Differential Labeling of Glycoproteins with Alkynyl Fucose Analogs
Abstract
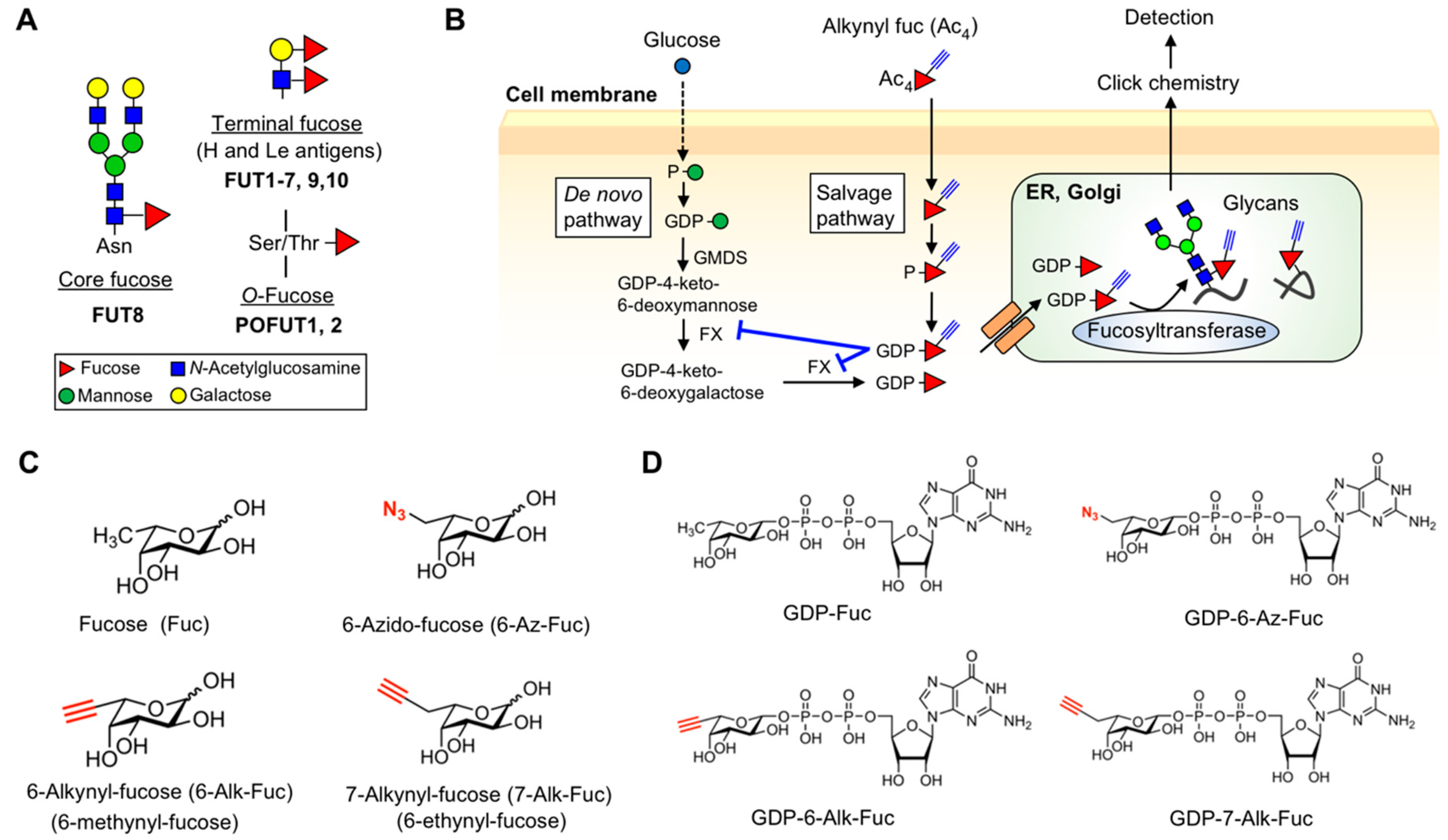
1. Introduction
2. Results
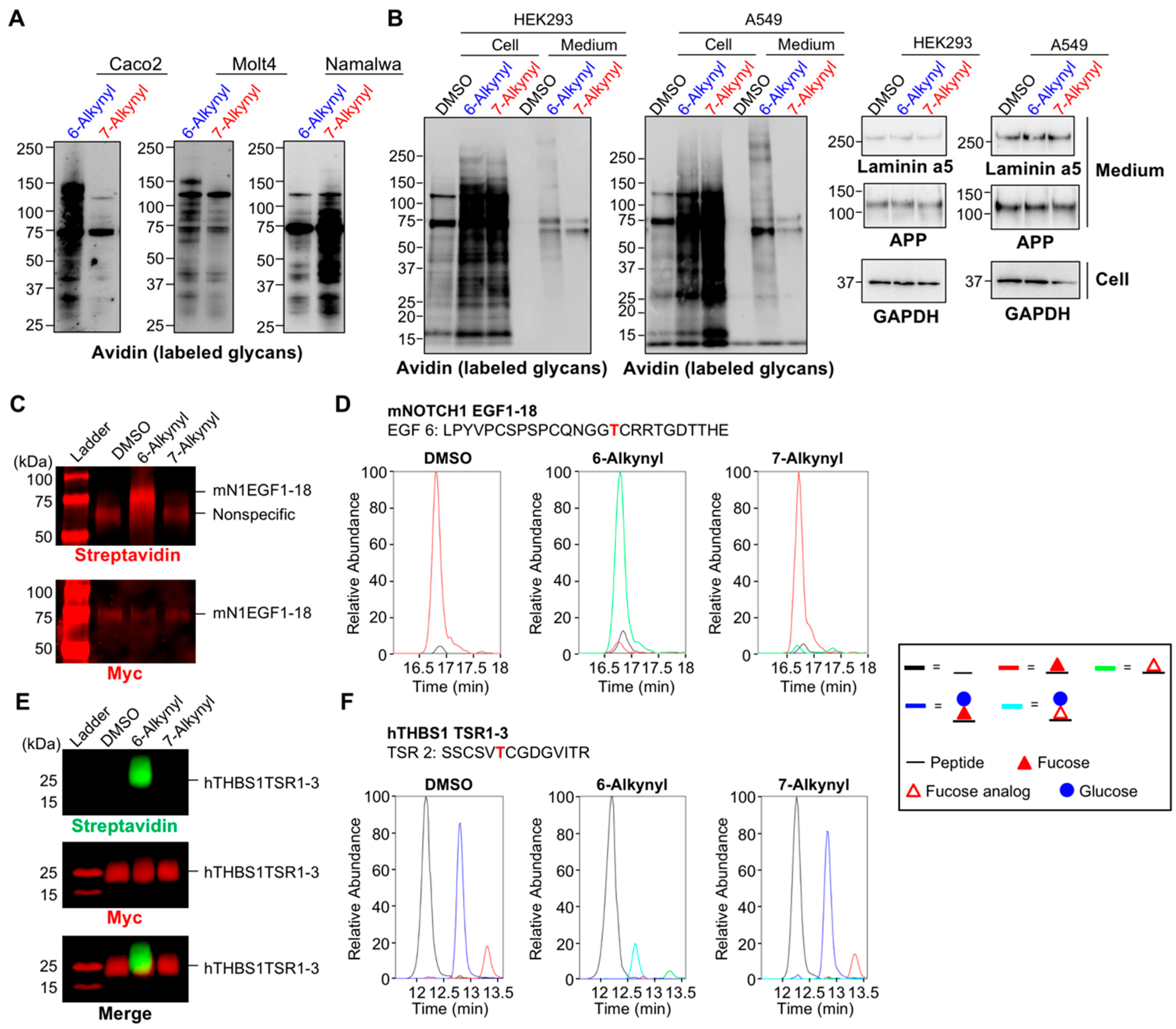
2.1. Differential Incorporation of 6-Alk-Fuc and 7-Alk-Fuc into Glycoproteins
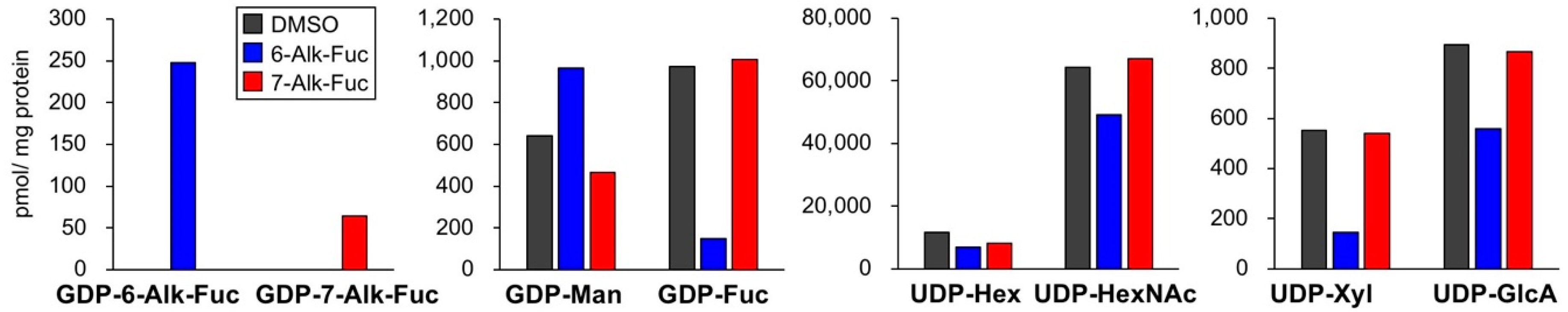
2.2. Differences in the Levels of GDP-6-Alk-Fuc and GDP-7-Alk-Fuc in Cells
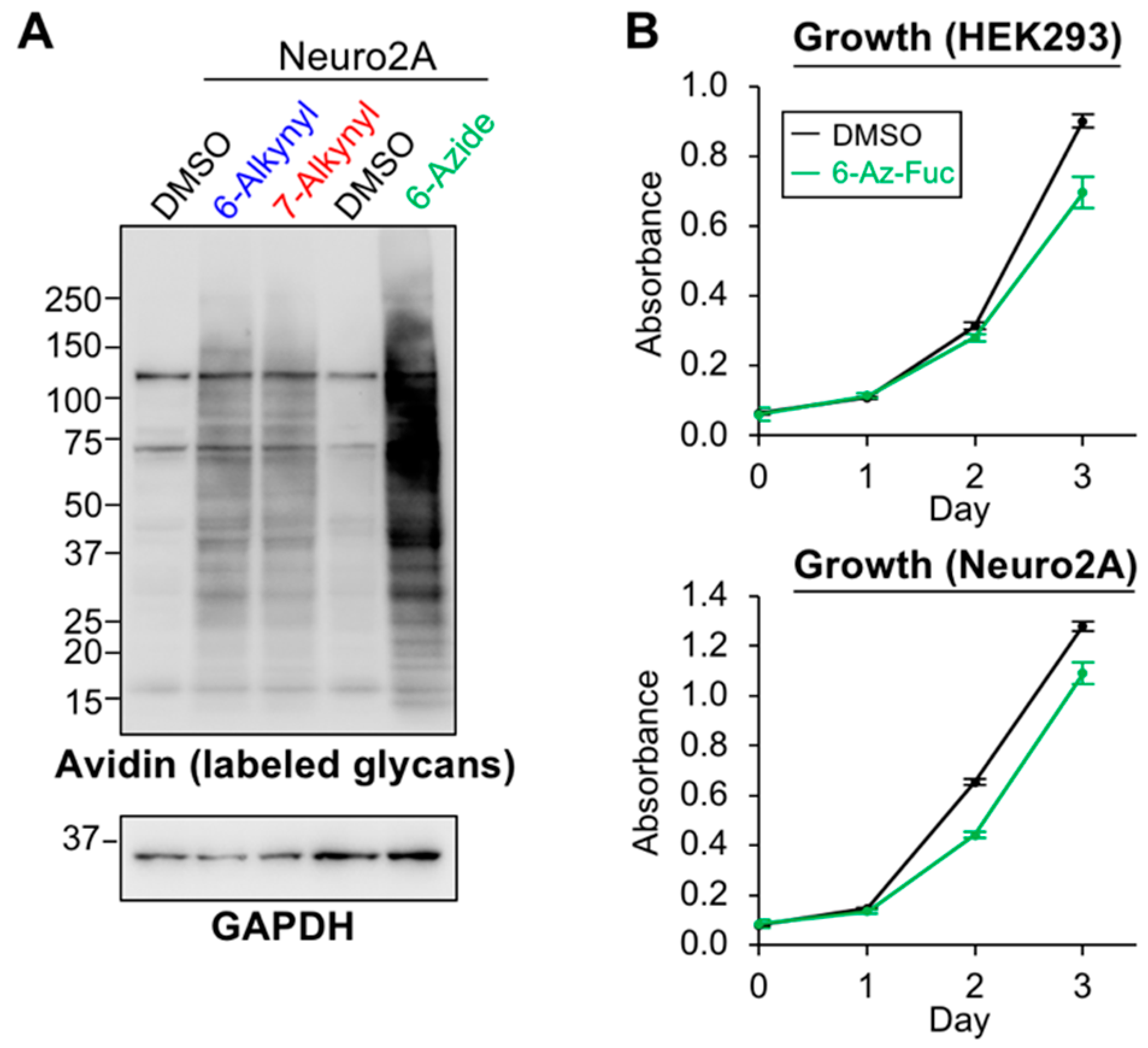
2.3. Labeling Efficiency and Cytotoxicity of 6-Az-Fuc
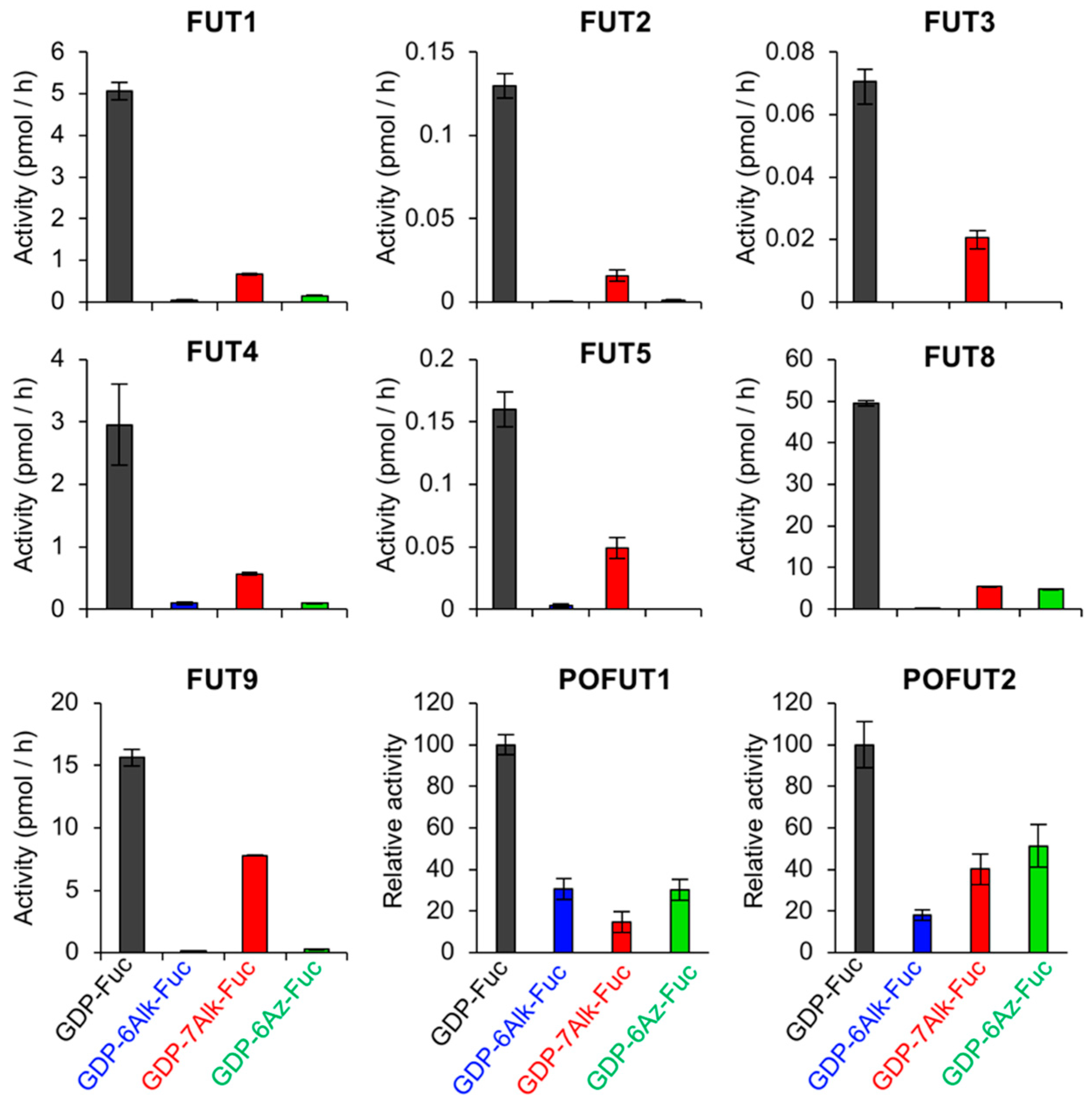
2.4. Tolerance of Fucosyltransferases with Fucose Analogs
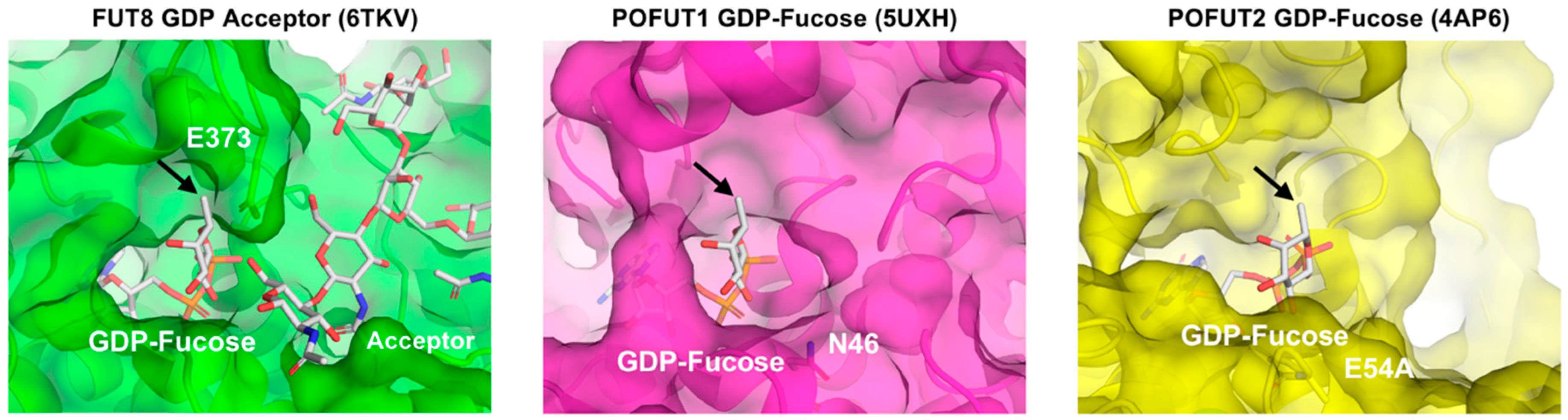
2.5. Structural Model of Fucosyltransferases and GDP-Fuc Analogs
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Plasmid Construction
4.4. Labeling and Detection of Fucosylated Glycans in Cells and Media
4.4.1. Labeling
4.4.2. SDS-PAGE, Detection of Biotinylated Proteins, and Western Blot
4.5. Labeling and Detection of O-Fuc Glycans
4.5.1. Production of 6- or 7-Alk-Fuc-Modified mNOTCH1 EGF1-18 and hTHBS1 TSR1-3
4.5.2. Biotinylation and Detection of 6- or 7-Alk-Fuc-Modified mNOTCH1 EGF1-18 and hTHBS1 TSR1-3 Proteins
4.5.3. Mass Spectrometry of 6- or 7-Alk-Fuc-Modified mNOTCH1 EGF1-18 and hTHBS1 TSR1-3 Proteins
4.6. Nucleotide Sugar Analysis
4.7. Cell Proliferation Assay
4.8. Activity Assay of FUTs
4.9. Activity Assay of POFUTs
4.9.1. Preparation and Purification of EGF Repeats and THBS1-TSR3 Repeats
4.9.2. Expression and Purification of Recombinant POFUT1 and POFUT2 Proteins
4.9.3. Determination of POFUTs’ Activities towards GDP-Fuc Analogs
4.10. Structural Modeling of GDP-Fucose Analogs and Fucosyltransferases
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Alk | Alkynyl |
| Fuc | Fucose |
| POFUT | Protein O-fucosyltransferase |
| Le | Lewis |
| ADCC | Antibody-dependent cellular cytotoxicity |
| FUT | Fucosyltransferase |
| EGF | Epidermal growth factor |
| TSR | Thrombospondin type 1 repeat |
| Man | Mannose |
| FX | GDP-4-keto-6-deoxymannose 3,5-epimerase-4-reductase |
| APP | Amyloid precursor protein |
| THBS1 | Thrombospondin 1 |
| LNnT | Lacto-N-neotetraose |
| PA | Pyridylamine |
References
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef]
- Kizuka, Y.; Kitazume, S.; Fujinawa, R.; Saito, T.; Iwata, N.; Saido, T.C.; Nakano, M.; Yamaguchi, Y.; Hashimoto, Y.; Staufenbiel, M.; et al. An aberrant sugar modification of BACE1 blocks its lysosomal targeting in Alzheimer’s disease. EMBO Mol. Med. 2015, 7, 175–189. [Google Scholar] [CrossRef]
