Intratumoral Distribution of Lactate and the Monocarboxylate Transporters 1 and 4 in Human Glioblastoma Multiforme and Their Relationships to Tumor Progression-Associated Markers
Abstract
1. Introduction
2. Results
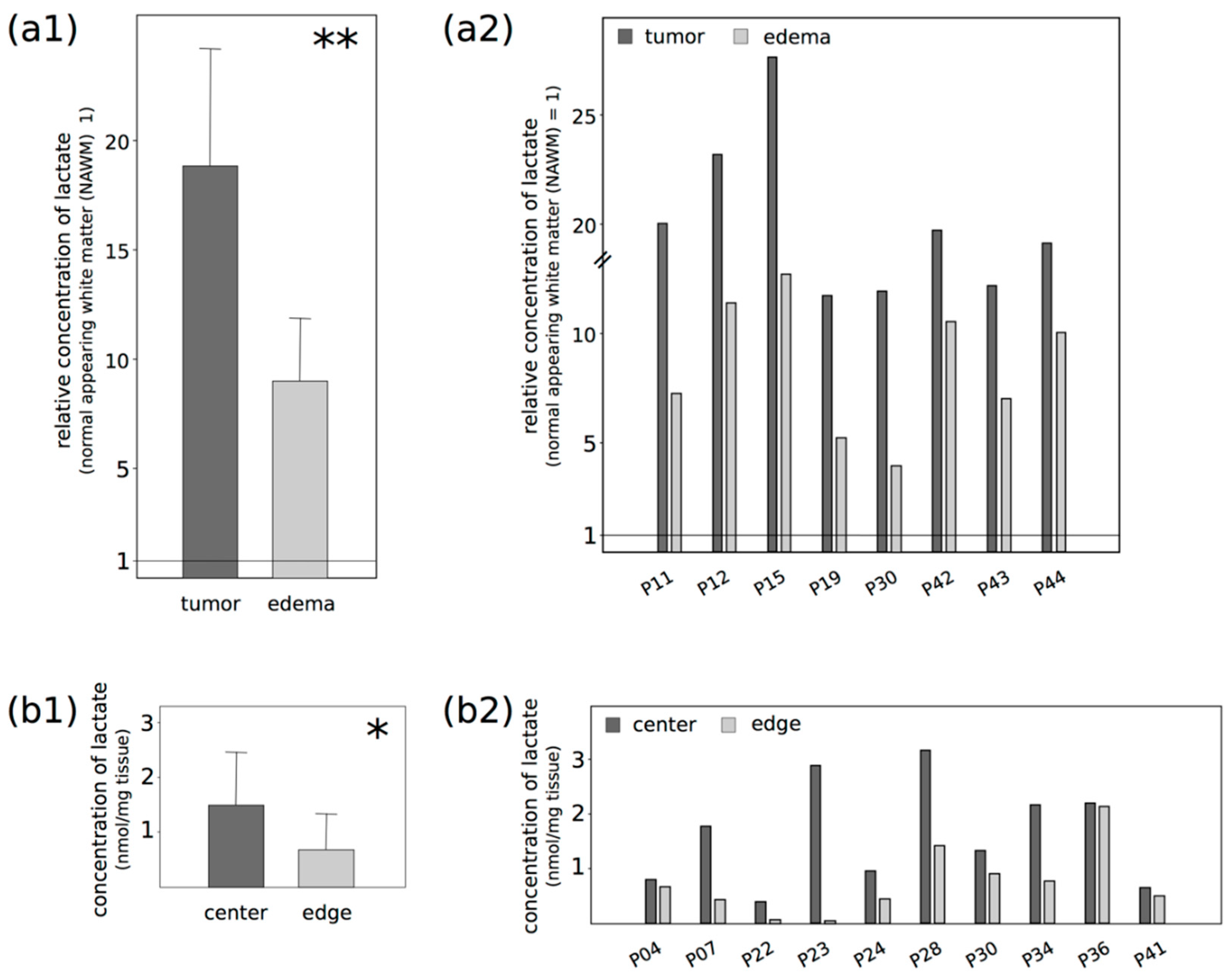
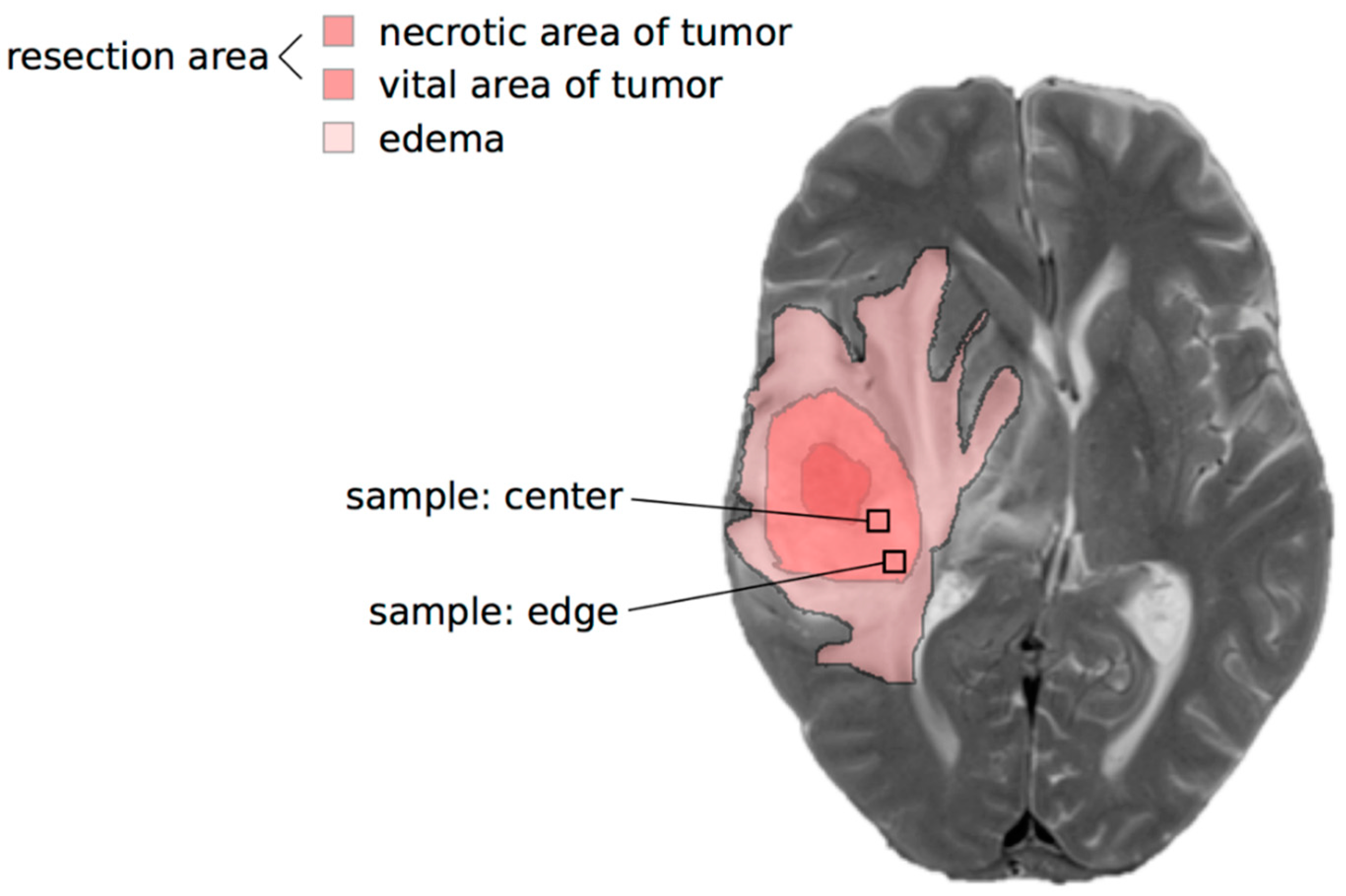
2.1. Lactate Concentrations Are Higher in the Contrast-Enhancing Part of the GBM with an Accumulation at the Center of the Vital Parts of the Tumor
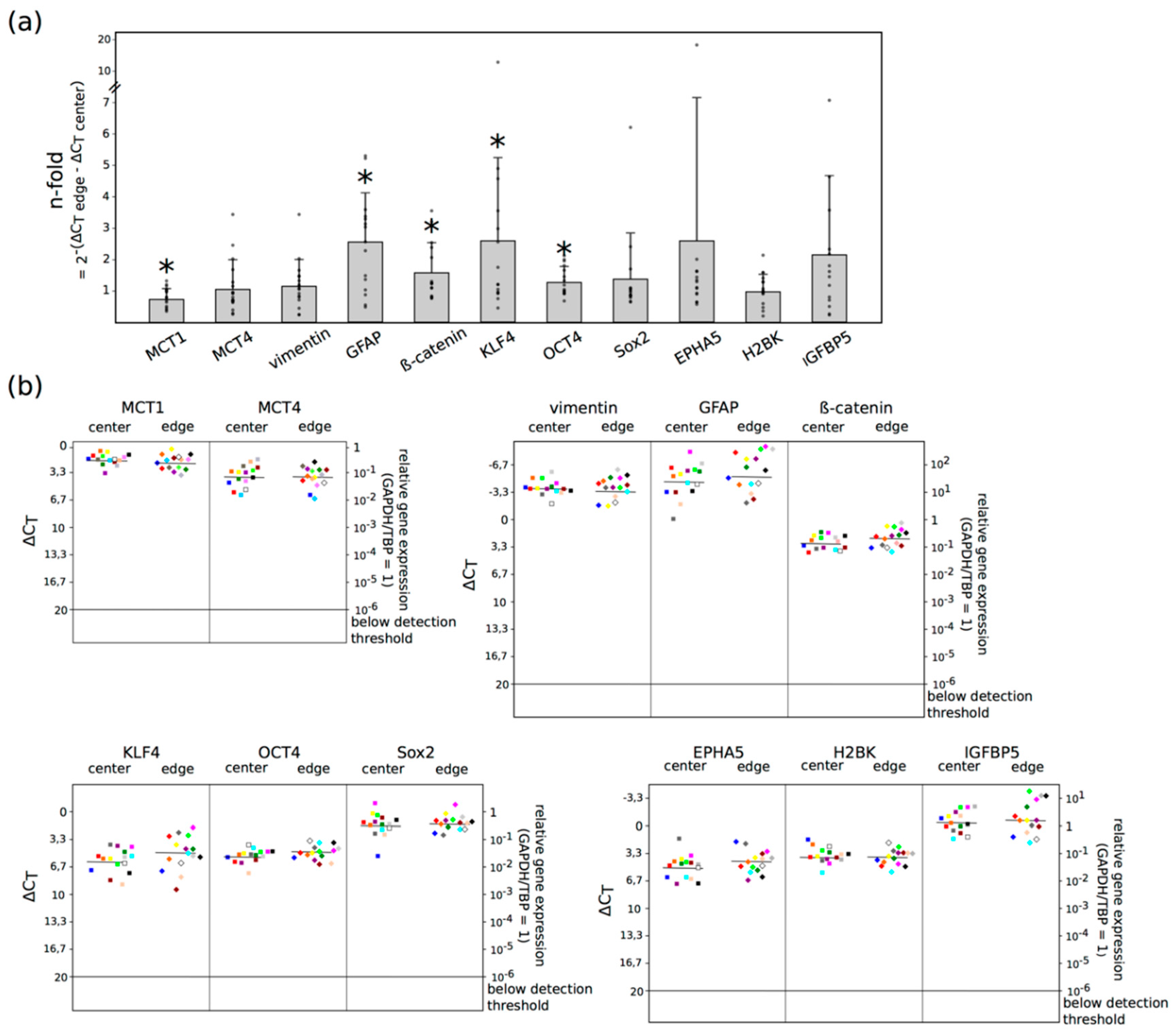
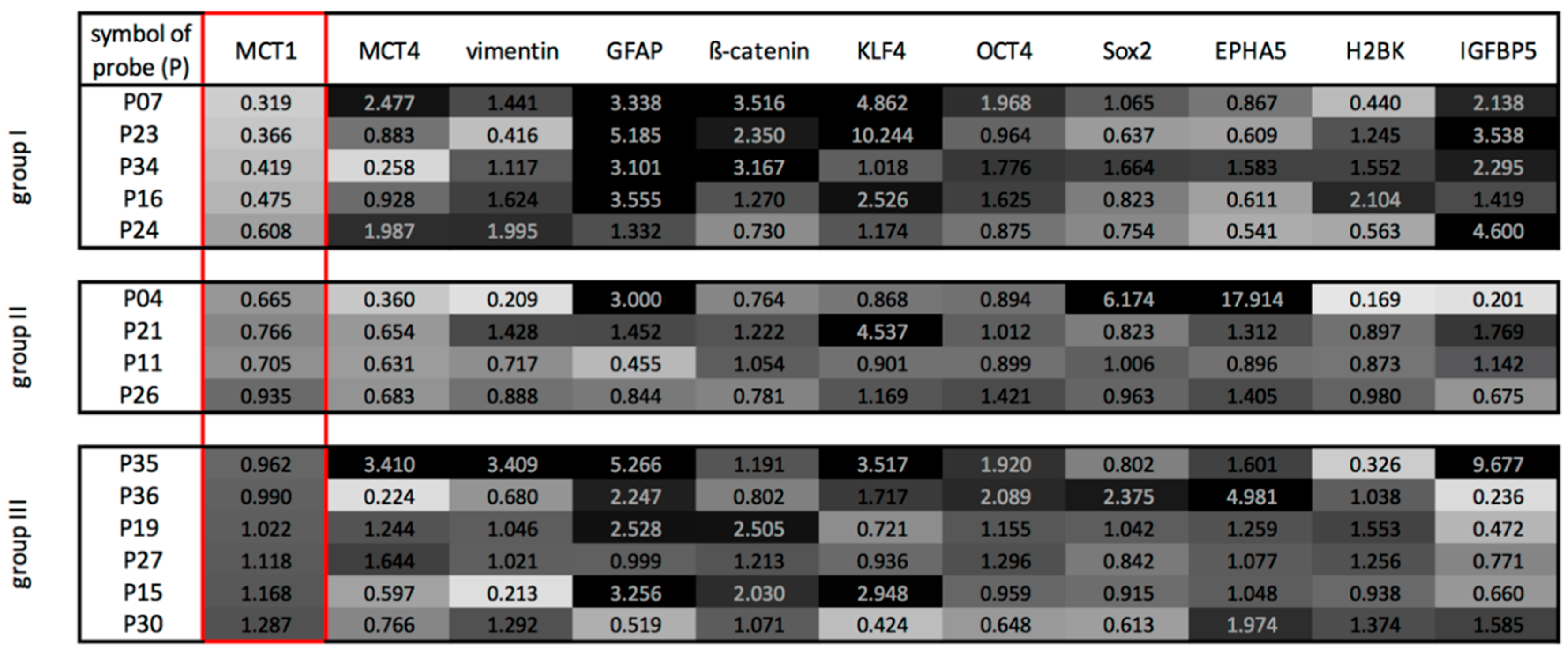
2.2. Three Main GBM Groups Were Distinguished, Based on Their RelatIve n-Fold mRNA Expression Differences in the Center and at the Edge of the Tumor
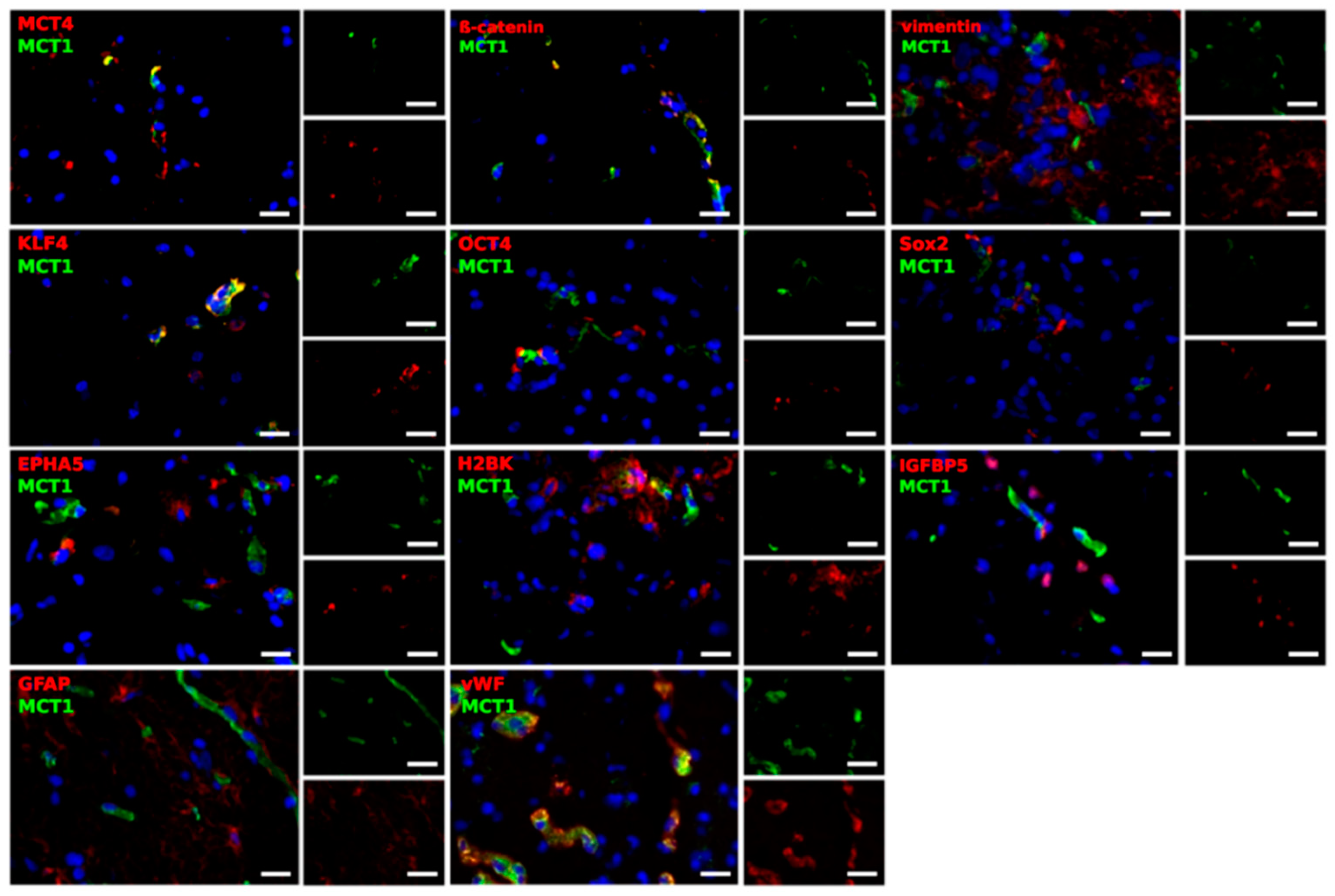
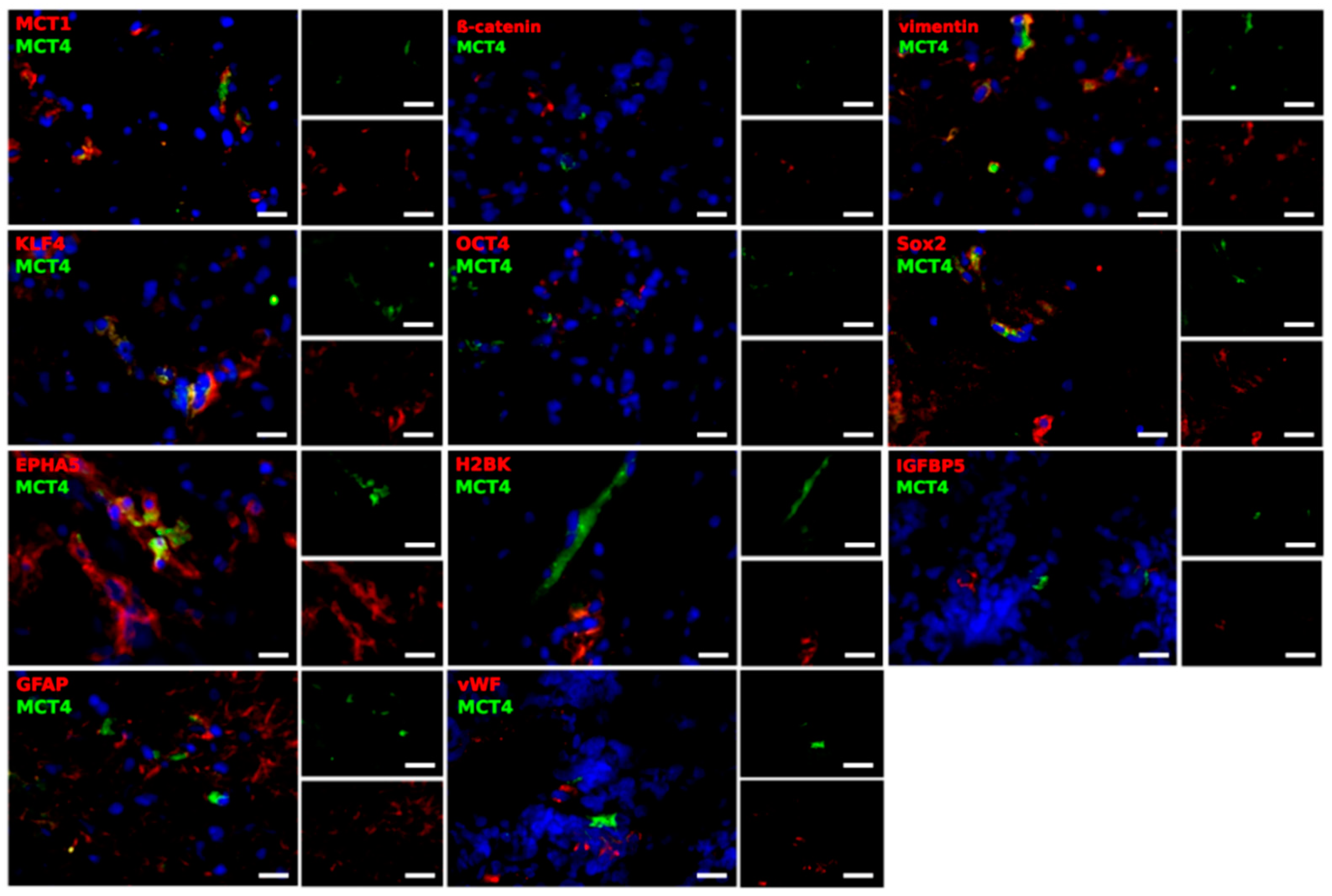
2.3. Different Cell Types Are Sources of the MCT Marker Expression
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Tumor Specimens and Ethical Consideration
5.2. Multi Voxel Magnetic Resonance Spectroscopic Imaging
5.3. Lactate ELISA
5.4. Quantitative Reverse Transcription and Real-Time PCR
5.5. Immunofluorescence
5.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | analysis of variance |
| CD147 | cluster of differentiation 147 |
| corr | correlation |
| CSI | chemical shift imaging |
| Ct | cycle threshold |
| DNase | deoxyribonuclease |
| ELISA | enzyme-linked immunosorbent assay |
| EMT | epithelial to mesenchymal transition |
| EPHA5 | ephrin receptor A5 |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| GBM | glioblastoma |
| GCS | glioma stem cells |
| GFAP | glial acidic fibrillary protein |
| H2BK | histone cluster 1 H2B family member k |
| IDH | isocitrate dehydrogenase |
| IGFBP5 | insulin-like growth factor-binding protein 5 |
| KLF4 | krüppel-like factor 4 |
| LCModel | linear combination model |
| MCT | monocarboxylate transporters |
| MGMT | O(6)-methylguanine-DNA methyltransferase |
| mRNA | messenger ribonucleic acid |
| MRSI | magnetic resonance spectroscopic imaging |
| NAA | N-Acetylaspartat |
| NAWM | normal appearing white matter |
| OCT4 | octamer binding transcription factor 4 |
| OD | optical density |
| PRESS | point-resolved spectroscopy |
| qRT-PCR | quantitative reverse transcription PCR |
| SD | standard deviation |
