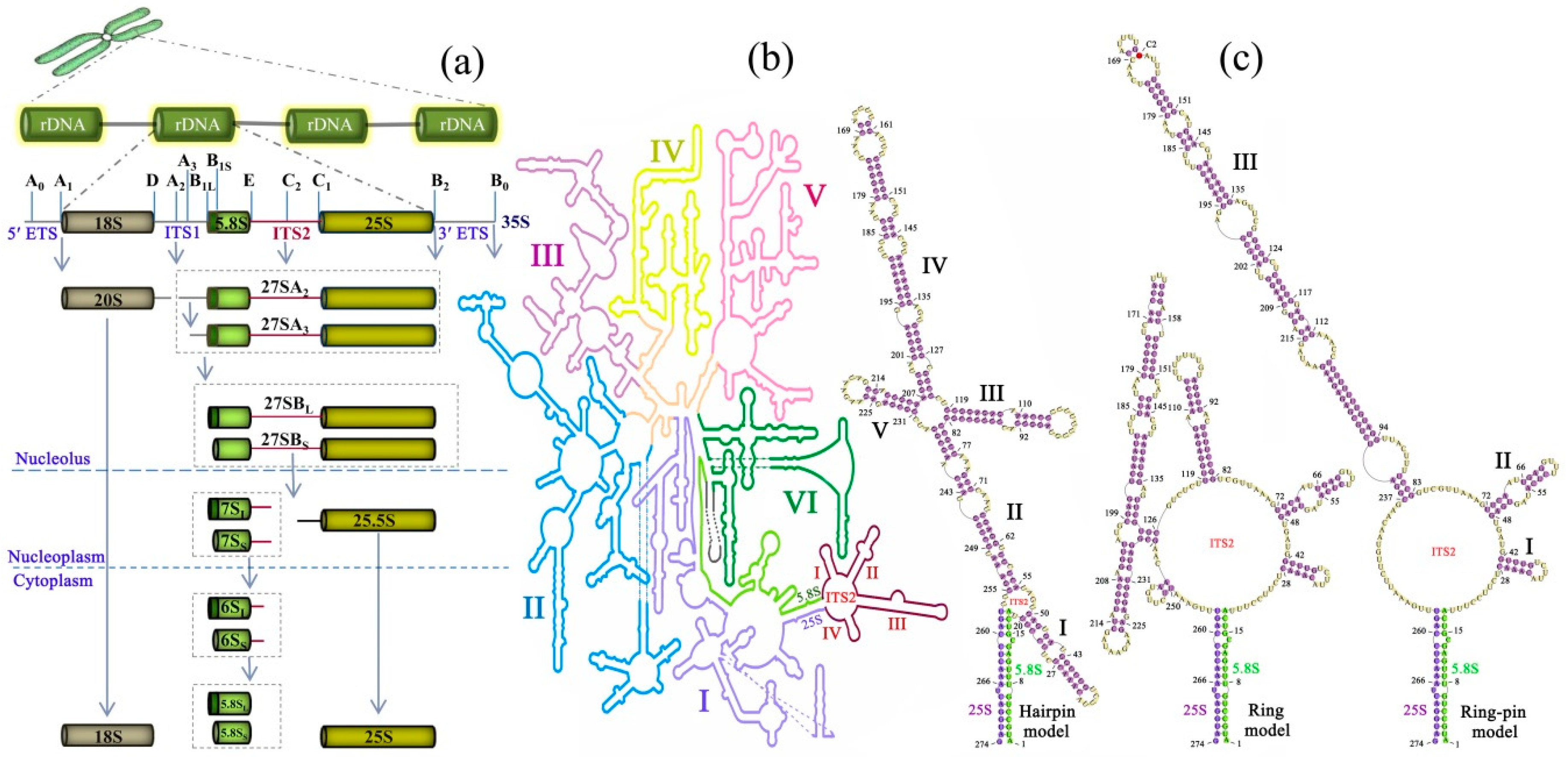
1. ITS2 Processing in Ribosome Biogenesis
2. ITS2 Function in Ribosome Biogenesis
3. Characteristics of ITS2 Secondary Structure
4. Dynamic Model of ITS2 Secondary Structure
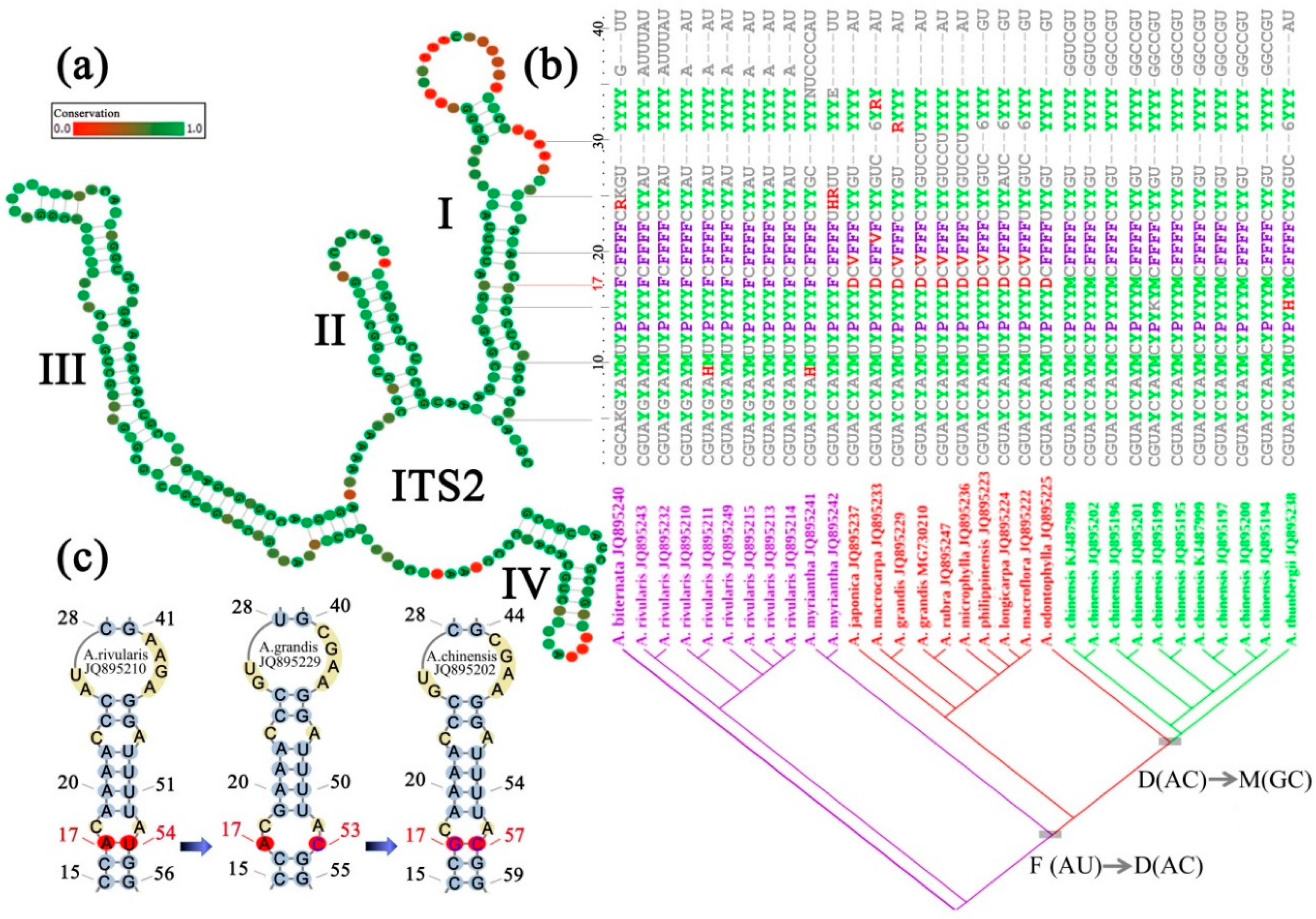
5. ITS2 Conserved Structure and Sequence-Structure Alignment
6. Structure-Guiding Alignment for Phylogeny
7. CBC in ITS2 Secondary Structure
8. CBCs Character Weighting
9. RNA Substitution Models
10. Species Delimitation Based on ITS2-CBC
11. Determining CBCs and Their Substitution Processes
12. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFs | assembly factors |
| cryo-EM | cryo–electron microscopy |
| CBC | compensatory base change |
| ETS | external spacer |
| ITS | internal transcribed spacers |
| LSU | large subunit |
| MEF | minimum free energy |
| RPs | ribosomal proteins |
| rRNA | ribosomal RNA |
| SSU | small subunit |
References
- Klinge, S.; Woolford, J.L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Bio. 2019, 20, 116–131. [Google Scholar] [CrossRef]
- Salim, D.; Bradford, W.D.; Freeland, A.; Cady, G.; Wang, J.; Pruitt, S.C.; Gerton, J.L. DNA replication stress restricts ribosomal DNA copy number. PLoS Genet. 2017, 13, e1007006. [Google Scholar] [CrossRef] [PubMed]
- Fromm, L.; Falk, S.; Flemming, D.; Schuller, J.M.; Thoms, M.; Conti, E.; Hurt, E. Reconstitution of the complete pathway of ITS2 processing at the pre-ribosome. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gasse, L.; Flemming, D.; Hurt, E. Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol. Cell 2015, 60, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Pillon, M.C.; Sobhany, M.; Borgnia, M.J.; Williams, J.G.; Stanley, R.E. Grc3 programs the essential endoribonuclease Las1 for specific RNA cleavage. Proc. Natl. Acad. Sci. USA 2017, 114, 5530–5538. [Google Scholar] [CrossRef]
- Thomson, E.; Tollervey, D. The final step in 5.8 S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol. Cell Biol. 2010, 30, 976–984. [Google Scholar] [CrossRef]
- Burlacu, E.; Lackmann, F.; Aguilar, L.C.; Belikov, S.; van Nues, R.; Trahan, C.; Hector, R.D.; Dominelli-Whiteley, N.; Cockroft, S.L.; Wieslander, L.; et al. High-throughput RNA structure probing reveals critical folding events during early 60S ribosome assembly in yeast. Nat. Commun. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Biedka, S.; Micic, J.; Wilson, D.; Brown, H.; Diorio-Toth, L.; Woolford, J.L. Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre-60S ribosomes. J. Cell Biol. 2018, 217, 2503–2518. [Google Scholar] [CrossRef]
- Baßler, J.; Hurt, E. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 2019, 88, 281–306. [Google Scholar] [CrossRef]