- Ohtsubo, K.; Chen, M.Z.; Olefsky, J.M.; Marth, J.D. Pathway to diabetes through attenuation of pancreatic beta cell glycosylation and glucose transport. Nat. Med. 2011, 17, 1067–1075. [Google Scholar] [CrossRef]
- Granovsky, M.; Fata, J.; Pawling, J.; Muller, W.J.; Khokha, R.; Dennis, J.W. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat. Med. 2000, 6, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Rudman, N.; Gornik, O.; Lauc, G. Altered N-glycosylation profiles as potential biomarkers and drug targets in diabetes. FEBS Lett. 2019, 593, 1598–1615. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Al-Shareffi, E.; Haltiwanger, R.S. Biological functions of fucose in mammals. Glycobiology 2017, 27, 601–618. [Google Scholar] [CrossRef]
- Becker, D.J.; Lowe, J.B. Fucose: Biosynthesis and biological function in mammals. Glycobiology 2003, 13, 41R–53R. [Google Scholar] [CrossRef]
- Wang, X.; Inoue, S.; Gu, J.; Miyoshi, E.; Noda, K.; Li, W.; Mizuno-Horikawa, Y.; Nakano, M.; Asahi, M.; Takahashi, M.; et al. Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. USA 2005, 102, 15791–15796. [Google Scholar] [CrossRef]
- Fukuda, T.; Hashimoto, H.; Okayasu, N.; Kameyama, A.; Onogi, H.; Nakagawasai, O.; Nakazawa, T.; Kurosawa, T.; Hao, Y.; Isaji, T.; et al. Alpha1,6-fucosyltransferase-deficient mice exhibit multiple behavioral abnormalities associated with a schizophrenia-like phenotype: Importance of the balance between the dopamine and serotonin systems. J. Biol. Chem. 2011, 286, 18434–18443. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Shinzaki, S.; Iijima, H.; Wakamatsu, K.; Iwamoto, C.; Sobajima, T.; Kuwahara, R.; Hiyama, S.; Hayashi, Y.; Takamatsu, S.; et al. Core fucosylation on T cells, required for activation of T-cell receptor signaling and induction of colitis in mice, is increased in patients with inflammatory bowel disease. Gastroenterology 2016, 150, 1620–1632. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, R.; Ma, B.; Yang, Y.; Jiao, X.; Liu, Y.; Cao, H.; Dong, W.; Liu, L.; Ma, K.; et al. Core fucosylation of IgG B cell receptor is required for antigen recognition and antibody production. J. Immunol. 2015, 194, 2596–2606. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, N.; Kizuka, Y. Glycans and cancer: Role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv. Cancer Res. 2015, 126, 11–51. [Google Scholar]
