Characterizing the Relapse Potential in Different Luminal Subtypes of Breast Cancers with Functional Proteomics
Abstract
1. Introduction
2. Results
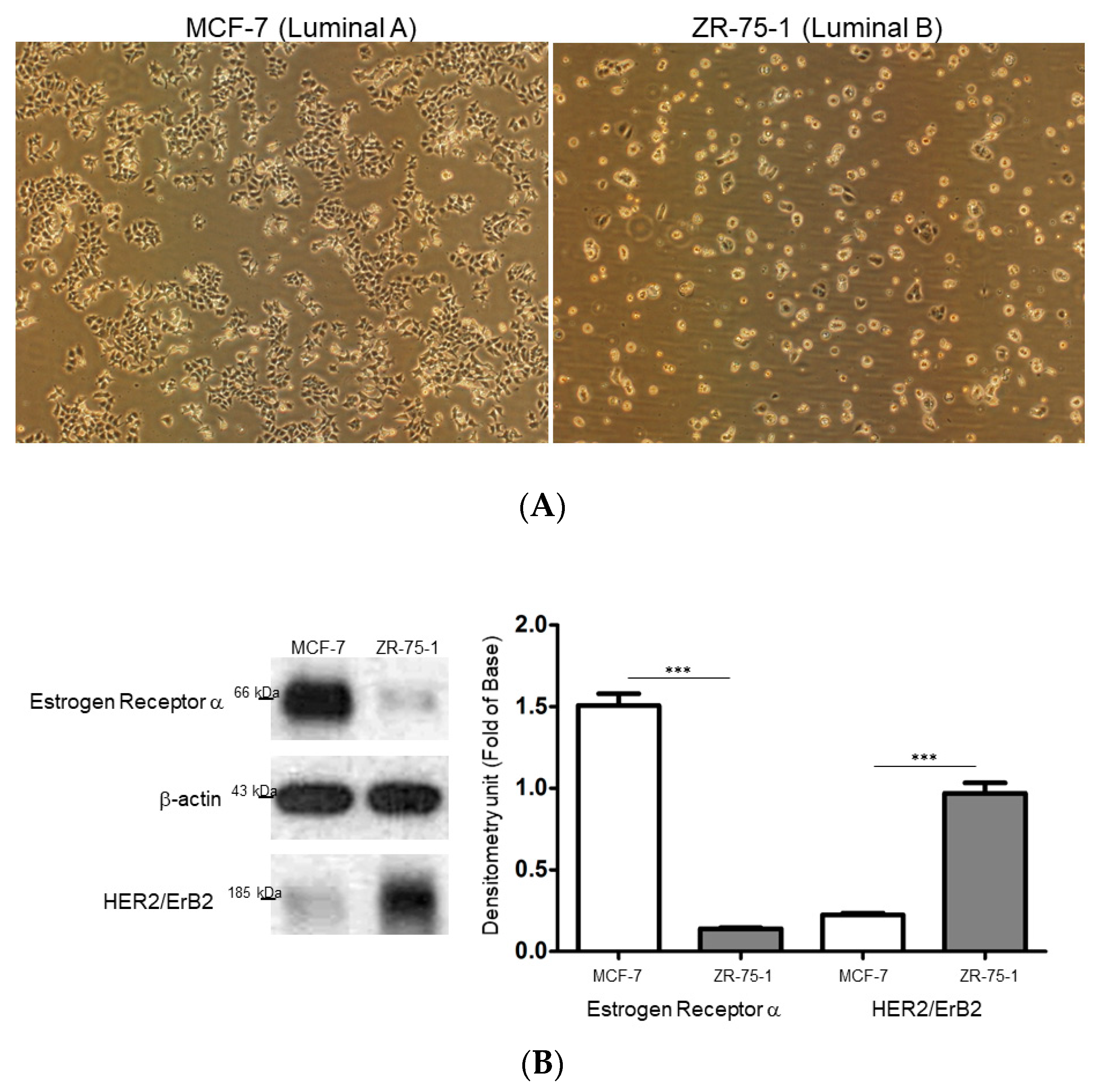
2.1. Comparison of the Different Breast Cancer Cell Models Reflecting Breast Cancer Heterogeneity
2.2. Exploring Target Proteins with Functional Proteome Tools
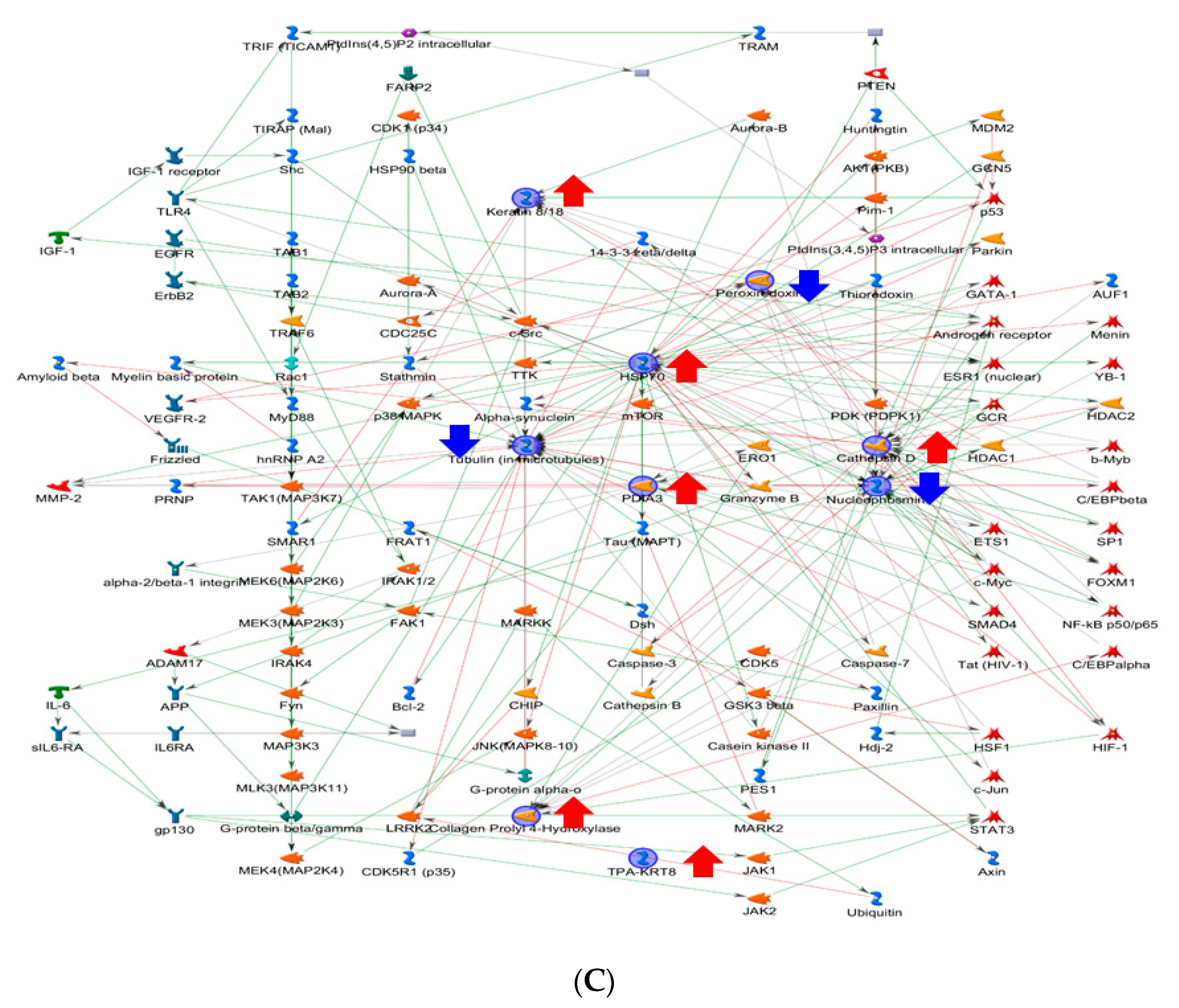
2.3. Functional Network Analysis
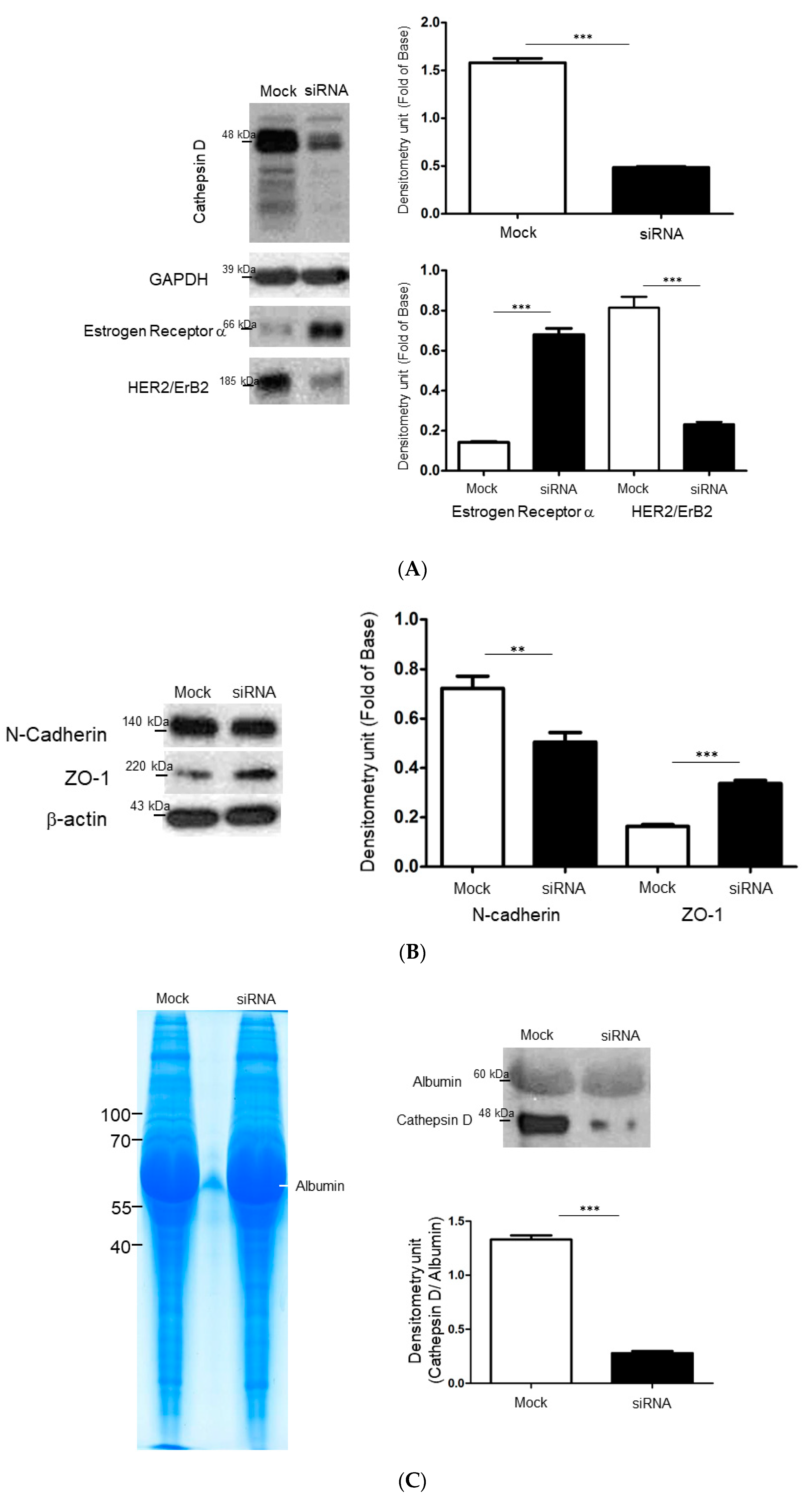
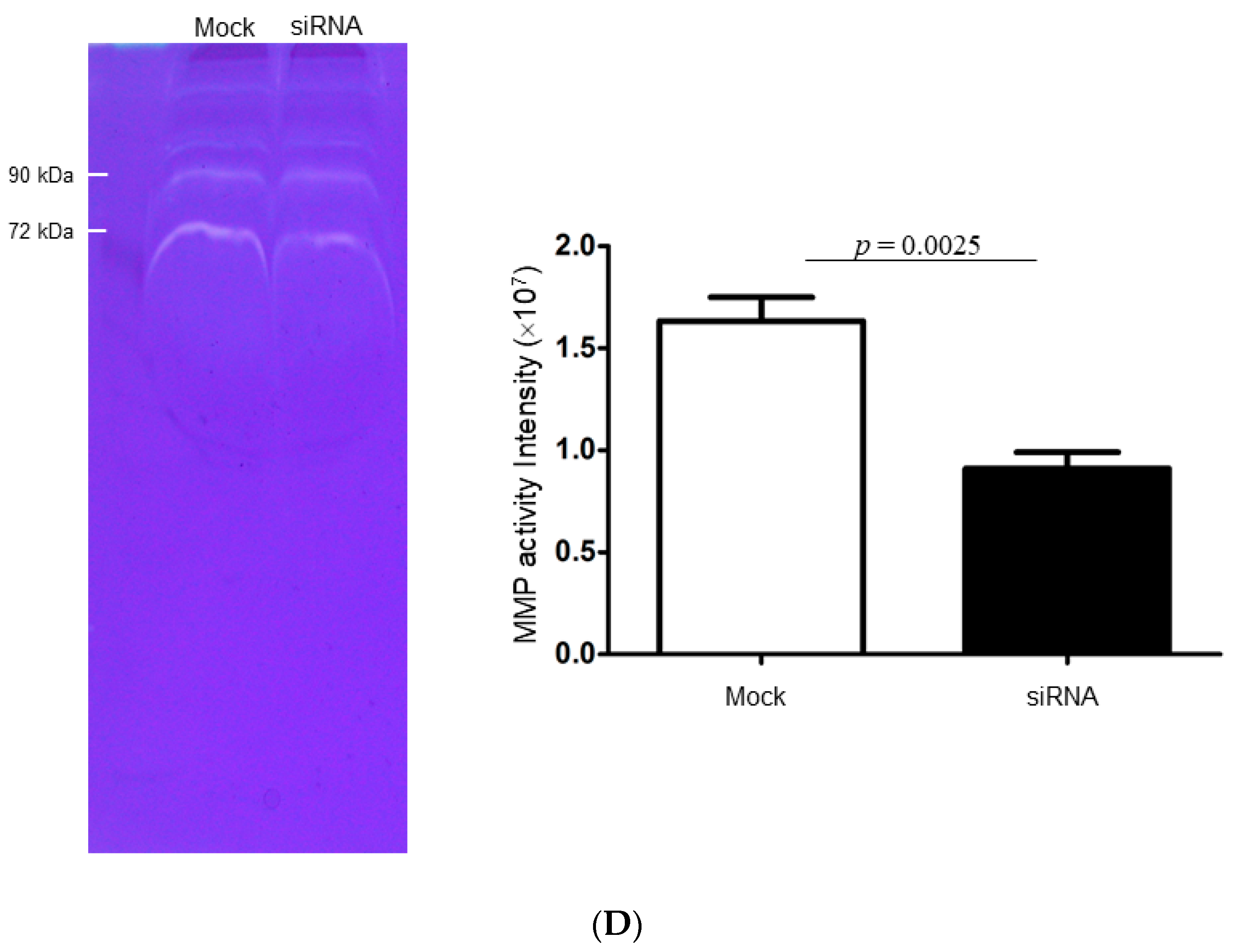
2.4. Identify the Cathepsin d-Dependent Regulation of Proteins in ZR-75-1 Cells
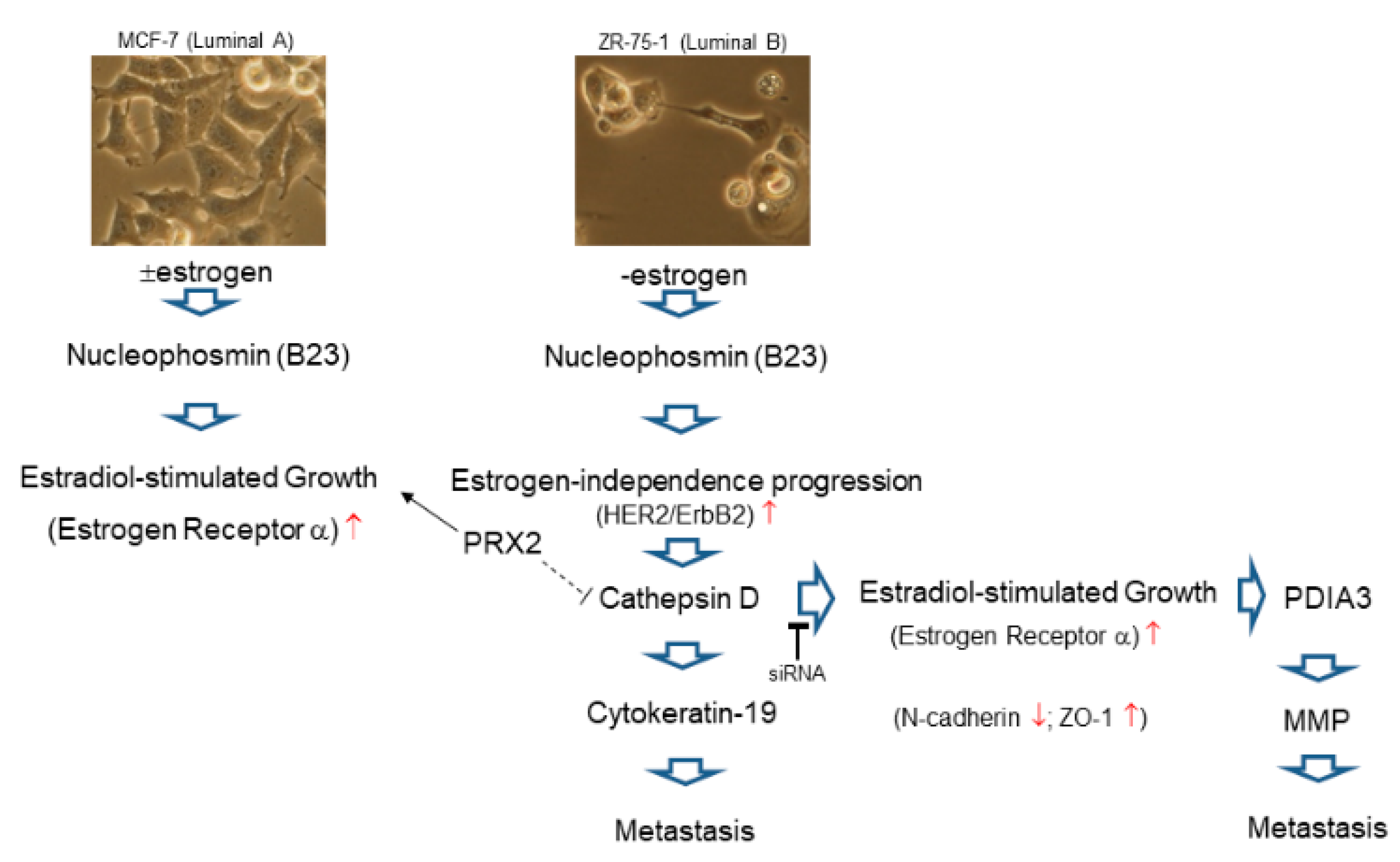
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Culture
5.2. Two-Dimensional Electrophoresis (2-DE) and Image Analysis
5.3. In-Gel Digestion of Proteins and Mass Spectrometric Analysis
5.4. Biological Network Analysis Using MetaCoreTM
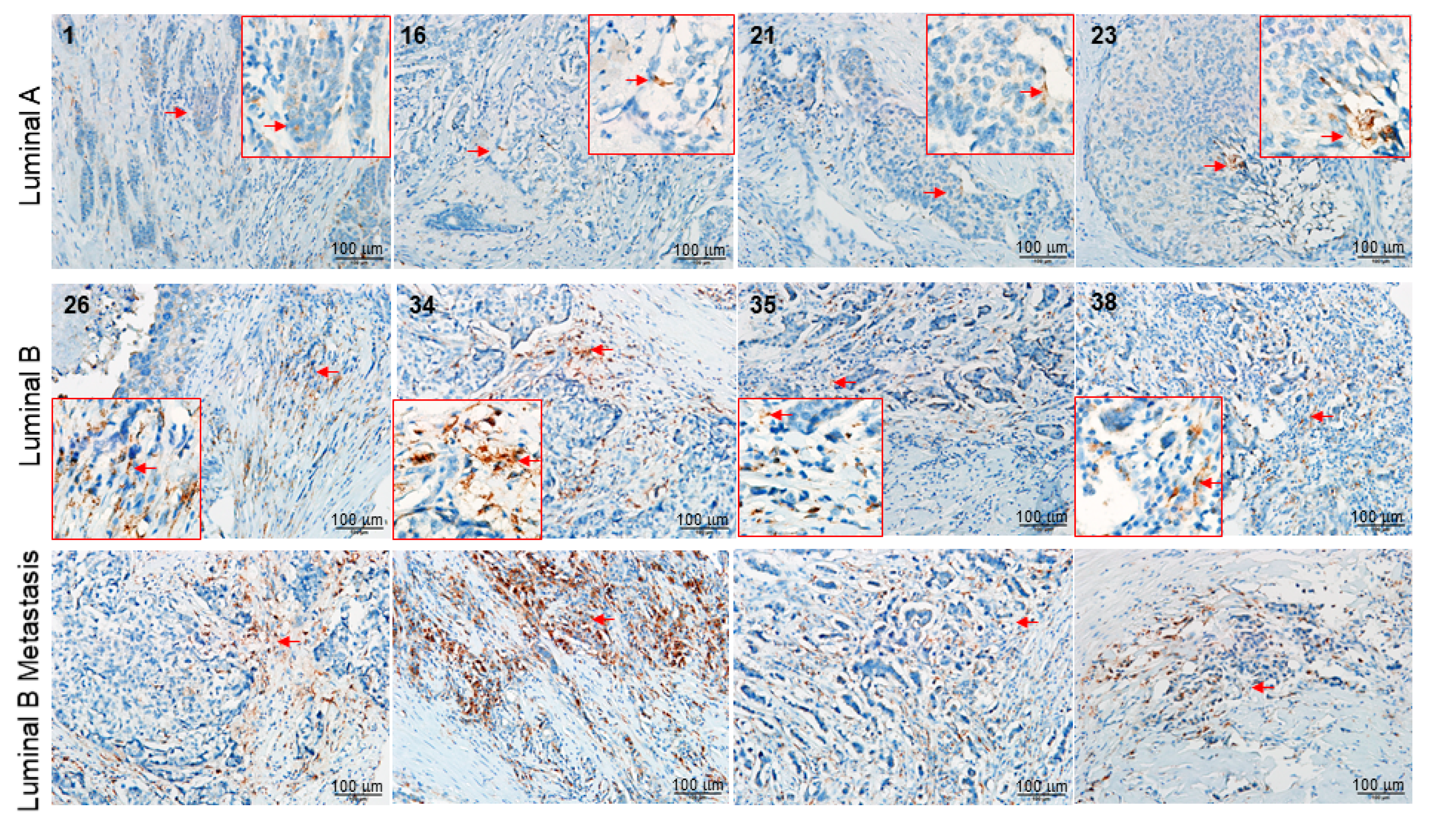
5.5. Tissue Array
5.6. Western Blot Analysis
5.7. Gene Silencing by Small Interfering RNA
5.8. Gelatin Zymography
5.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ER | estrogen receptor |
| HER2 | human epidermal growth factor receptor 2 |
| 2-DE | Two-dimensional electrophoresis |
| MALDI-TOF-MS | Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry |
| PMF | Peptide mass fingerprinting |
| EMT | epithelial-to-mesenchymal transition |
| IPG | Immobilized pH gradient |
| IHC | Immunohistochemistry |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| TBST | Tris-buffered saline Tween-20 |
| PVDF | polyvinylidene difluoride |
| ECM | extracellular matrix |
| MMP | matrix metalloproteinase |
| PRX2 | peroxiredoxin 2 |
References
