Regulation of Female Folliculogenesis by Tsp1a in Nile Tilapia (Oreochromis niloticus)
Abstract
1. Introduction
2. Results
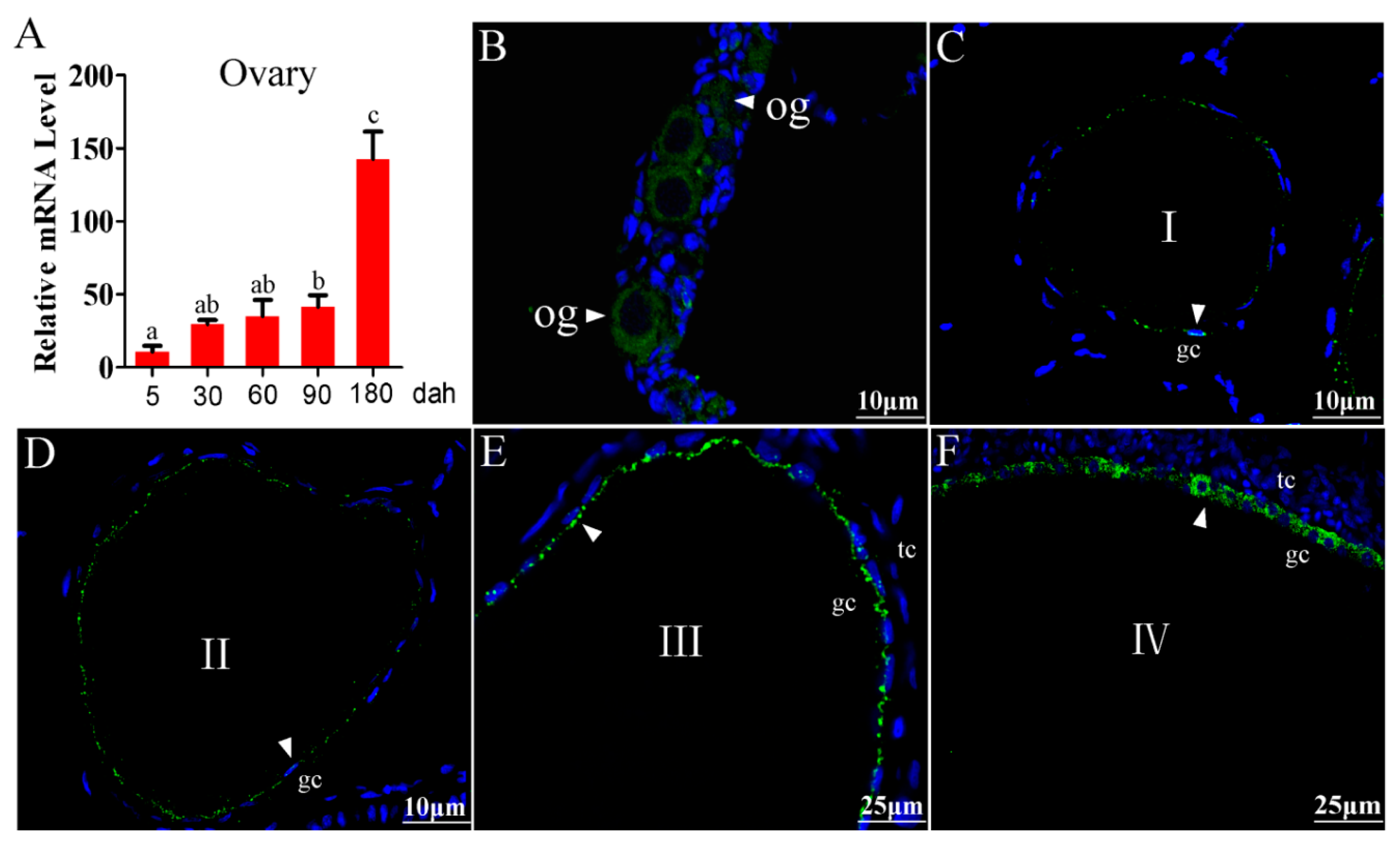
2.1. Cellular Localization of Tsp1a in Tilapia Ovaries
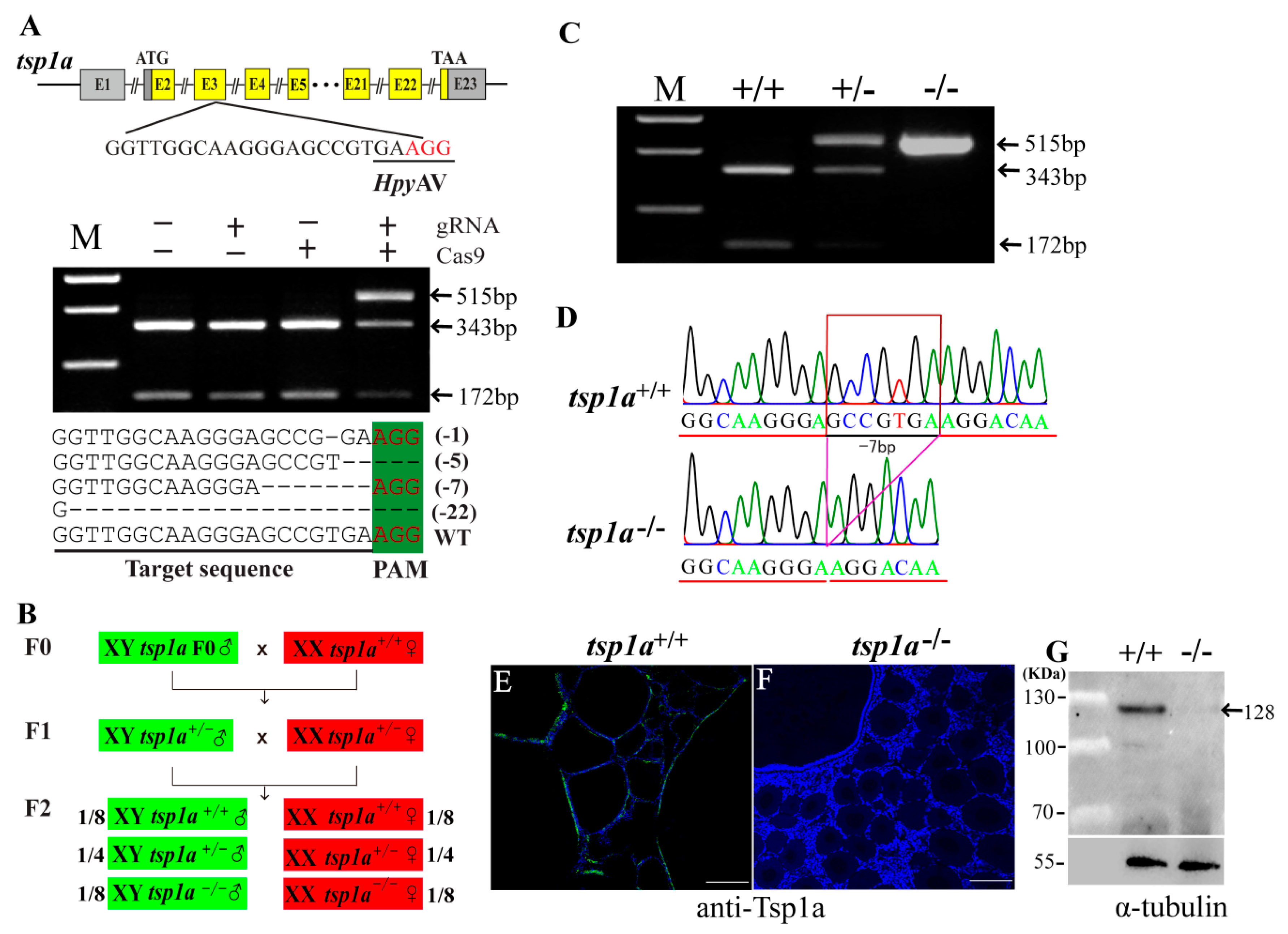
2.2. Generation of tsp1a Homozygous Mutants
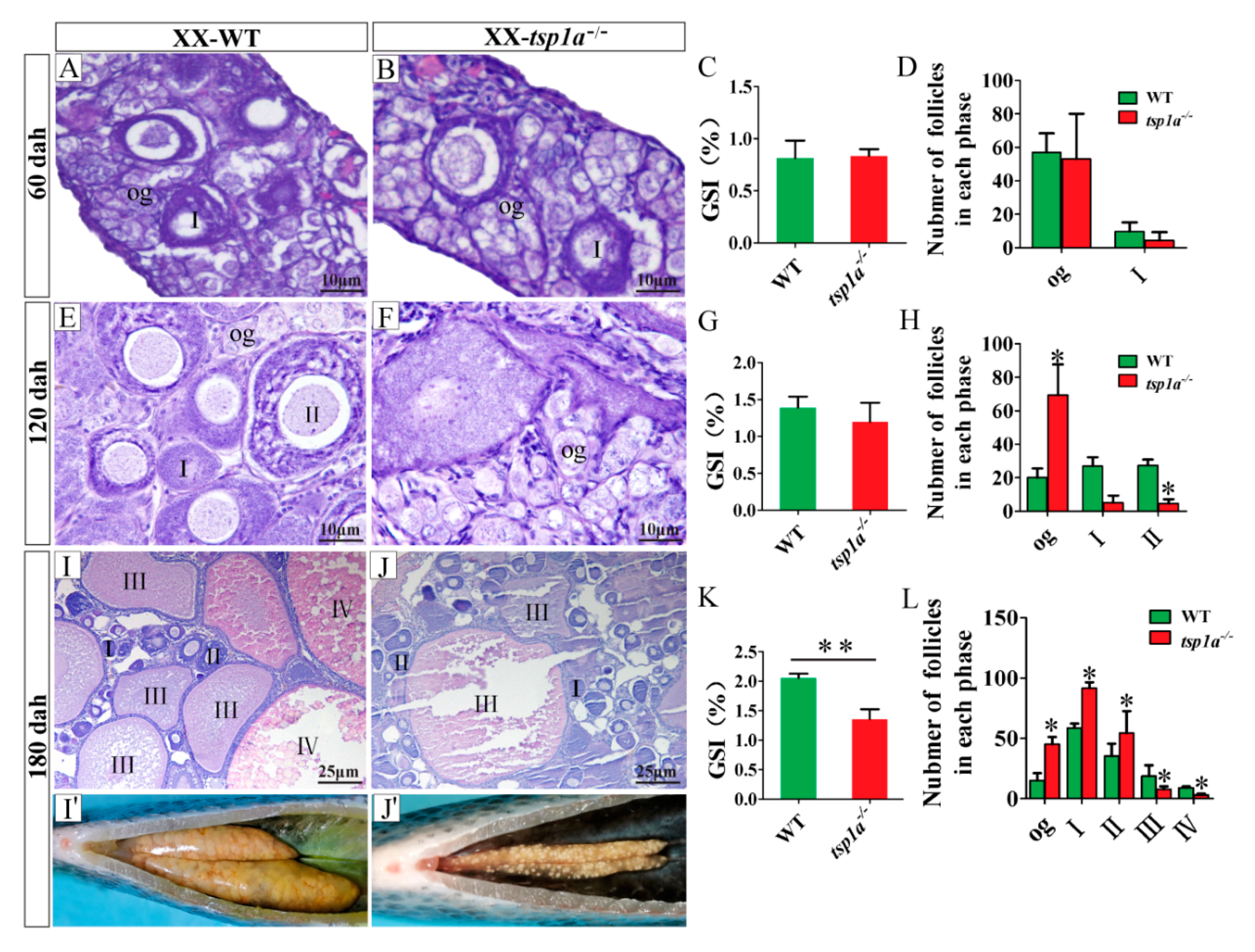
2.3. Homozygous Mutation of tsp1a Causes Increased Oogonia, Reduced Phase III and Phase IV Follicles and Retarded Follicle Growth
2.4. Loss of tsp1a Alters Expression Pattern of Genes Related to Ovary Development
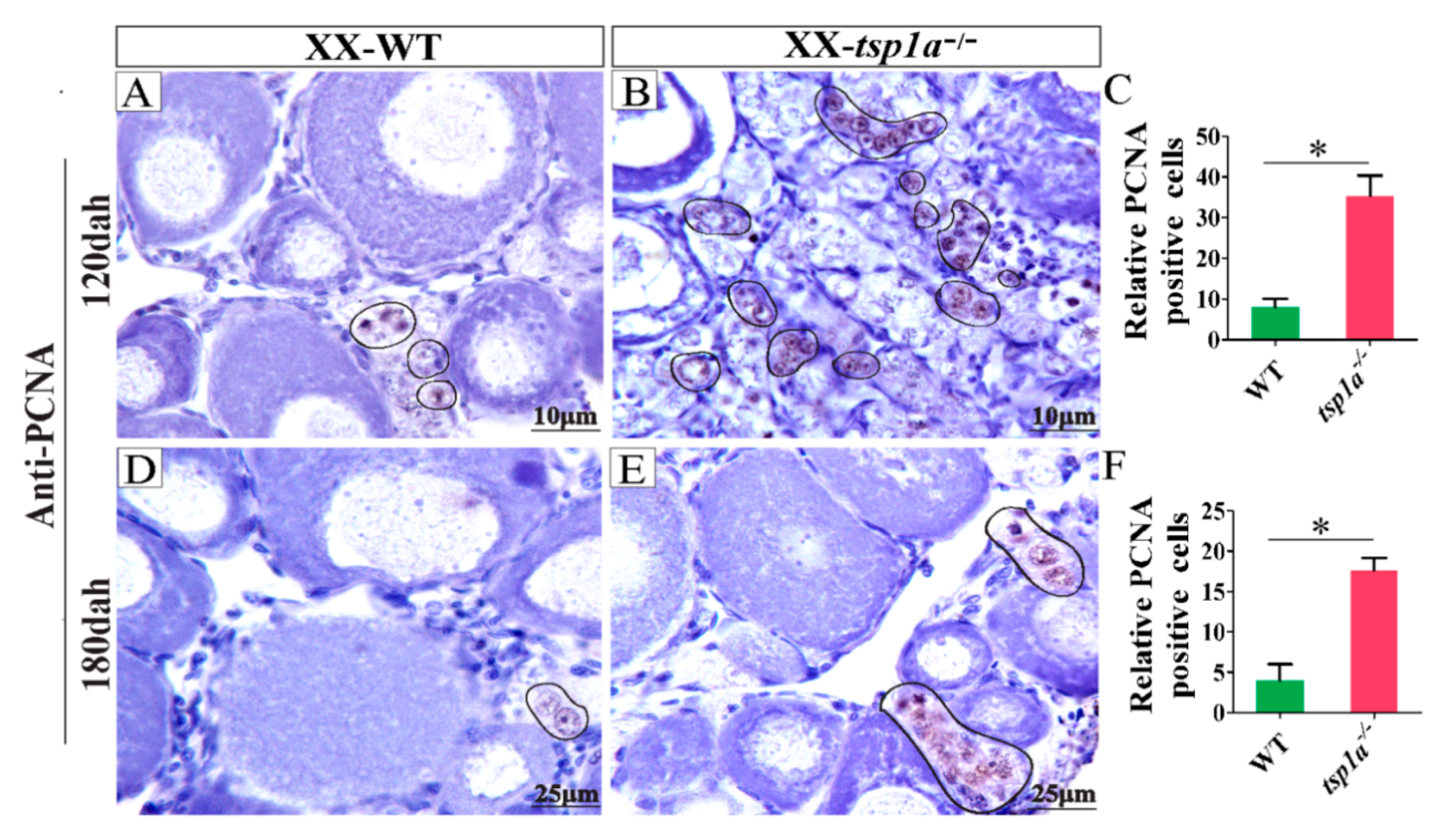
2.5. Up-Regulation of PCNA Expression in tsp1a−/− Ovaries in Comparison with WT Ovaries
2.6. Effects of tsp1a Homozygous Mutation on the Expression of Cyp19a1a and Serum E2 Level
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Establishment of tsp1a Homozygous Mutants by CRISPR/Cas9
4.3. Sampling and Histological Analysis
4.4. Immunofluorescence (IF) and Immunohistochemistry (IHC)
4.5. Transcriptome Sequencing and Analysis
4.6. Follicle Counting
4.7. Measurement of Serum E2 Level
4.8. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.9. Western Blot
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| E2 | 17β-estradiol |
| ECM | extracellular matrix |
| TSP1 | thrombospondin1 |
| qRT-PCR | quantitative reverse transcription polymerase chain reaction |
| IF | immunofluorescence |
