Hepatitis B Virus preS/S Truncation Mutant rtM204I/sW196* Increases Carcinogenesis through Deregulated HIF1A, MGST2, and TGFbi
Abstract
1. Introduction
2. Results
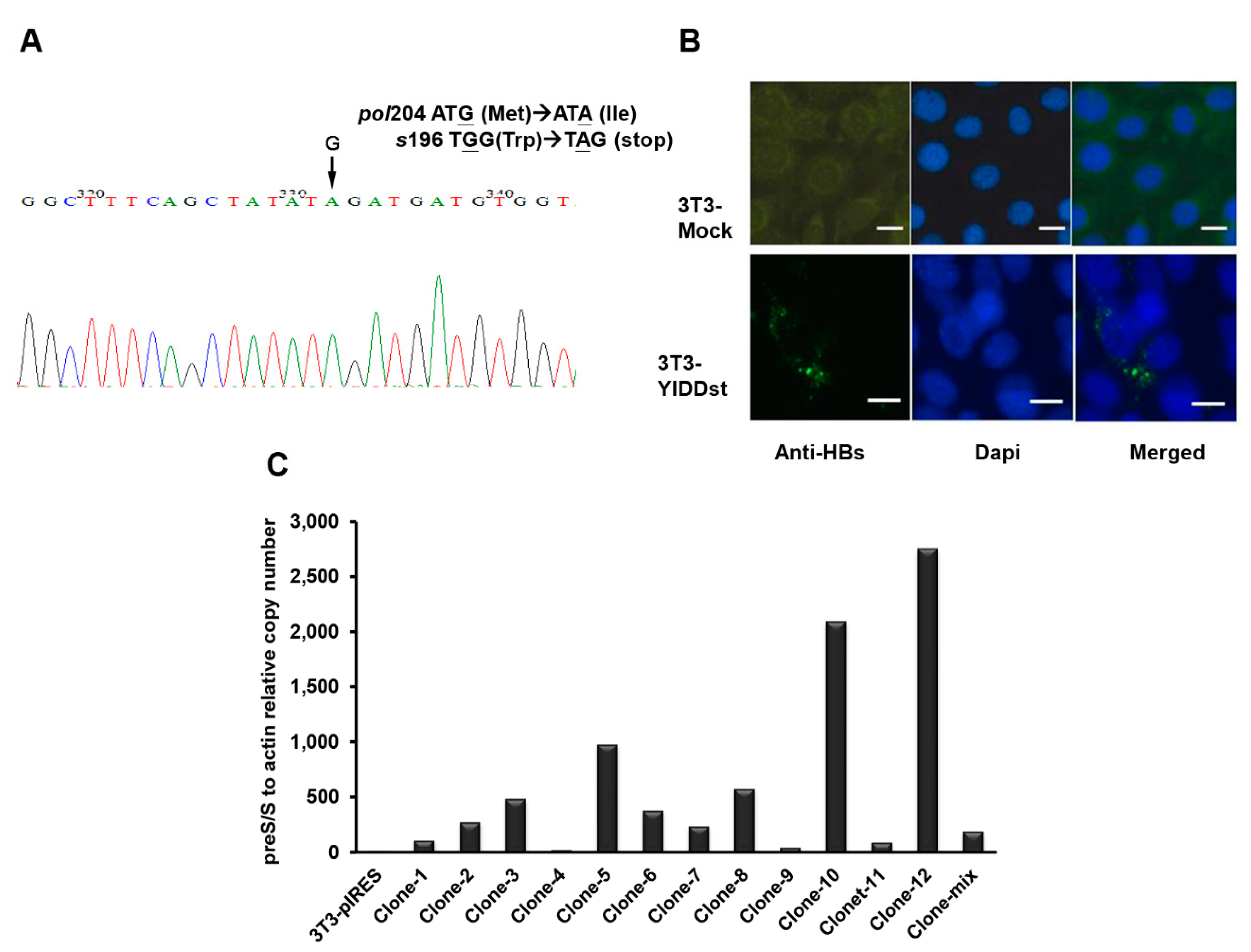
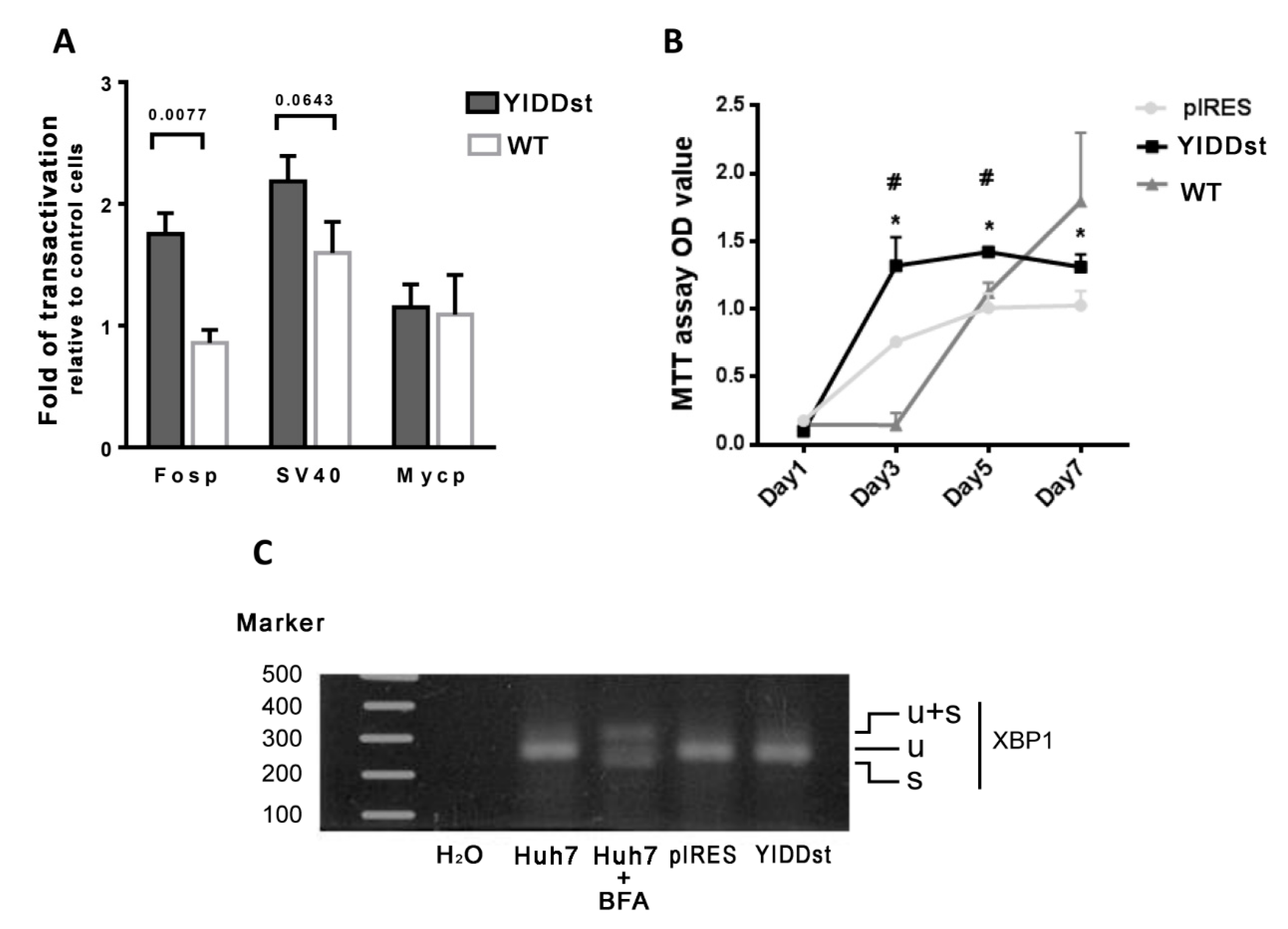
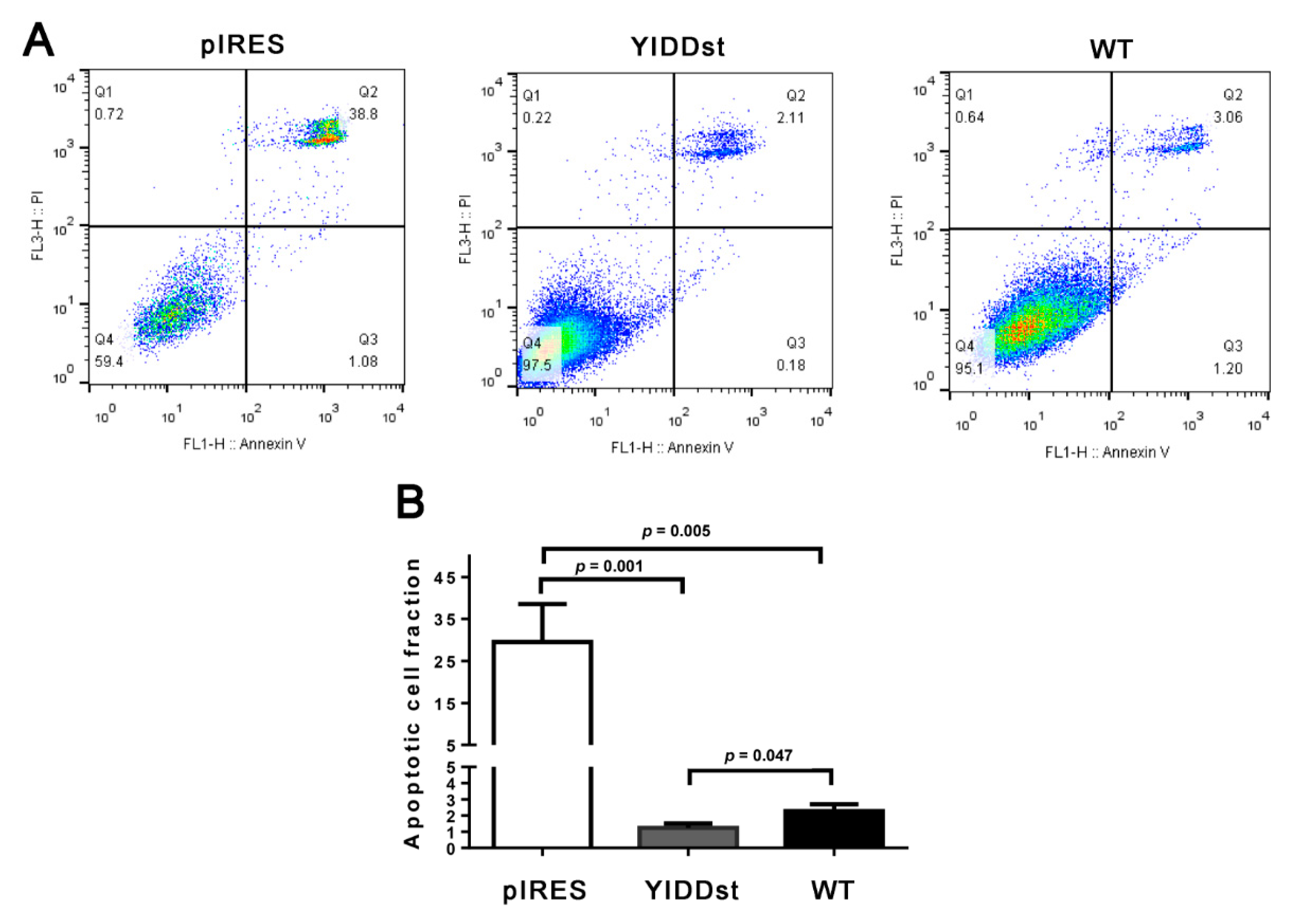
2.1. rtM204I/sW196* Resulted in an Enhanced Transactivation of Proto-Oncogenes, Increased Cell Proliferation, and Decreased Apoptosis, but Did Not Elicit an Endoplasmic Reticulum (ER) Stress Response
2.2. rtM204I/sW196* Resulted in More Anchorage-Independent Growth and Enhanced Mouse Xenograft Tumorigenesis
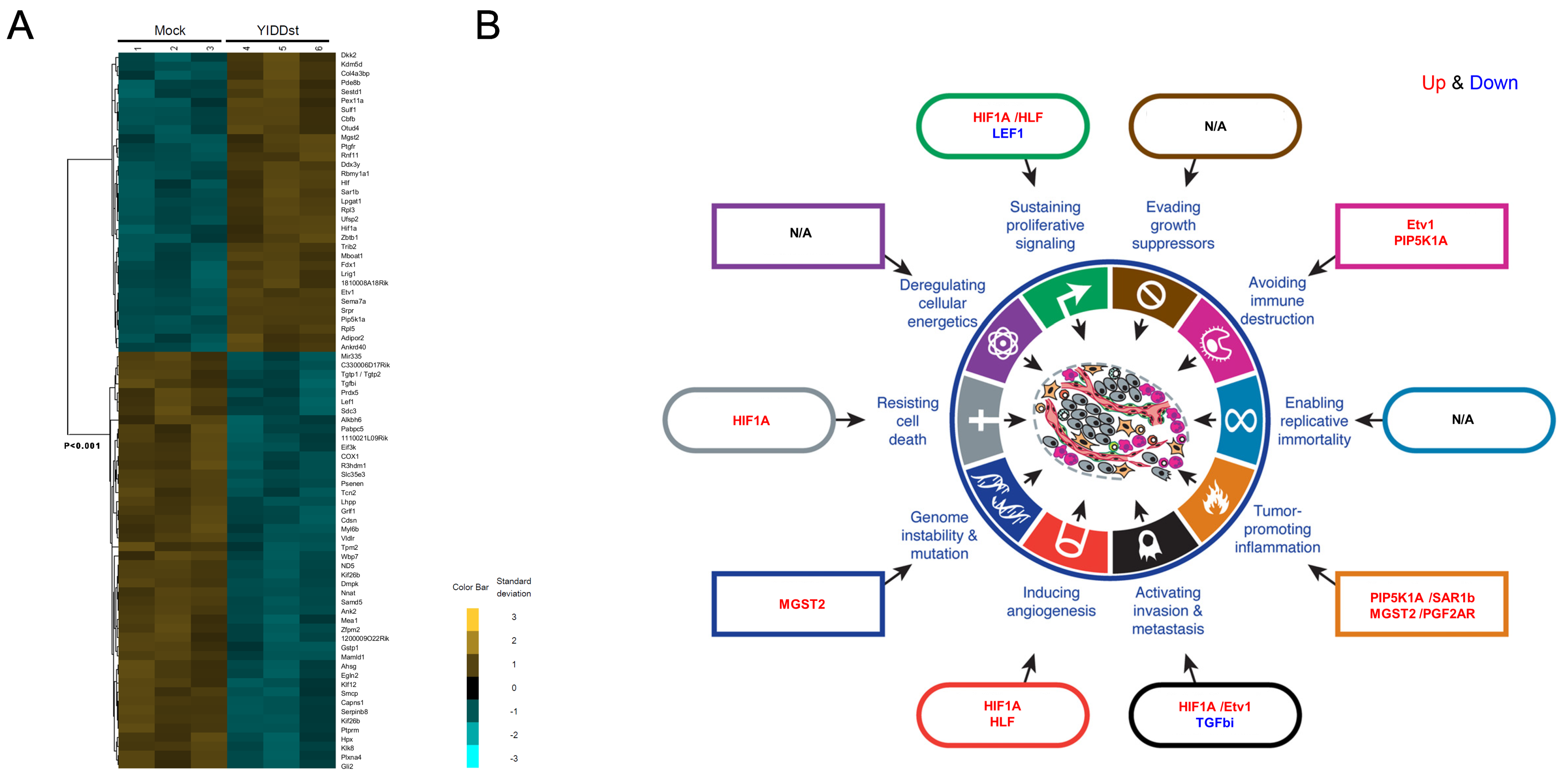
2.3. Genome-Wide Exploration of Expression Alterations in YIDDst-Transformed Cells
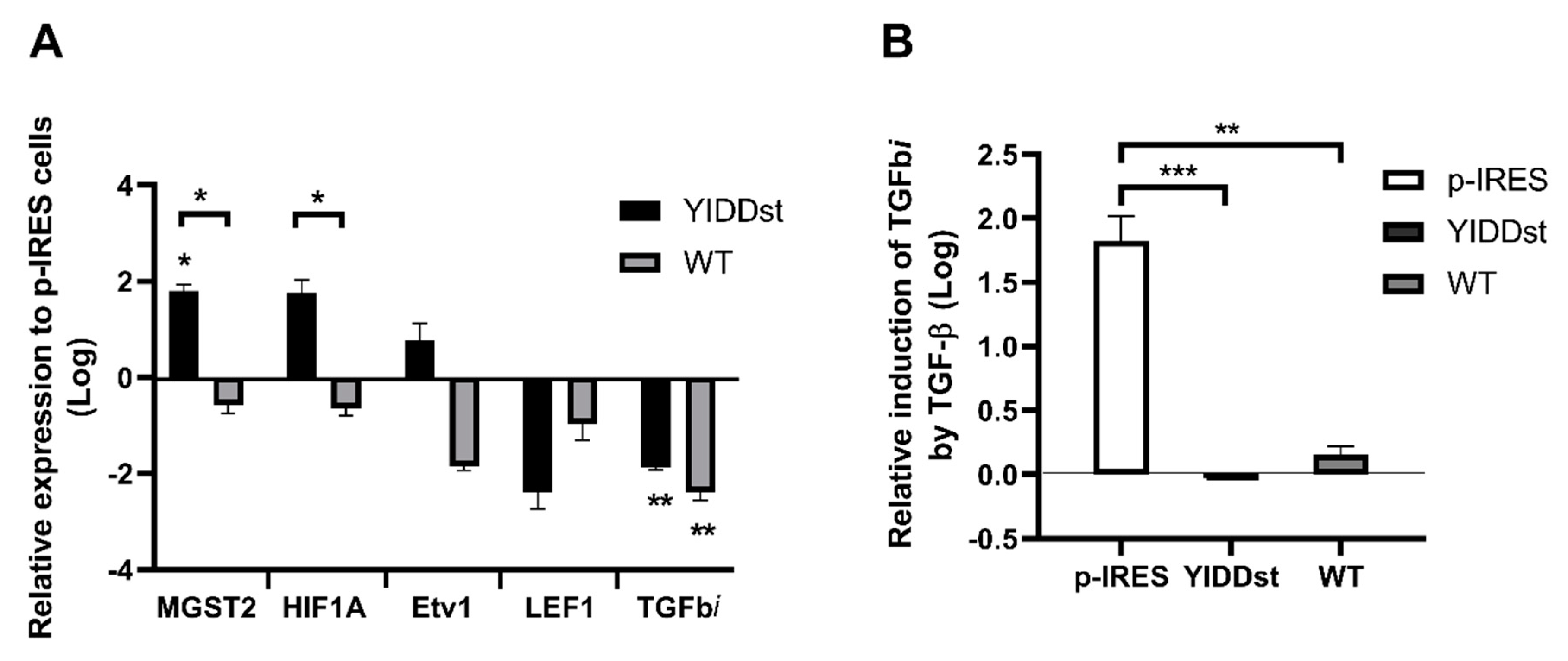
2.4. Validation of the Differentially Expressed Transcripts in Transformants with WT, Mutant, or Mock-Control by RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction and Site-Directed Mutagenesis
4.2. Cell Culture, Plasmid Transfection, and Immunofluorescence Analysis
4.3. Transactivation Analysis with Dual-Luciferase Assay
4.4. Cell Proliferation Assay
4.5. FITC Annexin V Apoptosis Assay by Flow Cytometry
4.6. Assessment of ER Stress Response
4.7. Anchorage-Independent Growth by Soft Agar Assay
4.8. Mouse Xenograft Tumorigenicity Assay
4.9. cDNA Microarray
4.10. Real-Time RT-PCR Validation of Up- and Down-Regulated Genes
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NA | nucleos(t)ide analog |
| rt | reverse-transcriptase |
| ER | endoplasmic reticulum |
| RT-qPCR | reverse transcription quantitative polymerase chain reaction |
| XBP1 | X box-binding protein-1 |