- Voduc, K.D.; Cheang, M.C.; Tyldesley, S.; Gelmon, K.; Nielsen, T.O.; Kennecke, H. Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 2010, 28, 1684–1691. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Hu, P.H.; Tu, J.H.; Yu, N.S. Luminal B breast cancer: Patterns of recurrence and clinical outcome. Oncotarget 2016, 7, 65024–65033. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, M.; Sung, H.; Bcr, D.; Guida, J.; Tang, T.S.; Pfeiffer, R.M.; Yang, X.R. Breast cancer risk factors, survival and recurrence, and tumor molecular subtype: Analysis of 3012 women from an indigenous Asian population. Breast Cancer Res. 2018, 20, 114. [Google Scholar] [CrossRef]
- Calhoun, B.C.; Collins, L.C. Predictive markers in breast cancer: An update on ER and HER2 testing and reporting. Semin. Diagn. Pathol. 2015, 32, 362–369. [Google Scholar] [CrossRef]
- Beca, F.; Polyak, K. Intratumor Heterogeneity in Breast Cancer. Adv. Exp. Med. Biol. 2016, 882, 169–189. [Google Scholar] [PubMed]
- Weigelt, B.; Geyer, F.C.; Reis-Filho, J.S. Histological types of breast cancer: How special are they? Mol. Oncol. 2010, 4, 192–208. [Google Scholar] [CrossRef]
- Rivenbark, A.G.; O’Connor, S.M.; Coleman, W.B. Molecular and cellular heterogeneity in breast cancer: Challenges for personalized medicine. Am. J. Pathol. 2013, 183, 1113–1124. [Google Scholar] [CrossRef]
- Yap, Y.S.; Lu, Y.S.; Tamura, K.; Lee, J.E.; Ko, E.Y.; Park, Y.H.; Cao, A.Y.; Lin, C.H.; Toi, M.; Wu, J.; et al. Insights into Breast Cancer in the East vs. the West: A Review. JAMA Oncol. 2019, 5, 1489–1496. [Google Scholar] [CrossRef]