| Sox2 | sex determining region Y-box 2 |
| TBP | TATA-box-binding protein |
| TCA | trichloroacetic acid |
| TE | echo time |
| TMZ | temozolomide |
| TR | repetition time |
| UKE | Universitätsklinikum Hamburg-Eppendorf |
| vWF | von Willebrand factor |
| WHO | World Health Organization |
References
- Thakkar, J.P.; Dolecek, T.A.; Horbinski, C.; Ostrom, Q.T.; Lightner, D.D.; Barnholtz-Sloan, J.S.; Villano, J.L. Epidemiologic and Molecular Prognostic Review of Glioblastoma. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1985–1996. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. (Berl.) 2007, 114, 97–109. [Google Scholar] [CrossRef]
- Vallée, A.; Guillevin, R.; Vallée, J.-N. Vasculogenesis and Angiogenesis Initiation under Normoxic Conditions through Wnt/β-Catenin Pathway in Gliomas. Rev. Neurosci. 2017, 29, 71–91. [Google Scholar] [CrossRef]
- Miranda-Gonçalves, V.; Honavar, M.; Pinheiro, C.; Martinho, O.; Pires, M.M.; Pinheiro, C.; Cordeiro, M.; Bebiano, G.; Costa, P.; Palmeirim, I.; et al. Monocarboxylate Transporters (MCTs) in Gliomas: Expression and Exploitation as Therapeutic Targets. Neuro Oncol. 2013, 15, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.S.; Lim, K.J.; Price, A.C.; Orr, B.A.; Eberhart, C.G.; Bar, E.E. Inhibition of Monocarboxylate Transporter-4 Depletes Stem-like Glioblastoma Cells and Inhibits HIF Transcriptional Response in a Lactate-Independent Manner. Oncogene 2014, 33, 4433–4441. [Google Scholar] [CrossRef] [PubMed]
- Takada, T.; Takata, K.; Ashihara, E. Inhibition of Monocarboxylate Transporter 1 Suppresses the Proliferation of Glioblastoma Stem Cells. J. Physiol. Sci. 2016, 66, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Adamski, V.; Hempelmann, A.; Flüh, C.; Lucius, R.; Synowitz, M.; Hattermann, K.; Held-Feindt, J. Dormant Glioblastoma Cells Acquire Stem Cell Characteristics and Are Differentially Affected by Temozolomide and AT101 Treatment. Oncotarget 2017, 8, 108064–108078. [Google Scholar] [CrossRef]
- Middeldorp, J.; Hol, E.M. GFAP in Health and Disease. Prog. Neurobiol. 2011, 93, 421–443. [Google Scholar] [CrossRef]
- Velasco, M.E.; Dahl, D.; Roessmann, U.; Gambetti, P. Immunohistochemical Localization of Glial Fibrillary Acidic Protein in Human Glial Neoplasms. Cancer 1980, 45, 484–494. [Google Scholar] [CrossRef]