| PAM | protospacer adjacent motif |
| GSI | gonadosomatic index |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| DEGs | differentially expressed genes |
| FDR | false discovery rate |
| IHC | immunohistochemistry |
| DAPI | 4′,6′-diamidine-2-phenylindole-dihydrochloride |
| GO | Gene Ontology |
| Dah | days after hatching |
| WT | wild type |
References
- Lubzens, E.; Young, G.; Bobe, J.; Cerdà, J. Oogenesis in teleosts: How fish eggs are formed. Gen. Comp. Endocrinol. 2010, 165, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Taranger, G.L.; Carrillo, M.; Schulz, R.W.; Fontaine, P.; Zanuy, S.; Felip, A.; Weltzien, F.A.; Dufour, S.; Karlsen, O.; Norberg, B.; et al. Control of puberty in farmed fish. Gen. Comp. Endocrinol. 2010, 165, 483–515. [Google Scholar] [CrossRef]
- Osz, K.; Ross, M.; Petrik, J. The thrombospondin-1 receptor CD36 is an important mediator of ovarian angiogenesis and folliculogenesis. Reprod. Biol. Endocrinol. 2014, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Yeung, C.K.; Wang, G.; Yao, Y.; Liang, J.; Chung, C.Y.T.; Chuai, M.; Lee, K.K.; Yang, X. BRE modulates granulosa cell death to affect ovarian follicle development and atresia in the mouse. Cell Death Dis. 2017, 8, e2697. [Google Scholar] [CrossRef]