- Yu, N.Y.; Iftimi, A.; Yau, C.; Tobin, N.P.; van ‘t Veer, L.; Hoadley, K.A.; Benz, C.C.; Nordenskjöld, B.; Fornander, T.; Stål, O.; et al. Assessment of Long-term Distant Recurrence-Free Survival Associated with Tamoxifen Therapy in Postmenopausal Patients With Luminal A or Luminal B Breast Cancer. JAMA Oncol. 2019, 5, 1304–1309. [Google Scholar] [CrossRef]
- Palomeras, S.; Ruiz-Martínez, S.; Puig, T. Targeting Breast Cancer Stem Cells to Overcome Treatment Resistance. Molecules 2018, 23, 2193. [Google Scholar] [CrossRef]
- Byler, S.; Goldgar, S.; Heerboth, S.; Leary, M.; Housman, G.; Moulton, K.; Sarkar, S. Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res. 2014, 34, 1071–1077. [Google Scholar] [PubMed]
- Huang, J.; Li, H.; Ren, G. Epithelial-mesenchymal transition and drug resistance in breast cancer (Review). Int. J. Oncol. 2015, 47, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Veeraraghavan, J.; Ma, J.; Hu, Y.; Wang, X.S. Recurrent and pathological gene fusions in breast cancer: Current advances in genomic discovery and clinical implications. Breast Cancer Res. Treat. 2016, 158, 219–232. [Google Scholar] [CrossRef]
- Wood, S.L.; Westbrook, J.A.; Brown, J.E. Omic-profiling in breast cancer metastasis to bone: Implications for mechanisms, biomarkers and treatment. Cancer Treat. Rev. 2014, 40, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.; Francavilla, C. ‘Omics Approaches to Explore the Breast Cancer Landscape. Front. Cell Dev. Biol. 2020, 7, 395. [Google Scholar] [CrossRef] [PubMed]
- Osin, P.; Shipley, J.; Lu, Y.J.; Crook, T.; Gusterson, B.A. Experimental pathology and breast cancer genetics: New technologies. Recent Results Cancer Res. 1998, 152, 35–48. [Google Scholar] [PubMed]