- Agrawal, P.; Fontanals-Cirera, B.; Sokolova, E.; Jacob, S.; Vaiana, C.A.; Argibay, D.; Davalos, V.; McDermott, M.; Nayak, S.; Darvishian, F.; et al. A systems biology approach identifies FUT8 as a driver of melanoma metastasis. Cancer Cell 2017, 31, 804–819. [Google Scholar] [CrossRef]
- Shinkawa, T.; Nakamura, K.; Yamane, N.; Shoji-Hosaka, E.; Kanda, Y.; Sakurada, M.; Uchida, K.; Anazawa, H.; Satoh, M.; Yamasaki, M.; et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2003, 278, 3466–3473. [Google Scholar] [CrossRef]
- Shields, R.L.; Lai, J.; Keck, R.; O’Connell, L.Y.; Hong, K.; Meng, Y.G.; Weikert, S.H.; Presta, L.G. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002, 277, 26733–26740. [Google Scholar] [CrossRef]
- Ley, K.; Kansas, G.S. Selectins in T-cell recruitment to non-lymphoid tissues and sites of inflammation. Nat. Rev. Immunol. 2004, 4, 325–335. [Google Scholar] [CrossRef]
- Rosen, S.D.; Bertozzi, C.R. The selectins and their ligands. Curr. Opin. Cell Biol. 1994, 6, 663–673. [Google Scholar] [CrossRef]
- Homeister, J.W.; Thall, A.D.; Petryniak, B.; Maly, P.; Rogers, C.E.; Smith, P.L.; Kelly, R.J.; Gersten, K.M.; Askari, S.W.; Cheng, G.; et al. The alpha(1,3)fucosyltransferases FucT-IV and FucT-VII exert collaborative control over selectin-dependent leukocyte recruitment and lymphocyte homing. Immunity 2001, 15, 115–126. [Google Scholar] [CrossRef]
- Matsumura, R.; Hirakawa, J.; Sato, K.; Ikeda, T.; Nagai, M.; Fukuda, M.; Imai, Y.; Kawashima, H. Novel antibodies reactive with sialyl lewis X in both humans and mice define its critical role in leukocyte trafficking and contact hypersensitivity responses. J. Biol. Chem. 2015, 290, 15313–15326. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J. The glycosylation landscape of pancreatic cancer. Oncol. Lett. 2019, 17, 2569–2575. [Google Scholar] [CrossRef] [PubMed]
- Urata, Y.; Takeuchi, H. Effects of Notch glycosylation on health and diseases. Dev. Growth Differ. 2020, 62, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Holdener, B.C.; Haltiwanger, R.S. Protein O-fucosylation: Structure and function. Curr. Opin. Struct. Biol. 2019, 56, 78–86. [Google Scholar] [CrossRef]
- Luo, Y.; Haltiwanger, R.S. O-fucosylation of notch occurs in the endoplasmic reticulum. J. Biol. Chem. 2005, 280, 11289–11294. [Google Scholar] [CrossRef]
- Luo, Y.; Koles, K.; Vorndam, W.; Haltiwanger, R.S.; Panin, V.M. Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. J. Biol. Chem. 2006, 281, 9393–9399. [Google Scholar] [CrossRef]