- Petrik, J.J.; Gentry, P.A.; Feige, J.J.; LaMarre, J. Expression and localization of thrombospondin-1 and -2 and their cell-surface receptor, CD36, during rat follicular development and formation of the corpus luteum1. Biol. Reprod. 2002, 67, 1522–1531. [Google Scholar] [CrossRef][Green Version]
- Kliem, H.; Welter, H.; Kraetzl, W.D.; Steffl, M.; Meyer, H.H.; Schams, D.; Berisha, B. Expression and localisation of extracellular matrix degrading proteases and their inhibitors during the oestrous cycle and after induced luteolysis in the bovine corpus luteum. Reproduction 2007, 134, 535–547. [Google Scholar] [CrossRef][Green Version]
- Briley, S.M.; Jasti, S.; McCracken, J.M.; Hornick, J.E.; Fegley, B.; Pritchard, M.T.; Duncan, F.E. Reproductive age-associated fibrosis in the stroma of the mammalian ovary. Reproduction 2016, 152, 245–260. [Google Scholar] [CrossRef]
- Watters, K.M.; Bajwa, P.; Kenny, H.A. Organotypic 3D models of the ovarian cancer tumor microenvironment. Cancers 2018, 10, 265. [Google Scholar] [CrossRef]
- Pors, S.E.; Ramløse, M.; Nikiforov, D.; Lundsgaard, K.; Cheng, J.; Andersen, C.Y.; Kristensen, S.G. Initial steps in reconstruction of the human ovary: Survival of pre-antral stage follicles in a decellularized human ovarian scaffold. Hum. Reprod. 2019, 34, 1523–1535. [Google Scholar] [CrossRef]
- MacDonald, J.A.; Takai, Y.; Ishihara, O.; Seki, H.; Woods, D.C.; Tilly, J.L. Extracellular matrix signaling activates differentiation of adult ovary-derived oogonial stem cells in a species-specific manner. Fertil. Steril. 