- Kaufhold, S.; Bonavida, B. Central Role of Snail1 in the Regulation of EMT and Resistance in Cancer: A Target for Therapeutic Intervention. J. Exp. Clin. Cancer Res. 2014, 33, 62. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-Catenin Signaling: Components, Mechanisms, and Diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Marchiq, I.; Pouysségur, J. Hypoxia, Cancer Metabolism and the Therapeutic Benefit of Targeting Lactate/H+ Symporters. J. Mol. Med. 2016, 94, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Hamans, B.; Navis, A.C.; Wright, A.; Wesseling, P.; Heerschap, A.; Leenders, W. Multivoxel 1H MR Spectroscopy Is Superior to Contrast-Enhanced MRI for Response Assessment after Anti-Angiogenic Treatment of Orthotopic Human Glioma Xenografts and Provides Handles for Metabolic Targeting. Neuro Oncol. 2013, 15, 1615–1624. [Google Scholar] [CrossRef]
- Lange, T.; Dydak, U.; Roberts, T.P.L.; Rowley, H.A.; Bjeljac, M.; Boesiger, P. Pitfalls in Lactate Measurements at 3T. AJNR Am. J. Neuroradiol. 2006, 27, 895–901. [Google Scholar] [PubMed]
- Ratai, E.-M.; Zhang, Z.; Fink, J.; Muzi, M.; Hanna, L.; Greco, E.; Richards, T.; Kim, D.; Andronesi, O.C.; Mintz, A.; et al. ACRIN 6684: Multicenter, Phase II Assessment of Tumor Hypoxia in Newly Diagnosed Glioblastoma Using Magnetic Resonance Spectroscopy. PLoS ONE 2018, 13, e0198548. [Google Scholar] [CrossRef]
- Roldan-Valadez, E.; Rios, C.; Motola-Kuba, D.; Matus-Santos, J.; Villa, A.R.; Moreno-Jimenez, S. Choline-to- N -Acetyl Aspartate and Lipids-Lactate-to-Creatine Ratios Together with Age Assemble a Significant Cox’s Proportional-Hazards Regression Model for Prediction of Survival in High-Grade Gliomas. Br. J. Radiol. 2016, 89, 20150502. [Google Scholar] [CrossRef]
- Heiland, D.H.; Mader, I.; Schlosser, P.; Pfeifer, D.; Carro, M.S.; Lange, T.; Schwarzwald, R.; Vasilikos, I.; Urbach, H.; Weyerbrock, A. Integrative Network-Based Analysis of Magnetic Resonance Spectroscopy and Genome Wide Expression in Glioblastoma Multiforme. Sci. Rep. 2016, 6, 29052. [Google Scholar] [CrossRef]