- Shi, S.; Stanley, P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. USA 2003, 100, 5234–5239. [Google Scholar] [CrossRef]
- Li, M.; Cheng, R.; Liang, J.; Yan, H.; Zhang, H.; Yang, L.; Li, C.; Jiao, Q.; Lu, Z.; He, J.; et al. Mutations in POFUT1, encoding protein O-fucosyltransferase 1, cause generalized Dowling-Degos disease. Am. J. Hum. Genet. 2013, 92, 895–903. [Google Scholar] [CrossRef]
- Ma, L.; Dong, P.; Liu, L.; Gao, Q.; Duan, M.; Zhang, S.; Chen, S.; Xue, R.; Wang, X. Overexpression of protein O-fucosyltransferase 1 accelerates hepatocellular carcinoma progression via the Notch signaling pathway. Biochem. Biophys. Res. Commun. 2016, 473, 503–510. [Google Scholar] [CrossRef]
- Benz, B.A.; Nandadasa, S.; Takeuchi, M.; Grady, R.C.; Takeuchi, H.; LoPilato, R.K.; Kakuda, S.; Somerville, R.P.T.; Apte, S.S.; Haltiwanger, R.S.; et al. Genetic and biochemical evidence that gastrulation defects in Pofut2 mutants result from defects in ADAMTS9 secretion. Dev. Biol. 2016, 416, 111–122. [Google Scholar] [CrossRef]
- Du, J.; Takeuchi, H.; Leonhard-Melief, C.; Shroyer, K.R.; Dlugosz, M.; Haltiwanger, R.S.; Holdener, B.C. O-fucosylation of thrombospondin type 1 repeats restricts epithelial to mesenchymal transition (EMT) and maintains epiblast pluripotency during mouse gastrulation. Dev. Biol. 2010, 346, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Moremen, K.W.; Haltiwanger, R.S. Emerging structural insights into glycosyltransferase-mediated synthesis of glycans. Nat. Chem. Biol. 2019, 15, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Bennett, E.P.; Mandel, U.; Clausen, H.; Gerken, T.A.; Fritz, T.A.; Tabak, L.A. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 2012, 22, 736–756. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, S.T.; Bertozzi, C.R. Imaging the glycome. Proc. Natl. Acad. Sci. USA 2009, 106, 12–17. [Google Scholar] [CrossRef]
- Chang, P.V.; Chen, X.; Smyrniotis, C.; Xenakis, A.; Hu, T.; Bertozzi, C.R.; Wu, P. Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angew. Chem. Int. Ed. Engl. 2009, 48, 4030–4033. [Google Scholar] [CrossRef]
- Zaro, B.W.; Yang, Y.Y.; Hang, H.C.; Pratt, M.R. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc. Natl. Acad. Sci. USA 2011, 108, 8146–8151. [Google Scholar] [CrossRef]