2019, 111, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.C.; Lawler, J. The thrombospondins. Int. J. Biochem. Cell Biol. 2004, 36, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.C.; Lawler, J. The Thrombospondins. Cold Spring Harb. Perspect. Biol. 2011, 3, a009712. [Google Scholar] [CrossRef]
- Berisha, B.; Schams, D.; Rodler, D.; Sinowatz, F.; Pfaffl, M.W. Expression and localization of members of the thrombospondin family during final follicle maturation and corpus luteum formation and function in the bovine ovary. J. Reprod. Dev. 2016, 62, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.H.; Wilson, H.; Silvestri, A.; Fraser, H.M. Thrombospondin-1 expression is increased during follicular atresia in the primate ovary. Endocrinology 2008, 149, 185–192. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, F.R.; Zhou, L.Y.; Nagahama, Y.; Wang, D.S. Duplication and distinct expression patterns of two thrombospondin-1 isoforms in teleost fishes. Gene Expr. Patterns 2009, 9, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Lawler, J.; Sunday, M.; Thibert, V.; Duquette, M.; George, E.L.; Rayburn, H.; Hynes, R.O. Thrombospondin-1 is required for normal murine pulmonary homeostasis and its absence causes pneumonia. J. Clin. Investig. 1998, 101, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Agah, A.; Kyriakides, T.R.; Lawler, J.; Bornstein, P. The lack of thrombospondin-1 (TSP1) dictates the course of wound healing in double-TSP1/TSP2-null mice. Am. J. Pathol. 2002, 161, 831–839. [Google Scholar] [CrossRef]