- Naser Al Deen, N.; Nassar, F.; Nasr, R.; Talhouk, R. Cross-Roads to Drug Resistance and Metastasis in Breast Cancer: miRNAs Regulatory Function and Biomarker Capability. Adv. Exp. Med. Biol. 2019, 1152, 335–364. [Google Scholar]
- Mueller, C.; Haymond, A.; Davis, J.B.; Williams, A.; Espina, V. Protein biomarkers for subtyping breast cancer and implications for future research. Expert Rev. Proteom. 2018, 15, 131–152. [Google Scholar] [CrossRef]
- Bambang, F.; Lu, D.; Li, H.; Chiu, L.L.; Lau, Q.C.; Koay, E.; Zhang, D. Cytokeratin 19 Regulates Endoplasmic Reticulum Stress and Inhibits ERp29 Expression via p38 MAPK/XBP-1 Signaling in Breast Cancer Cells. Exp. Cell Res. 2009, 315, 1964–1974. [Google Scholar] [CrossRef]
- Carta, F.; Demuro, P.P.; Zanini, C.; Santona, A.; Castiglia, D.; D’Atri, S.; Ascierto, P.A.; Napolitano, M.; Cossu, A.; Tadolini, B.; et al. Analysis of candidate genes through a proteomics-based approach in primary cell lines from malignant melanomas and their metastases. Melanoma Res. 2005, 15, 235–244. [Google Scholar] [CrossRef]
- Lee, D.J.; Kang, D.H.; Choi, M.; Choi, Y.J.; Lee, J.Y.; Park, J.H.; Park, Y.J.; Lee, K.W.; Kang, S.W. Peroxiredoxin-2 represses melanoma metastasis by increasing E-Cadherin/β-Catenin complexes in adherens junctions. Cancer Res. 2013, 73, 4744–4757. [Google Scholar] [CrossRef] [PubMed]
- Karhemo, P.R.; Rivinoja, A.; Lundin, J.; Hyvönen, M.; Chernenko, A.; Lammi, J.; Sihto, H.; Lundin, M.; Heikkilä, P.; Joensuu, H.; et al. An Extensive Tumor Array Analysis Supports Tumor Suppressive Role for Nucleophosmin in Breast Cancer. Am. J. Pathol. 2011, 179, 1004–1014. [Google Scholar] [CrossRef]