- Vanbeselaere, J.; Vicogne, D.; Matthijs, G.; Biot, C.; Foulquier, F.; Guerardel, Y. Alkynyl monosaccharide analogues as a tool for evaluating Golgi glycosylation efficiency: Application to congenital disorders of glycosylation (CDG). Chem. Commun. (Camb.) 2013, 49, 11293–11295. [Google Scholar] [CrossRef]
- Sawa, M.; Hsu, T.L.; Itoh, T.; Sugiyama, M.; Hanson, S.R.; Vogt, P.K.; Wong, C.H. Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 12371–12376. [Google Scholar] [CrossRef]
- Rabuka, D.; Hubbard, S.C.; Laughlin, S.T.; Argade, S.P.; Bertozzi, C.R. A chemical reporter strategy to probe glycoprotein fucosylation. J. Am. Chem. Soc. 2006, 128, 12078–12079. [Google Scholar] [CrossRef]
- Hsu, T.L.; Hanson, S.R.; Kishikawa, K.; Wang, S.K.; Sawa, M.; Wong, C.H. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc. Natl. Acad. Sci. USA 2007, 104, 2614–2619. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Funayama, S.; Shogomori, H.; Nakano, M.; Nakajima, K.; Oka, R.; Kitazume, S.; Yamaguchi, Y.; Sano, M.; Korekane, H.; et al. High-sensitivity and low-toxicity fucose probe for glycan imaging and biomarker discovery. Cell Chem. Biol. 2016, 23, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Nakano, M.; Yamaguchi, Y.; Nakajima, K.; Oka, R.; Sato, K.; Ren, C.T.; Hsu, T.L.; Wong, C.H.; Taniguchi, N. An alkynyl-fucose halts hepatoma cell migration and invasion by inhibiting GDP-fucose-synthesizing enzyme FX, TSTA3. Cell Chem. Biol. 2017, 24, 1467–1478. [Google Scholar] [CrossRef] [PubMed]
- Rillahan, C.D.; Antonopoulos, A.; Lefort, C.T.; Sonon, R.; Azadi, P.; Ley, K.; Dell, A.; Haslam, S.M.; Paulson, J.C. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat. Chem. Biol. 2012, 8, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Zandberg, W.F.; Kumarasamy, J.; Pinto, B.M.; Vocadlo, D.J. Metabolic inhibition of sialyl-Lewis X biosynthesis by 5-thiofucose remodels the cell surface and impairs selectin-mediated cell adhesion. J. Biol. Chem. 2012, 287, 40021–40030. [Google Scholar] [CrossRef]
- Okeley, N.M.; Alley, S.C.; Anderson, M.E.; Boursalian, T.E.; Burke, P.J.; Emmerton, K.M.; Jeffrey, S.C.; Klussman, K.; Law, C.L.; Sussman, D.; et al. Development of orally active inhibitors of protein and cellular fucosylation. Proc. Natl. Acad. Sci. USA 2013, 110, 5404–5409. [Google Scholar] [CrossRef]
- Allen, J.G.; Mujacic, M.; Frohn, M.J.; Pickrell, A.J.; Kodama, P.; Bagal, D.; San Miguel, T.; Sickmier, E.A.; Osgood, S.; Swietlow, A.; et al. Facile modulation of antibody fucosylation with small molecule fucostatin inhibitors and cocrystal structure with GDP-mannose 4,6-dehydratase. ACS Chem. Biol. 2016, 11, 2734–2743. [Google Scholar] [CrossRef]