- Wu, S.; Tutuncuoglu, B.; Yan, K.; Brown, H.; Zhang, Y.; Tan, D.; Gamalinda, M.; Yuan, Y.; Li, Z.; Jakovljevic, J.; et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 2016, 534, 133–137. [Google Scholar] [CrossRef]
- Sanghai, Z.A.; Miller, L.; Molloy, K.R.; Barandun, J.; Hunziker, M.; Chaker-Margot, M.; Wang, J.J.; Chait, B.T.; Klinge, S. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 2018, 556, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Côté, C.A.; Peculis, B.A. Role of the ITS2-proximal stem and evidence for indirect recognition of processing sites in pre-rRNA processing in yeast. Nucleic Acids Res. 2001, 29, 2106–2116. [Google Scholar] [CrossRef] [PubMed]
- Woolford, J.L.; Baserga, S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013, 195, 643–681. [Google Scholar] [CrossRef] [PubMed]
- Kofler, L.; Prattes, M.; Bergler, H. From snapshots to flipbook—Resolving the dynamics of ribosome biogenesis with chemical probes. Int. J. Mol. Sci. 2020, 21, 2998. [Google Scholar] [CrossRef]
- Yeh, L.C.C.; Lee, J.C. Structural analysis of the internal transcribed spacer 2 of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol. 1990, 211, 699–712. [Google Scholar] [CrossRef]
- van der Sande, C.A.; Kwa, M.; van Nues, R.W.; van Heerikhuizen, H.; Raué, H.A.; Planta, R.J. Functional analysis of internal transcribed spacer 2 of Saccharomyces cerevisiae ribosomal DNA. J. Mol. Biol. 1992, 223, 899–910. [Google Scholar] [CrossRef]
- van Nues, R.W.; Rientjes, J.M.; Morré, S.A.; Mollee, E.; Planta, R.J.; Venema, J.; Raué, H.A. Evolutionarily conserved structural elements are critical for processing of internal transcribed spacer 2 from Saccharomyces cerevisiae precursor ribosomal RNA. J. Mol. Biol. 1995, 250, 24–36. [Google Scholar] [CrossRef]
- Ankenbrand, M.J.; Keller, A.; Wolf, M.; Schultz, J.; Förster, F. ITS2 database V: Twice as much. Mol. Biol. Evol. 2015, 32, 3030–3032. [Google Scholar] [CrossRef]
- Wolf, M.; Achtziger, M.; Schultz, J.; Dandekar, T.; Müller, T. Homology modeling revealed more than 20,000 rRNA internal transcribed spacer 2 (ITS2) secondary structures. RNA 2005, 11, 1616–1623. [Google Scholar] [CrossRef]
- Joseph, N.; Krauskopf, E.; Vera, M.I.; Michot, B. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res. 1999, 27, 4533–4540. [Google Scholar] [CrossRef]
- Coleman, A.W. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 2003, 19, 370–375. [Google Scholar] [CrossRef]
- Schultz, J.; Maisel, S.; Gerlach, D.; Müller, T.; Wolf, M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007, 35, 3322–3329. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends Genet. 2015, 31, 157–163. [Google Scholar] [CrossRef]
- Côté, C.A.; Greer, C.L.; Peculis, B.A. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA 2002, 8, 786–797. [Google Scholar]