| MGST2 | microsomal glutathione s-transferase 2 |
| HIF1A | hypoxia-inducible factor 1 subunit alpha |
| TGFbi | transforming growth factor beta-induced |
| 3TC | lamivudine |
| HBV | hepatitis B virus |
| ALT | alanine aminotransferases |
| HBeAg | hepatitis e antigen |
| HCC | hepatocellular carcinoma |
| CHB | chronic hepatitis B |
| MTT | 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| HLF | hepatic leukemia factor |
| LEF | lymphoid enhancer-binding factor |
| Etv1 | ETS variant transcription factor 1 |
| PIP5K1A | phosphatidylinositol-4-phosphate 5-kinase type 1 alpha |
| SAR1b | secretion associated Ras-related GTPase 1B |
| PGF2AR | prostaglandin F 2 alpha receptor |
| LTC4 | leukotriene C4 |
References
- Lai, C.L.; Chien, R.N.; Leung, N.W.; Chang, T.T.; Guan, R.; Tai, D.I.; Ng, K.Y.; Wu, P.C.; Dent, J.C.; Barber, J.; et al. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N. Engl. J. Med. 1998, 339, 61–68. [Google Scholar] [CrossRef]
- Liaw, Y.F.; Leung, N.W.; Chang, T.T.; Guan, R.; Tai, D.I.; Ng, K.Y.; Chien, R.N.; Dent, J.; Roman, L.; Edmundson, S.; et al. Effects of extended lamivudine therapy in Asian patients with chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. Gastroenterology 2000, 119, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Leung, N.W.; Lai, C.L.; Chang, T.T.; Guan, R.; Lee, C.M.; Ng, K.Y.; Lim, S.G.; Wu, P.C.; Dent, J.C.; Edmundson, S. Extended lamivudine treatment in patients with chronic hepatitis B enhances hepatitis B e antigen seroconversion rates: Results after 3 years of therapy. Hepatology 2001, 33, 1527–1532. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.T.; Lai, C.L.; Chien, R.N.; Guan, R.; Lim, S.G.; Lee, C.M.; Ng, K.Y.; Nicholls, G.J.; Dent, J.C.; Leung, N.W. Four years of lamivudine treatment in Chinese patients with chronic hepatitis B. J. Gastroenterol. Hepatol. 2004, 19, 1276–1282. [Google Scholar] [CrossRef] [PubMed]
- Liaw, Y.F. Results of lamivudine trials in Asia. J. Hepatol. 2003, 39, 111–115. [Google Scholar] [CrossRef]
- Liaw, Y.F.; Sung, J.J.; Chow, W.C.; Farrell, G.; Lee, C.Z.; Yuen, H.; Tanwandee, T.; Tao, Q.M.; Shue, K.; Keene, O.N.; et al. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N. Engl. J. Med. 2004, 351, 1521–1531. [Google Scholar] [CrossRef]
- Papatheodoridis, G.V.; Dimou, E.; Dimakopoulos, K.; Manolakopoulos, S.; Rapti, I.; Kitis, G.; Tzourmakliotis, D.; Manesis, E.; Hadziyannis, S.J. Outcome of hepatitis B e antigen-negative chronic hepatitis B on long-term nucleos(t)ide analog therapy starting with lamivudine. Hepatology 2005, 42, 121–129. [Google Scholar] [CrossRef]
- Hosaka, T.; Suzuki, F.; Kobayashi, M.; Seko, Y.; Kawamura, Y.; Sezaki, H.; Akuta, N.; Suzuki, Y.; Saitoh, S.; Arase, Y.; et al. Long-term entecavir treatment reduces hepatocellular carcinoma incidence in patients with hepatitis B virus infection. Hepatology 2013, 58, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Lin, J.T.; Ho, H.J.; Su, C.W.; Lee, T.Y.; Wang, S.Y.; Wu, C.; Wu, J.C. Association of nucleos(t)ide analogue therapy with reduced risk of hepatocellular carcinoma in patients with chronic hepatitis B—A nationwide cohort study. Gastroenterology 2014, 147, 143–151. [Google Scholar] [CrossRef]