- Rochefort, H.; Garcia, M.; Glondu, M.; Laurent, V.; Liaudet, E.; Rey, J.M.; Roger, P. Cathepsin D in breast cancer: Mechanisms and clinical applications, a 1999 overview. Clin. Chim. Acta 2000, 291, 157–170. [Google Scholar] [CrossRef]
- Dian, D.; Heublein, S.; Wiest, I.; Barthell, L.; Friese, K.; Jeschke, U. Significance of the tumor protease cathepsin D for the biology of breast cancer. Histol. Histopathol. 2014, 29, 433–438. [Google Scholar]
- Dubey, V.; Luqman, S. Cathepsin D as a Promising Target for the Discovery of Novel Anticancer Agents. Curr. Cancer Drug Targets 2017, 17, 404–422. [Google Scholar] [CrossRef] [PubMed]
- Liaudet-Coopman, E.; Beaujouin, M.; Derocq, D.; Garcia, M.; Glondu-Lassis, M.; Laurent-Matha, V.; Prébois, C.; Rochefort, H.; Vignon, F. Cathepsin D: Newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006, 237, 167–179. [Google Scholar] [CrossRef]
- Rochefort, H. Estrogens, cathepsin D and metastasis in cancers of the breast and ovary: Invasion or proliferation? C. R. Seances Soc. Biol. Fil. 1998, 192, 241–251. [Google Scholar] [PubMed]
- Rochefort, H.; Platet, N.; Hayashido, Y.; Derocq, D.; Lucas, A.; Cunat, S.; Garcia, M. Estrogen receptor mediated inhibition of cancer cell invasion and motility: An overview. J. Steroid Biochem. Mol. Biol. 1998, 65, 163–168. [Google Scholar] [CrossRef]
- Chu, P.Y.; Hou, M.F.; Lai, J.C.; Chen, L.F.; Lin, C.S. Cell Reprogramming in Tumorigenesis and Its Therapeutic Implications for Breast Cancer. Int. J. Mol. Sci. 2019, 20, 1827. [Google Scholar] [CrossRef]
- Garcia, M.; Platet, N.; Liaudet, E.; Laurent, V.; Derocq, D.; Brouillet, J.P.; Rochefort, H. Biological and Clinical Significance of Cathepsin D in Breast Cancer Metastasis. Stem Cells 1996, 14, 642–650. [Google Scholar] [CrossRef]