- Simmons, M.P.; Ochoterena, H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 2000, 49, 369–381. [Google Scholar] [CrossRef]
- Will, S.; Reiche, K.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 2007, 3, e65. [Google Scholar] [CrossRef]
- Tan, Z.; Fu, Y.; Sharma, G.; Mathews, D.H. TurboFold II: RNA structural alignment and secondary structure prediction informed by multiple homologs. Nucleic Acids Res. 2017, 45, 11570–11581. [Google Scholar] [CrossRef]
- Seemann, S.E.; Mirza, A.H.; Hansen, C.; Bang-Berthelsen, C.H.; Garde, C.; Christensen-Dalsgaard, M.; Torarinsson1, E.; Yao, Z.Z.; Workman, C.T.; Pociot, F.; et al. The identification and functional annotation of RNA structures conserved in vertebrates. Genome Res. 2017, 27, 1371–1383. [Google Scholar] [CrossRef]
- Glouzon, J.P.S.; Ouangraoua, A. aliFreeFold: An alignment-free approach to predict secondary structure from homologous RNA sequences. Bioinformatics 2018, 34, 70–78. [Google Scholar] [CrossRef]
- Selig, C.; Wolf, M.; Müller, T.; Dandekar, T.; Schultz, J. The ITS2 Database II: Homology modelling RNA structure for molecular systematics. Nucleic Acids Res. 2007, 36, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Seibel, P.N.; Müller, T.; Dandekar, T.; Schultz, J.; Wolf, M. 4SALE–a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform. 2006, 7, 1–7. [Google Scholar]
- Wolf, M.; Koetschan, C.; Mueller, T. ITS2, 18S, 16S or any other RNA—Simply aligning sequences and their individual secondary structures simultaneously by an automatic approach. Gene 2014, 546, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Bernier, C.R.; Petrov, A.S.; Kovacs, N.A.; Penev, P.I.; Williams, L.D. Translation: The universal structural core of life. Mol. Biol. Evol. 2018, 35, 2065–2076. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.A. Multiple sequence alignment for phylogenetic purposes. Aust. Syst. Bot. 2006, 19, 479–539. [Google Scholar] [CrossRef]
- Cvicek, V.; Goddard III, W.A.; Abrol, R. Structure-based sequence alignment of the transmembrane domains of all human GPCRs: Phylogenetic, structural and functional implications. PLoS Comput. Biol. 2016, 12, e1004805. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Yao, H.; Song, J.; Liu, C.; Luo, K.; Han, J.; Li, Y.; Pang, X.; Xu, H.; Zhu, Y.; Xiao, P.; et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE 2010, 5, e13102. [Google Scholar] [CrossRef]
- Qin, Y.; Li, M.; Cao, Y.; Gao, Y.; Zhang, W. Molecular thresholds of ITS2 and their implications for molecular evolution and species identification in seed plants. Sci. Rep-UK 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, S.; Zhao, H.; Huang, L. Using the ITS2 sequence-structure as a DNA mini-barcode: A case study in authenticating the traditional medicine “Fang Feng”. Biochem. Syst. Ecol. 2016, 69, 188–194. [Google Scholar] [CrossRef]