- Andreone, P.; Gramenzi, A.; Cursaro, C.; Biselli, M.; Cammà, C.; Trevisani, F.; Bernardi, M. High risk of hepatocellular carcinoma in anti-HBe positive liver cirrhosis patients developing lamivudine resistance. J. Viral. Hepat. 2004, 11, 439–442. [Google Scholar] [CrossRef]
- Sung, J.J.Y.; Tsoi, K.K.F.; Wong, V.W.S.; Li, K.C.T.; Chan, H.L.Y. Meta-analysis: Treatment of hepatitis B infection reduces risk of hepatocellular carcinoma. Aliment. Pharmacol. Ther. 2008, 28, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, W.; Chen, Y.-H.; Fan, C.L.; Dong, P.L.; Wei, F.L.; Li, B.; Chen, D.X.; Ding, H.G. Antiviral drug resistance increases hepatocellular carcinoma: A prospective decompensated cirrhosis cohort study. World J. Gastroenterol. 2013, 19, 8373–8381. [Google Scholar] [CrossRef]
- Zoulim, F.; Locarnini, S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 2009, 137, 1593–1608. [Google Scholar] [CrossRef]
- Lai, M.W.; Yeh, C.T. The oncogenic potential of hepatitis B virus rtA181T/surface truncation mutant. Antivir. Ther. 2008, 13, 875–879. [Google Scholar]
- Caselmann, W.H.; Meyer, M.; Kekule, A.S.; Lauer, U.; Hofschneider, P.H.; Koshy, R. A trans-activator function is generated by integration of hepatitis B virus preS/S sequences in human hepatocellular carcinoma DNA. Proc. Natl. Acad. Sci. USA 1990, 87, 2970–2974. [Google Scholar] [CrossRef]
- Kekule, A.; Lauer, U.; Meyer, M.; Caselmann, W.; Hofschneider, P.; Koshy, R. The preS2/S region of integrated hepatitis B virus DNA encodes a transcriptional transactivator. Nature 1990, 343, 457–461. [Google Scholar] [CrossRef]