- Frangogiannis, N.G.; Ren, G.; Dewald, O.; Zymek, P.; Haudek, S.; Koerting, A.; Winkelmann, K.; Michael, L.H.; Lawler, J.; Entman, M.L. Critical role of endogenous thrombospondin-1 in preventing expansion of healing myocardial infarcts. Circulation 2005, 111, 2935–2942. [Google Scholar] [CrossRef]
- Daniel, C.; Schaub, K.; Amann, K.; Lawler, J.; Hugo, C. Thrombospondin-1 is an endogenous activator of TGF-β in experimental diabetic nephropathyin vivo. Diabetes 2007, 56, 2982–2989. [Google Scholar] [CrossRef]
- Malek, M.H.; Olfert, I.M. Global deletion of thrombospondin-1 increases cardiac and skeletal muscle capillarity and exercise capacity in mice. Exp. Physiol. 2009, 94, 749–760. [Google Scholar] [CrossRef]
- Greenaway, J.; Lawler, J.; Moorehead, R.; Bornstein, P.; LaMarre, J.; Petrik, J. Thrombospondin-1 inhibits VEGF levels in the ovary directly by binding and internalization via the low density lipoprotein receptor-related protein-1 (LRP-1). J. Cell. Physiol. 2007, 210, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Greenaway, J.; Gentry, P.A.; Feige, J.J.; LaMarre, J.; Petrik, J.J. Thrombospondin and vascular endothelial growth factor are cyclically expressed in an inverse pattern during bovine ovarian follicle development. Biol. Reprod. 