- Vallée, A.; Vallée, J.-N.; Guillevin, R.; Lecarpentier, Y. Interactions Between the Canonical WNT/Beta-Catenin Pathway and PPAR Gamma on Neuroinflammation, Demyelination, and Remyelination in Multiple Sclerosis. Cell. Mol. Neurobiol. 2018, 38, 783–795. [Google Scholar] [CrossRef]
- Kahlert, U.D.; Maciaczyk, D.; Doostkam, S.; Orr, B.A.; Simons, B.; Bogiel, T.; Reithmeier, T.; Prinz, M.; Schubert, J.; Niedermann, G.; et al. Activation of Canonical WNT/β-Catenin Signaling Enhances in Vitro Motility of Glioblastoma Cells by Activation of ZEB1 and Other Activators of Epithelial-to-Mesenchymal Transition. Cancer Lett. 2012, 325, 42–53. [Google Scholar] [CrossRef]
- Payen, V.L.; Mina, E.; Van Hée, V.F.; Porporato, P.E.; Sonveaux, P. Monocarboxylate Transporters in Cancer. Mol. Metab. 2020, 33, 48–66. [Google Scholar] [CrossRef]
- Park, S.J.; Smith, C.P.; Wilbur, R.R.; Cain, C.P.; Kallu, S.R.; Valasapalli, S.; Sahoo, A.; Guda, M.R.; Tsung, A.J.; Velpula, K.K. An Overview of MCT1 and MCT4 in GBM: Small Molecule Transporters with Large Implications. Am. J. Cancer Res. 2018, 8, 1967–1976. [Google Scholar] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ontañon, P.; Orgaz, J.L.; Aldaz, B.; Elosegui-Artola, A.; Martino, J.; Berciano, M.T.; Montero, J.A.; Grande, L.; Nogueira, L.; Diaz-Moralli, S.; et al. Cellular Plasticity Confers Migratory and Invasive Advantages to a Population of Glioblastoma-Initiating Cells That Infiltrate Peritumoral Tissue. Stem Cells 2013, 31, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Sundar, S.J.; Hsieh, J.K.; Manjila, S.; Lathia, J.D.; Sloan, A. The Role of Cancer Stem Cells in Glioblastoma. Neurosurg. Focus 2014, 37, E6. [Google Scholar] [CrossRef]
- Diehl, K.; Dinges, L.-A.; Helm, O.; Ammar, N.; Plundrich, D.; Arlt, A.; Röcken, C.; Sebens, S.; Schäfer, H. Nuclear Factor E2-Related Factor-2 Has a Differential Impact on MCT1 and MCT4 Lactate Carrier Expression in Colonic Epithelial Cells: A Condition Favoring Metabolic Symbiosis between Colorectal Cancer and Stromal Cells. Oncogene 2018, 37, 39–51. [Google Scholar] [CrossRef]
- Fiaschi, T.; Marini, A.; Giannoni, E.; Taddei, M.L.; Gandellini, P.; De Donatis, A.; Lanciotti, M.; Serni, S.; Cirri, P.; Chiarugi, P. Reciprocal Metabolic Reprogramming through Lactate Shuttle Coordinately Influences Tumor-Stroma Interplay. Cancer Res. 2012, 72, 5130–5140. [Google Scholar] [CrossRef]