- Lai, M.W.; Huang, S.F.; Hsu, C.W.; Chang, M.H.; Liaw, Y.F.; Yeh, C.T. Identification of nonsense mutations in hepatitis B virus S gene in patients with hepatocellular carcinoma developed after lamivudine therapy. Antivir. Ther. 2009, 14, 249–261. [Google Scholar]
- Liang, K.H.; Hsu, C.W.; Chang, M.L.; Chen, Y.C.; Lai, M.W.; Yeh, C.T. Peginterferon is superior to nucleos(t)ide analogues for prevention of hepatocellular carcinoma in chronic hepatitis B. J. Infect. Dis. 2016, 213, 966–974. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Lai, M.W.; Liang, K.H.; Lin, W.R.; Huang, Y.H.; Huang, S.F.; Chen, T.C.; Yeh, C.T. Hepatocarcinogenesis in transgenic mice carrying hepatitis B virus pre-S/S gene with the sW172* mutation. Oncogenesis 2016, 5, e273. [Google Scholar] [CrossRef][Green Version]
- Huang, S.F.; Chen, Y.T.; Lee, W.C.; Chang, I.C.; Chiu, Y.T.; Chang, Y.; Tu, H.C.; Yuh, C.H.; Matsuura, I.; Shih, L.Y. Identification of transforming hepatitis B virus S gene nonsense mutations derived from freely replicative viruses in hepatocellular carcinoma. PLoS ONE 2014, 9, e89753. [Google Scholar] [CrossRef]
- Lee, S.A.; Kim, K.; Kim, H.; Kim, B.J. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J. Hepatol. 2012, 56, 63–69. [Google Scholar] [CrossRef]
- Jiang, S.S.; Huang, S.F.; Huang, M.S.; Chen, Y.T.; Jhong, H.J.; Chang, I.C.; Chen, Y.T.; Chang, J.W.; Chen, W.L.; Lee, W.C.; et al. Dysregulation of the TGFBI gene is involved in the oncogenic activity of the nonsense mutation of hepatitis B virus surface gene sW182*. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2014, 1842, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Colledge, D.; Soppe, S.; Yuen, L.; Selleck, L.; Walsh, R.; Locarnini, S.; Warner, N. Stop codons in the hepatitis B surface proteins are enriched during antiviral therapy and are associated with host cell apoptosis. Virology 2017, 501, 70–78. [Google Scholar] [CrossRef] [PubMed]