- Schneider, M.; Kumar, V.; Nordstrom, L.U.; Feng, L.; Takeuchi, H.; Hao, H.; Luca, V.C.; Garcia, K.C.; Stanley, P.; Wu, P.; et al. Inhibition of delta-induced Notch signaling using fucose analogs. Nat. Chem. Biol. 2018, 14, 65–71. [Google Scholar] [CrossRef]
- Al-Shareffi, E.; Chaubard, J.L.; Leonhard-Melief, C.; Wang, S.K.; Wong, C.H.; Haltiwanger, R.S. 6-alkynyl fucose is a bioorthogonal analog for O-fucosylation of epidermal growth factor-like repeats and thrombospondin type-1 repeats by protein O-fucosyltransferases 1 and 2. Glycobiology 2013, 23, 188–198. [Google Scholar] [CrossRef]
- Nakano, M.; Mishra, S.K.; Tokoro, Y.; Sato, K.; Nakajima, K.; Yamaguchi, Y.; Taniguchi, N.; Kizuka, Y. Bisecting GlcNAc is a general suppressor of terminal modification of N-glycan. Mol. Cell Proteom. 2019, 18, 2044–2057. [Google Scholar] [CrossRef]
- Tomida, S.; Takata, M.; Hirata, T.; Nagae, M.; Nakano, M.; Kizuka, Y. The SH3 domain in the fucosyltransferase FUT8 controls FUT8 activity and localization and is essential for core fucosylation. J. Biol. Chem. 2020, 295, 7992–8004. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.I.; Keusch, J.J.; Klein, D.; Hess, D.; Hofsteenge, J.; Gut, H. Structure of human POFUT2: Insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 2012, 31, 3183–3197. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, A.; Ceballos-Laita, L.; Serna, S.; Artschwager, R.; Reichardt, N.C.; Corzana, F.; Hurtado-Guerrero, R. Structural basis for substrate specificity and catalysis of alpha1,6-fucosyltransferase. Nat. Commun. 2020, 11, 973:1–973:9. [Google Scholar] [CrossRef]
- McMillan, B.J.; Zimmerman, B.; Egan, E.D.; Lofgren, M.; Xu, X.; Hesser, A.; Blacklow, S.C. Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations. Glycobiology 2017, 27, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Jarva, M.A.; Dramicanin, M.; Lingford, J.P.; Mao, R.; John, A.; Jarman, K.E.; Grinter, R.; Goddard-Borger, E.D. Structural basis of substrate recognition and catalysis by fucosyltransferase 8. J. Biol. Chem. 2020, 295, 6677–6688. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Han, K.; Pak, J.E.; Satkunarajah, M.; Zhou, D.; Rini, J.M. Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat. Chem. Biol. 2017, 13, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Hadley, B.; Maggioni, A.; Ashikov, A.; Day, C.J.; Haselhorst, T.; Tiralongo, J. Structure and function of nucleotide sugar transporters: Current progress. Comput. Struct. Biotechnol. J. 2014, 10, 23–32. [Google Scholar] [CrossRef]