- Balmaceda-Aguilera, C.; Cortés-Campos, C.; Cifuentes, M.; Peruzzo, B.; Mack, L.; Tapia, J.C.; Oyarce, K.; García, M.A.; Nualart, F. Glucose Transporter 1 and Monocarboxylate Transporters 1, 2, and 4 Localization within the Glial Cells of Shark Blood-Brain-Barriers. PLoS ONE 2012, 7, e32409. [Google Scholar] [CrossRef]
- Wilhelmsson, U.; Eliasson, C.; Bjerkvig, R.; Pekny, M. Loss of GFAP Expression in High-Grade Astrocytomas Does Not Contribute to Tumor Development or Progression. Oncogene 2003, 22, 3407–3411. [Google Scholar] [CrossRef]
- Nishikawa, M.; Inoue, A.; Ohnishi, T.; Kohno, S.; Ohue, S.; Matsumoto, S.; Suehiro, S.; Yamashita, D.; Ozaki, S.; Watanabe, H.; et al. Significance of Glioma Stem-Like Cells in the Tumor Periphery That Express High Levels of CD44 in Tumor Invasion, Early Progression, and Poor Prognosis in Glioblastoma. Stem Cells Int. 2018, 2018, 1–15. [Google Scholar] [CrossRef]
- Glas, M.; Rath, B.H.; Simon, M.; Reinartz, R.; Schramme, A.; Trageser, D.; Eisenreich, R.; Leinhaas, A.; Keller, M.; Schildhaus, H.-U.; et al. Residual Tumor Cells Are Unique Cellular Targets in Glioblastoma. Ann. Neurol. 2010, 68, 264–269. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-Cell RNA-Seq Highlights Intratumoral Heterogeneity in Primary Glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Galeffi, F.; Turner, D.A. Exploiting Metabolic Differences in Glioma Therapy. Curr. Drug Discov. Technol. 2012, 9, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Puchalski, R.B.; Shah, N.; Miller, J.; Dalley, R.; Nomura, S.R.; Yoon, J.-G.; Smith, K.A.; Lankerovich, M.; Bertagnolli, D.; Bickley, K.; et al. An Anatomic Transcriptional Atlas of Human Glioblastoma. Science 2018, 360, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Aubry, M.; de Tayrac, M.; Etcheverry, A.; Clavreul, A.; Saikali, S.; Menei, P.; Mosser, J. From the Core to beyond the Margin: A Genomic Picture of Glioblastoma Intratumor Heterogeneity. Oncotarget 2015, 6, 12094–12109. [Google Scholar] [CrossRef]
- Darmanis, S.; Sloan, S.A.; Croote, D.; Mignardi, M.; Chernikova, S.; Samghababi, P.; Zhang, Y.; Neff, N.; Kowarsky, M.; Caneda, C.; et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017, 21, 1399–1410. [Google Scholar] [CrossRef]