- “Expression of MGST2 in cancer”-Summary, The Human Protein Atlas. Available online: https://v15.proteinatlas.org/ENSG00000085871-MGST2 (accessed on 1 June 2020).
- Dvash, E.; Har-Tal, M.; Barak, S.; Meir, O.; Rubinstein, M. Leukotriene C4 is the major trigger of stress-induced oxidative DNA damage. Nat. Commun. 2015, 6, 10112. [Google Scholar] [CrossRef]
- Tien Kuo, M.; Savaraj, N. Roles of reactive oxygen species in hepatocarcinogenesis and drug resistance gene expression in liver cancers. Mol. Carcinog. 2006, 45, 701–709. [Google Scholar] [CrossRef]
- Wang, D.; Dubois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193. [Google Scholar] [CrossRef]
- “HIF1A protein expression summary”, The Human Protein Atlas. Available online: https://v15.proteinatlas.org/ENSG00000100644-HIF1A (accessed on 1 June 2020).
- Lim, J.H.; Kim, D.G.; Yu, D.Y.; Kang, H.M.; Noh, K.H.; Kim, D.S.; Park, D.; Chang, T.K.; Im, D.S.; Jung, C.R. Stabilization of E2-EPF UCP protein is implicated in hepatitis B virus-associated hepatocellular carcinoma progression. Cell. Mol. Life Sci. 2019, 76, 2647–2662. [Google Scholar] [CrossRef]
- Liu, L.P.; Hu, B.G.; Ye, C.; Ho, R.L.; Chen, G.G.; Lai, P.B. HBx mutants differentially affect the activation of hypoxia-inducible factor-1alpha in hepatocellular carcinoma. Br. J. Cancer 2014, 110, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Caselmann, W.; Renner, M.; Schluter, V.; Hofschneider, P.; Koshy, R.; Meyer, M. The hepatitis B virus MHBst167 protein is a pleiotropic transactivator mediating its effect via ubiquitous cellular transcription factors. J. Gen. Virol. 