- Ishikawa, H.O.; Ayukawa, T.; Nakayama, M.; Higashi, S.; Kamiyama, S.; Nishihara, S.; Aoki, K.; Ishida, N.; Sanai, Y.; Matsuno, K. Two pathways for importing GDP-fucose into the endoplasmic reticulum lumen function redundantly in the O-fucosylation of Notch in Drosophila. J. Biol. Chem. 2010, 285, 4122–4129. [Google Scholar] [CrossRef]
- Ashikov, A.; Routier, F.; Fuhlrott, J.; Helmus, Y.; Wild, M.; Gerardy-Schahn, R.; Bakker, H. The human solute carrier gene SLC35B4 encodes a bifunctional nucleotide sugar transporter with specificity for UDP-xylose and UDP-N-acetylglucosamine. J. Biol. Chem. 2005, 280, 27230–27235. [Google Scholar] [CrossRef]
- Lu, L.; Hou, X.; Shi, S.; Korner, C.; Stanley, P. Slc35c2 promotes Notch1 fucosylation and is required for optimal Notch signaling in mammalian cells. J. Biol. Chem. 2010, 285, 36245–36254. [Google Scholar] [CrossRef]
- Rana, N.A.; Nita-Lazar, A.; Takeuchi, H.; Kakuda, S.; Luther, K.B.; Haltiwanger, R.S. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J. Biol. Chem. 2011, 286, 31623–31637. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zheng, T.; Lopez-Aguilar, A.; Feng, L.; Kopp, F.; Marlow, F.L.; Wu, P. Monitoring dynamic glycosylation in vivo using supersensitive click chemistry. Bioconj. Chem. 2014, 25, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Kakuda, S.; Haltiwanger, R.S. Analyzing the posttranslational modification status of Notch using mass spectrometry. Methods Mol. Biol. 2014, 1187, 209–221. [Google Scholar] [PubMed]
- Ito, J.; Herter, T.; Baidoo, E.E.; Lao, J.; Vega-Sanchez, M.E.; Michelle Smith-Moritz, A.; Adams, P.D.; Keasling, J.D.; Usadel, B.; Petzold, C.J.; et al. Analysis of plant nucleotide sugars by hydrophilic interaction liquid chromatography and tandem mass spectrometry. Anal. Biochem. 2014, 448, 14–22. [Google Scholar] [CrossRef]
- Nakajima, K.; Kitazume, S.; Angata, T.; Fujinawa, R.; Ohtsubo, K.; Miyoshi, E.; Taniguchi, N. Simultaneous determination of nucleotide sugars with ion-pair reversed-phase HPLC. Glycobiology 2010, 20, 865–871. [Google Scholar] [CrossRef]
- Rampal, R.; Li, A.S.; Moloney, D.J.; Georgiou, S.A.; Luther, K.B.; Nita-Lazar, A.; Haltiwanger, R.S. Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. J. Biol. Chem. 2005, 280, 42454–42463. [Google Scholar] [CrossRef]