- Pan, T.L.; Wang, P.W.; Huang, C.C.; Yeh, C.T.; Hu, T.H.; Yu, J.S. Network analysis and proteomic identification of vimentin as a key regulator associated with invasion and metastasis in human hepatocellular carcinoma cells. J. Proteomics 2012, 75, 4676–4692. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.W.; Lin, T.Y.; Hung, Y.C.; Chang, W.N.; Yang, P.M.; Chen, M.H.; Yeh, C.T.; Pan, T.L. Characterization of Fibrinogen as a Key Modulator in Patients with Wilson’s Diseases with Functional Proteomic Tools. Int. J. Mol. Sci. 2019, 20, 4528. [Google Scholar] [CrossRef] [PubMed]
- Yom, C.K.; Noh, D.Y.; Kim, W.H.; Kim, H.S. Clinical significance of high focal adhesion kinase gene copy number and overexpression in invasive breast cancer. Breast Cancer Res. Treat. 2011, 128, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.W.; Wu, T.H.; Lin, T.Y.; Chen, M.H.; Yeh, C.T.; Pan, T.L. Characterization of the Roles of Vimentin in Regulating the Proliferation and Migration of HSCs during Hepatic Fibrogenesis. Cells 2019, 8, 1184. [Google Scholar] [CrossRef]
- Pan, T.L.; Wang, P.W.; Leu, Y.L.; Wu, T.H.; Wu, T.S. Inhibitory effects of Scutellaria baicalensis extract on hepatic stellate cells through inducing G2/M cell cycle arrest and activating ERK-dependent apoptosis via Bax and caspase pathway. J. Ethnopharmacol. 2012, 139, 829–837. [Google Scholar] [CrossRef]
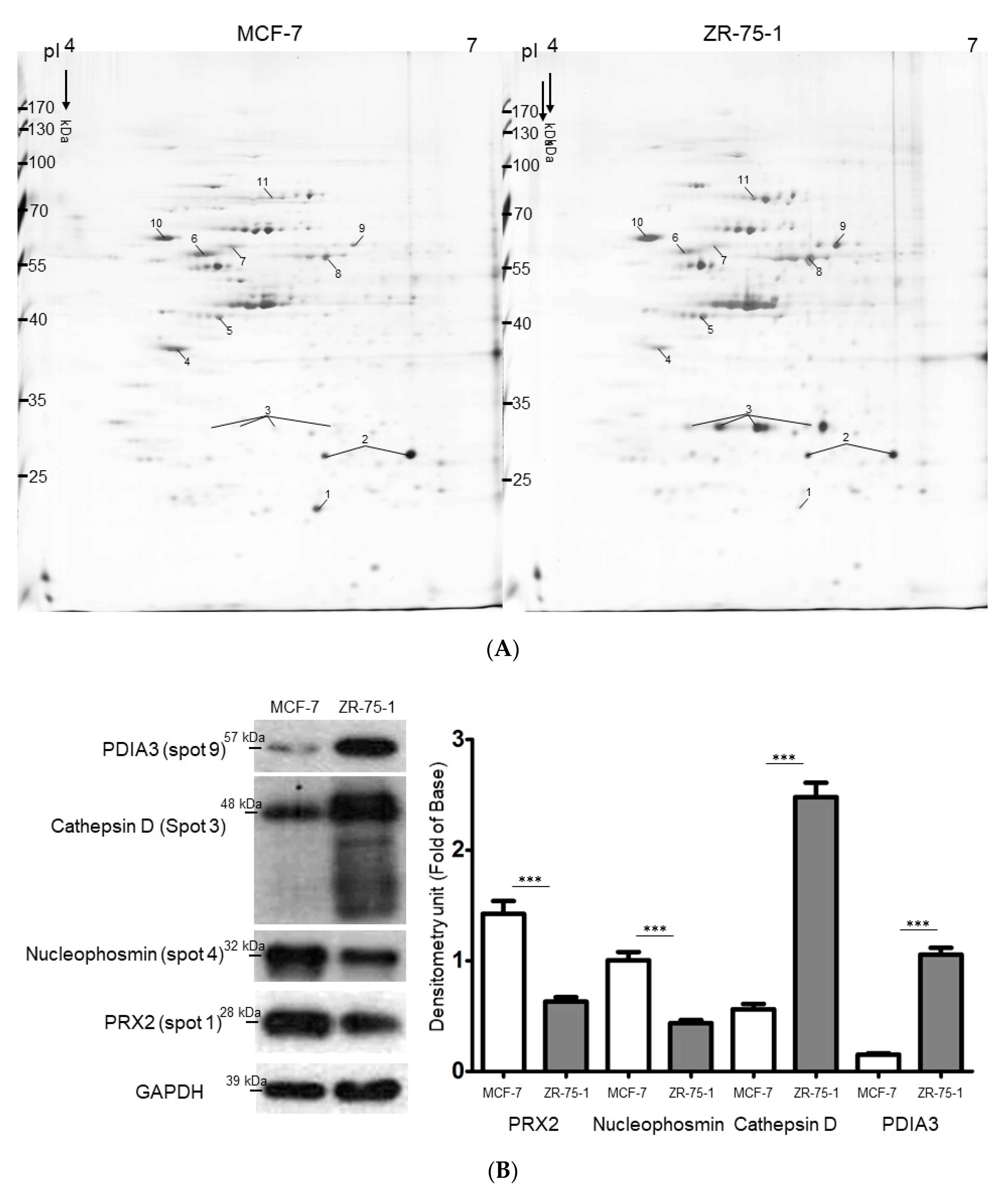
| Spot No. | Protein Name | Accession Number | Mw/pI | Score (Coverage) 1 | Match Fragment | Fold Change 2 (ZR-75-1 vs. MCF-7) | p-Value 3 | Function |
|---|---|---|---|---|---|---|---|---|