- Zhang, W.; Yuan, Y.; Yang, S.; Huang, J.; Huang, L. ITS2 secondary structure improves discrimination between medicinal “Mu Tong” species when using DNA barcoding. PLoS ONE 2015, 10, e0131185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cao, Y.; Zhang, W.; Simmons, M.P. Adenine· cytosine substitutions are an alternative pathway of compensatory mutation in angiosperm ITS2. RNA 2020, 26, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, H.; Zhao, F.; Jiang, L.; Peng, H.; Zhang, W.; Simmons, M.P. Alternative analyses of compensatory base changes in an ITS2 phylogeny of Corydalis (Papaveraceae). Ann. Bot. 2019, 124, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Antczak, M.; Zablocki, M.; Zok, T.; Rybarczyk, A.; Blazewicz, J.; Szachniuk, M. RNAvista: A webserver to assess RNA secondary structures with non-canonical base pairs. Bioinformatics 2019, 35, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Masquida, B.; Westhof, E. On the wobble GoU and related pairs. RNA 2000, 6, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Savill, N.J.; Hoyle, D.C.; Higgs, P.G. RNA sequence evolution with secondary structure constraints: Comparison of substitution rate models using maximum-likelihood methods. Genetics 2001, 157, 399–411. [Google Scholar]
- Golden, M.; Murrell, B.; Martin, D.; Pybus, O.G.; Hein, J. Evolutionary analyses of base-pairing interactions in DNA and RNA secondary structures. Mol. Biol. Evol. 2020, 37, 576–592. [Google Scholar] [CrossRef]
- Wheeler, W.C.; Honeycutt, R.L. Paired sequence difference in ribosomal RNAs: Evolutionary and phylogenetic implications. Mol. Biol. Evol. 1988, 5, 90–96. [Google Scholar]
- Dixon, M.T.; Hillis, D.M. Ribosomal RNA secondary structure: Compensatory mutations and implications for phylogenetic analysis. Mol. Biol. Evol. 1993, 10, 256–267. [Google Scholar]
- Gutell, R.R.; Cannone, J.J.; Shang, Z.; Du, Y.; Serra, M.J. A story: Unpaired adenosine bases in ribosomal RNAs. J. Mol. Biol. 2000, 304, 335–354. [Google Scholar] [CrossRef]
- Allen, J.E.; Whelan, S. Assessing the state of substitution models describing noncoding RNA evolution. Genome Biol. Evol. 2014, 6, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 2000, 151, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 2009, 50, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Philippi, N.; Dandekar, T.; Schultz, J.; Wolf, M. Distinguishing species. RNA 2007, 13, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Chen, S.; Song, J.; Ankenbrand, M.; Müller, T. Compensatory base changes in ITS2 secondary structures correlate with the biological species concept despite intragenomic variability in ITS2 sequences—A proof of concept. PLoS ONE 2013, 8, e66726. [Google Scholar] [CrossRef]