- Marchiq, I.; Albrengues, J.; Granja, S.; Gaggioli, C.; Pouysségur, J.; Simon, M.-P. Knock out of the BASIGIN/CD147 Chaperone of Lactate/H+ Symporters Disproves Its pro-Tumour Action via Extracellular Matrix Metalloproteases (MMPs) Induction. Oncotarget 2015, 6, 24636–24648. [Google Scholar] [CrossRef]
- Keith, B.; Simon, M.C. Hypoxia-Inducible Factors, Stem Cells, and Cancer. Cell 2007, 129, 465–472. [Google Scholar] [CrossRef]
- Pierre, K.; Pellerin, L. Monocarboxylate Transporters in the Central Nervous System: Distribution, Regulation and Function. J. Neurochem. 2005, 94, 1–14. [Google Scholar] [CrossRef]
- Nomura, A.; Dauer, P.; Gupta, V.; McGinn, O.; Arora, N.; Majumdar, K.; Iii, C.U.; Dalluge, J.; Dudeja, V.; Saluja, A.; et al. Microenvironment Mediated Alterations to Metabolic Pathways Confer Increased Chemo-Resistance in CD133+ Tumor Initiating Cells. Oncotarget 2016, 7, 56324–56337. [Google Scholar] [CrossRef]
- Voss, D.M.; Spina, R.; Carter, D.L.; Lim, K.S.; Jeffery, C.J.; Bar, E.E. Disruption of the Monocarboxylate Transporter-4-Basigin Interaction Inhibits the Hypoxic Response, Proliferation, and Tumor Progression. Sci. Rep. 2017, 7, 4292. [Google Scholar] [CrossRef]
- Sonveaux, P.; Végran, F.; Schroeder, T.; Wergin, M.C.; Verrax, J.; Rabbani, Z.N.; De Saedeleer, C.J.; Kennedy, K.M.; Diepart, C.; Jordan, B.F.; et al. Targeting Lactate-Fueled Respiration Selectively Kills Hypoxic Tumor Cells in Mice. J. Clin. Investig. 2008, 118, 3930–3942. [Google Scholar] [CrossRef] [PubMed]
- Colen, C.B.; Shen, Y.; Ghoddoussi, F.; Yu, P.; Francis, T.B.; Koch, B.J.; Monterey, M.D.; Galloway, M.P.; Sloan, A.E.; Mathupala, S.P. Metabolic Targeting of Lactate Efflux by Malignant Glioma Inhibits Invasiveness and Induces Necrosis: An In Vivo Study. Neoplasia 2011, 13, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, F.A.; Woitek, R.; McLean, M.A.; Gill, A.B.; Manzano Garcia, R.; Provenzano, E.; Riemer, F.; Kaggie, J.; Chhabra, A.; Ursprung, S.; et al. Imaging Breast Cancer Using Hyperpolarized Carbon-13 MRI. Proc. Natl. Acad. Sci. USA 2020, 117, 2092. [Google Scholar] [CrossRef] [PubMed]