- Luo, Y.; Nita-Lazar, A.; Haltiwanger, R.S. Two distinct pathways for O-fucosylation of epidermal growth factor-like or thrombospondin type 1 repeats. J. Biol. Chem. 2006, 281, 9385–9392. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Kantharia, J.; Sethi, M.K.; Bakker, H.; Haltiwanger, R.S. Site-specific O-glucosylation of the epidermal growth factor-like (EGF) repeats of notch: Efficiency of glycosylation is affected by proper folding and amino acid sequence of individual EGF repeats. J. Biol. Chem. 2012, 287, 33934–33944. [Google Scholar] [CrossRef]
- Takeuchi, H.; Wong, D.; Schneider, M.; Freeze, H.H.; Takeuchi, M.; Berardinelli, S.J.; Ito, A.; Lee, H.; Nelson, S.F.; Haltiwanger, R.S. Variant in human POFUT1 reduces enzymatic activity and likely causes a recessive microcephaly, global developmental delay with cardiac and vascular features. Glycobiology 2018, 28, 276–283. [Google Scholar] [CrossRef]
- Urata, Y.; Saiki, W.; Tsukamoto, Y.; Sago, H.; Hibi, H.; Okajima, T.; Takeuchi, H. Xylosyl extension of o-glucose glycans on the extracellular domain of NOTCH1 and NOTCH2 regulates Notch cell surface Trafficking. Cells 2020, 9, 1220. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004, 60 Pt 12 Pt 1, 2256–2268. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Takeuchi, H.; Hao, H.; Yonekawa, C.; Nakajima, K.; Nagae, M.; Okajima, T.; Haltiwanger, R.S.; Kizuka, Y. Differential Labeling of Glycoproteins with Alkynyl Fucose Analogs. Int. J. Mol. Sci. 2020, 21, 6007. https://doi.org/10.3390/ijms21176007
Ma C, Takeuchi H, Hao H, Yonekawa C, Nakajima K, Nagae M, Okajima T, Haltiwanger RS, Kizuka Y. Differential Labeling of Glycoproteins with Alkynyl Fucose Analogs. International Journal of Molecular Sciences. 2020; 21(17):6007. https://doi.org/10.3390/ijms21176007
Chicago/Turabian StyleMa, Chenyu, Hideyuki Takeuchi, Huilin Hao, Chizuko Yonekawa, Kazuki Nakajima, Masamichi Nagae, Tetsuya Okajima, Robert S. Haltiwanger, and Yasuhiko Kizuka. 2020. "Differential Labeling of Glycoproteins with Alkynyl Fucose Analogs" International Journal of Molecular Sciences 21, no. 17: 6007. https://doi.org/10.3390/ijms21176007
APA StyleMa, C., Takeuchi, H., Hao, H., Yonekawa, C., Nakajima, K., Nagae, M., Okajima, T., Haltiwanger, R. S., & Kizuka, Y. (2020). Differential Labeling of Glycoproteins with Alkynyl Fucose Analogs. International Journal of Molecular Sciences, 21(17), 6007. https://doi.org/10.3390/ijms21176007