- Lim, H.C.; Teng, S.T.; Leaw, C.P.; Lim, P.T. Three novel species in the Pseudo-nitzschia pseudodelicatissima complex: P. batesiana sp. nov.; P. lundholmiae sp. nov.; and P. fukuyoi sp. nov. (Bacillariophyceae) from the Strait of Malacca, Malaysia. J. Phycol. 2013, 49, 902–916. [Google Scholar]
- Samanta, B.; Ehrman, J.M.; Kaczmarska, I. A consensus secondary structure of ITS2 for the diatom Order Cymatosirales (Mediophyceae, Bacillariophyta) and reappraisal of the order based on DNA, morphology, and reproduction. Mol. Phylogenet. Evol. 2018, 129, 117–129. [Google Scholar] [CrossRef]
- Sundaresan, N.; Jagan, E.G.; Kathamuthu, G.; Pandi, M. Internal transcribed spacer 2 (ITS2) molecular morphometric analysis based species delimitation of foliar endophytic fungi from Aglaia elaeagnoidea, Flacourtia inermis and Premna serratifolia. PLoS ONE 2019, 14, e0215024. [Google Scholar] [CrossRef]
- Caisová, L.; Marin, B.; Melkonian, M. A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist 2013, 164, 482–496. [Google Scholar] [CrossRef]
- Caisová, L.; Marin, B.; Melkonian, M. A close-up view on ITS2 evolution and speciation-a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evol. Biol. 2011, 11, 262. [Google Scholar] [CrossRef]
- Will, S.; Joshi, T.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs. RNA 2012, 18, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Fitch, W.M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Biol. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Wolf, M.; Friedrich, J.; Dandekar, T.; Müller, T. CBCAnalyzer: Inferring phylogenies based on compensatory base changes in RNA secondary structures. Silico Biol. 2005, 5, 291–294. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Tian, W.; Gao, Z.; Wang, G.; Zhao, H. Phylogenetic Utility of rRNA ITS2 Sequence-Structure under Functional Constraint. Int. J. Mol. Sci. 2020, 21, 6395. https://doi.org/10.3390/ijms21176395
Zhang W, Tian W, Gao Z, Wang G, Zhao H. Phylogenetic Utility of rRNA ITS2 Sequence-Structure under Functional Constraint. International Journal of Molecular Sciences. 2020; 21(17):6395. https://doi.org/10.3390/ijms21176395
Chicago/Turabian StyleZhang, Wei, Wen Tian, Zhipeng Gao, Guoli Wang, and Hong Zhao. 2020. "Phylogenetic Utility of rRNA ITS2 Sequence-Structure under Functional Constraint" International Journal of Molecular Sciences 21, no. 17: 6395. https://doi.org/10.3390/ijms21176395
APA StyleZhang, W., Tian, W., Gao, Z., Wang, G., & Zhao, H. (2020). Phylogenetic Utility of rRNA ITS2 Sequence-Structure under Functional Constraint. International Journal of Molecular Sciences, 21(17), 6395. https://doi.org/10.3390/ijms21176395