- Radoul, M.; Chaumeil, M.M.; Eriksson, P.; Wang, A.S.; Phillips, J.J.; Ronen, S.M. MR Studies of Glioblastoma Models Treated with Dual PI3K/MTOR Inhibitor and Temozolomide:Metabolic Changes Are Associated with Enhanced Survival. Mol. Cancer Ther. 2016, 15, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Provencher, S.W. Estimation of Metabolite Concentrations from Localizedin Vivo Proton NMR Spectra. Magn. Reson. Med. 1993, 30, 672–679. [Google Scholar] [CrossRef]

| Symbol of Probe (P) | Age (Years) | Gender (Female ♀; Male ♂) | Symptoms Before Diagnosis (Days) | Primary Versus Recurrence | Time to Relapse (Months) | IDH Status | MGMT Status | 1p19q Status |
|---|---|---|---|---|---|---|---|---|
| P01 | 50 | ♂ | 3 | recurrence | 6 | wild-type | not methylated | co-deletion |
| P02 | 52 | ♂ | 28 | recurrence | 13 | wild-type | not methylated | no co-deletion |
| P04 | 43 | ♀ | 11 | primary | wild-type | methylated | no co-deletion | |
| P05 | 66 | ♂ | 3 | primary | wild-type | methylated | no co-deletion | |
| P06 | 78 | ♂ | 1 | primary | wild-type | methylated | no co-deletion | |
| P07 | 62 | ♂ | 3 | primary | wild-type | not methylated | no co-deletion | |
| P11 | 63 | ♀ | 0 | primary | wild-type | methylated | no co-deletion | |
| P12 | 71 | ♂ | 3 | primary | wild-type | methylated | no co-deletion | |
| P15 | 59 | ♂ | 3 | primary | wild-type | methylated | no co-deletion | |
| P16 | 78 | ♂ | 3 | primary | wild-type | not methylated | co-deletion | |
| P19 | 73 | ♂ | 60 | recurrence | 3 | wild-type | not methylated | no co-deletion |
| P21 | 56 | ♂ | 0 | primary | wild-type | methylated | no co-deletion | |
| P22 | 78 | ♂ | 2 | primary | wild-type | not methylated | no co-deletion | |
| P23 | 81 | ♀ | 180 | primary | wild-type | methylated | no co-deletion | |
| P24 | 57 | ♀ | 14 | primary | wild-type | methylated | no co-deletion | |
| P26 | 56 | ♂ | 180 | primary | wild-type | not methylated | not measured | |
| P27 | 52 | ♀ | 0 | recurrence | 17 | wild-type | methylated | no co-deletion |
| P28 | 79 | ♂ | 0 | primary | wild-type | methylated | not measured | |
| P29 | 63 | ♀ | 0 | recurrence | 3 | wild-type | methylated | no co-deletion |
| P30 | 63 | ♀ | 14 | primary | wild-type | unclear | not measured | |