1997, 78, 1487–1495. [Google Scholar] [CrossRef]
- Zhao, Y.; El-Gabry, M.; Hei, T.K. Loss of Betaig-h3 protein is frequent in primary lung carcinoma and related to tumorigenic phenotype in lung cancer cells. Mol. Carcinog. 2006, 45, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Calaf, G.M.; Echiburu-Chau, C.; Zhao, Y.; Hei, T.K. BigH3 protein expression as a marker for breast cancer. Int. J. Mol. Med. 2008, 21, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Dong, S.M.; Park, N.H. Frequent promoter hypermethylation of TGFBI in epithelial ovarian cancer. Gynecol. Oncol. 2010, 118, 58–63. [Google Scholar] [CrossRef]
- Buckhaults, P.; Rago, C.; Croix, B.S.; Romans, K.E.; Saha, S.; Zhang, L.; Vogelstein, B.; Kinzler, K.W. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001, 61, 6996–7001. [Google Scholar]
- Schneider, D.; Kleeff, J.; Berberat, P.O.; Zhu, Z.; Korc, M.; Friess, H.; Büchler, M.W. Induction and expression of βig-h3 in pancreatic cancer cells. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2002, 1588, 1–6. [Google Scholar] [CrossRef]
- Han, B.; Cai, H.; Chen, Y.; Hu, B.; Luo, H.; Wu, Y.; Wu, J. The role of TGFBI (βig-H3) in gastrointestinal tract tumorigenesis. Mol. Cancer 2015, 14, 64. [Google Scholar] [CrossRef]
- “Expression of TGFBI in liver cancer”, The Human Protein Atlas. Available online: https://v15.proteinatlas.org/ENSG00000120708-TGFBI (accessed on 1 June 2020).