| 1 | Peroxiredoxin-2 | P32119 | 22.049/5.66 | 111 (56%) | 15 | −3.59 ± 0.04 | 0.001 | Plays a role in cell protection against oxidative stress by detoxifying peroxides and as sensor of hydrogen peroxide-mediated signaling events. |
| 2 | Heat shock protein beta-1 | P04792 | 22.826/5.98 | 71 (55%) | 16 | −1.56 ± 0.08 | 0.045 | Plays a role in stress resistance and actin organization. |
| 3 | Cathepsin D | P07339 | 45.037/6.10 | 85 (39%) | 17 | +5.88 ± 0.02 | 0.001 | Involved in the pathogenesis of several diseases such as breast cancer and possibly Alzheimer disease. |
| 4 | Nucleophosmin (B23) | P06748 | 32.7264.14 | 20(11%) | 3 | −2.46 ± 0.03 | 0.038 | Involved in diverse cellular processes such as ribosome biogenesis, centrosome duplication, protein chaperoning, histone assembly, cell proliferation, and regulation of tumor suppressors p53/TP53 and ARF. |
| 5 | Cytokeratin-19 | P08727 | 44.079/5.04 | 255 (78%) | 38 | +1.58 ± 0.03 | 0.002 | Together with KRT8, helps to link the contractile apparatus to dystrophin at the costameres of striated muscle. |
| 6 | Tubulin beta chain | P07437 | 50.096/4.75 | 108 (44%) | 22 | −1.68 ± 0.01 | 0.049 | Tubulin is the major constituent of microtubules. |
| 7 | Tubulin α-1B chain | P68363 | 50.804/4.94 | 50 (34%) | 11 | −1.57 ± 0.05 | 0.042 | Tubulin is the major constituent of microtubules. |
| 8 | Cytokeratin-8 | P05787 | 53.529/5.52 | 170 (50%) | 31 | +3.28 ± 0.04. | 0.003 | Together with KRT19, helps to link the contractile apparatus to dystrophin at the costameres of striated muscle. |
| 9 | Protein disulfide-isomerase A3 | P30101 | 55.328/6.42 | 86 (45%) | 19 | +2.58 ± 0.01 | 0.018 | Catalyzes the rearrangement of -S-S- bonds in proteins. |