| P34 | 63 | ♂ | 30 | primary | wild-type | methylated | not measured | |
| P35 | 79 | ♂ | 0 | recurrence | 15 | wild-type | methylated | no co-deletion |
| P36 | 74 | ♂ | 21 | primary | wild-type | not methylated | not measured | |
| P41 | 69 | ♀ | 60 | primary | wild-type | not methylated | not measured | |
| P42 | 71 | ♂ | 30 | primary | wild-type | methylated | not measured | |
| P43 | 58 | ♀ | 2 | primary | wild-type | not methylated | not measured | |
| P44 | 75 | ♀ | 0 | primary | wild-type | methylated | not measured |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubelt, C.; Peters, S.; Ahmeti, H.; Huhndorf, M.; Huber, L.; Cohrs, G.; Hövener, J.-B.; Jansen, O.; Synowitz, M.; Held-Feindt, J. Intratumoral Distribution of Lactate and the Monocarboxylate Transporters 1 and 4 in Human Glioblastoma Multiforme and Their Relationships to Tumor Progression-Associated Markers. Int. J. Mol. Sci. 2020, 21, 6254. https://doi.org/10.3390/ijms21176254
Kubelt C, Peters S, Ahmeti H, Huhndorf M, Huber L, Cohrs G, Hövener J-B, Jansen O, Synowitz M, Held-Feindt J. Intratumoral Distribution of Lactate and the Monocarboxylate Transporters 1 and 4 in Human Glioblastoma Multiforme and Their Relationships to Tumor Progression-Associated Markers. International Journal of Molecular Sciences. 2020; 21(17):6254. https://doi.org/10.3390/ijms21176254
Chicago/Turabian StyleKubelt, Carolin, Sönke Peters, Hajrullah Ahmeti, Monika Huhndorf, Lukas Huber, Gesa Cohrs, Jan-Bernd Hövener, Olav Jansen, Michael Synowitz, and Janka Held-Feindt. 2020. "Intratumoral Distribution of Lactate and the Monocarboxylate Transporters 1 and 4 in Human Glioblastoma Multiforme and Their Relationships to Tumor Progression-Associated Markers" International Journal of Molecular Sciences 21, no. 17: 6254. https://doi.org/10.3390/ijms21176254
APA StyleKubelt, C., Peters, S., Ahmeti, H., Huhndorf, M., Huber, L., Cohrs, G., Hövener, J.-B., Jansen, O., Synowitz, M., & Held-Feindt, J. (2020). Intratumoral Distribution of Lactate and the Monocarboxylate Transporters 1 and 4 in Human Glioblastoma Multiforme and Their Relationships to Tumor Progression-Associated Markers. International Journal of Molecular Sciences, 21(17), 6254. https://doi.org/10.3390/ijms21176254