- Wang, M.L.; Wu, D.B.; Tao, Y.C.; Chen, L.L.; Liu, C.P.; Chen, E.Q.; Tang, H. The truncated mutant HBsAg expression increases the tumorigenesis of hepatitis B virus by regulating TGF-beta/Smad signaling pathway. Virol. J. 2018, 15, 61. [Google Scholar] [CrossRef] [PubMed]

| Transformants | Colony Counts in Soft Agar | Tumor Growth Proportion | Tumor Size (cm3) |
|---|---|---|---|
| pIRES | 0.33 ± 0.58 ***,* | 0/5 ‡,§ | - |
| pIRES-YIDDst | 101.07 ± 28.87 ***,† | 4/4 ††,‡,¶ | 5.95 ± 1.63 # |
| pIRES-WT | 49.67 ± 5.51 *,† | 2/10 §,¶ | 2.37 ± 0.03 # |
| Symbols | Gene Title | Forward Primer | Reverse Primer |
|---|---|---|---|
| MGST2 | Microsomal glutathione S-transferase 2 | GCCATCCACAGCATTACGAT | AAAACACAAGATTGCACCCC |
| HIF1A | Hypoxia-inducible factor 1-alpha | AAGTGGCAACTGATGAGCAA | GGCGAGAACGAGAAGAAAAA |
| Etv1 (ER81) | Ets Variant Gene 1 (Ets-Related Protein 81) | CTTGATTTTCAGTGGGAGGC | AACATGGCTTGCTGAAGCTC |
| LEF1 | Lymphoid Enhancer Binding Factor 1 | AGAAAAGTGCTCGTCGCTGT | AAATGGGTCCCTTTCTCCAC |
| TGFbi | Transforming growth factor beta-induced | CCTTTCTCTCCTGGGACCTT | TCATTGGCACCAACAAGAAA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, M.-W.; Liang, K.-H.; Yeh, C.-T. Hepatitis B Virus preS/S Truncation Mutant rtM204I/sW196* Increases Carcinogenesis through Deregulated HIF1A, MGST2, and TGFbi. Int. J. Mol. Sci. 2020, 21, 6366. https://doi.org/10.3390/ijms21176366
Lai M-W, Liang K-H, Yeh C-T. Hepatitis B Virus preS/S Truncation Mutant rtM204I/sW196* Increases Carcinogenesis through Deregulated HIF1A, MGST2, and TGFbi. International Journal of Molecular Sciences. 2020; 21(17):6366. https://doi.org/10.3390/ijms21176366
Chicago/Turabian StyleLai, Ming-Wei, Kung-Hao Liang, and Chau-Ting Yeh. 2020. "Hepatitis B Virus preS/S Truncation Mutant rtM204I/sW196* Increases Carcinogenesis through Deregulated HIF1A, MGST2, and TGFbi" International Journal of Molecular Sciences 21, no. 17: 6366. https://doi.org/10.3390/ijms21176366
APA StyleLai, M.-W., Liang, K.-H., & Yeh, C.-T. (2020). Hepatitis B Virus preS/S Truncation Mutant rtM204I/sW196* Increases Carcinogenesis through Deregulated HIF1A, MGST2, and TGFbi. International Journal of Molecular Sciences, 21(17), 6366. https://doi.org/10.3390/ijms21176366