| 10 | Protein disulfide-isomerase | P07237 | 57.48/4.76 | 70 (45%) | 23 | +2.35 ± 0.04 | 0.023 | This multifunctional protein catalyzes the formation, breakage and rearrangement of disulfide bonds. |
| 11 | Heat shock cognate 71 kDa protein | P11142 | 71.082/5.37 | 136 (50%) | 32 | +3.45 ± 0.04 | 0.029 | Molecular chaperone implicated in a wide variety of cellular processes, including protection of the proteome from stress, folding and transport of newly synthesized polypeptides, activation of proteolysis of misfolded proteins and the formation and dissociation of protein complexes. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, T.-Y.; Wang, P.-W.; Huang, C.-H.; Yang, P.-M.; Pan, T.-L. Characterizing the Relapse Potential in Different Luminal Subtypes of Breast Cancers with Functional Proteomics. Int. J. Mol. Sci. 2020, 21, 6077. https://doi.org/10.3390/ijms21176077
Lin T-Y, Wang P-W, Huang C-H, Yang P-M, Pan T-L. Characterizing the Relapse Potential in Different Luminal Subtypes of Breast Cancers with Functional Proteomics. International Journal of Molecular Sciences. 2020; 21(17):6077. https://doi.org/10.3390/ijms21176077
Chicago/Turabian StyleLin, Tung-Yi, Pei-Wen Wang, Chun-Hsun Huang, Pei-Ming Yang, and Tai-Long Pan. 2020. "Characterizing the Relapse Potential in Different Luminal Subtypes of Breast Cancers with Functional Proteomics" International Journal of Molecular Sciences 21, no. 17: 6077. https://doi.org/10.3390/ijms21176077
APA StyleLin, T.-Y., Wang, P.-W., Huang, C.-H., Yang, P.-M., & Pan, T.-L. (2020). Characterizing the Relapse Potential in Different Luminal Subtypes of Breast Cancers with Functional Proteomics. International Journal of Molecular Sciences, 21(17), 6077. https://doi.org/10.3390/ijms21176077