2005, 72, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Watkins, S.C.; Lynch, G.W.; Kane, L.P.; Slayter, H.S. Thrombospondin expression in traumatized skeletal muscle. Cell Tissue Res. 1990, 261, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Iruela-Arispe, M.L.; Liska, D.J.; Sage, E.H.; Bornstein, P. Differential expression of thrombospondin 1, 2, and 3 during murine development. Dev. Dyn. 1993, 197, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Jiang, D.N.; Zeng, S.; Hu, C.J.; Ye, K.; Yang, C.; Yang, S.J.; Li, M.H.; Wang, D.S. Screening and characterization of sex-linked DNA markers and marker-assisted selection in the Nile tilapia (Oreochromis niloticus). Aquaculture 2014, 433, 19–27. [Google Scholar] [CrossRef]
- Brawand, D.; Wagner, C.E.; Li, Y.I.; Malinsky, M.; Keller, I.; Fan, S.; Simakov, O.; Ng, A.Y.; Lim, Z.W.; Bezault, E.; et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature 2014, 513, 375–381. [Google Scholar] [CrossRef]
- Li, M.H.; Yang, H.H.; Zhao, J.; Fang, L.L.; Shi, H.J.; Li, M.R.; Sun, Y.L.; Zhang, X.B.; Jiang, D.; Zhou, L.N.; et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 2014, 197, 591–599. [Google Scholar] [CrossRef]
- Li, M.H.; Sun, Y.L.; Zhao, J.E.; Shi, H.J.; Zeng, S.; Ye, K.; Jiang, D.N.; Zhou, L.Y.; Sun, L.N.; Tao, W.J.; et al. A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLoS Genet. 2015, 11, e1005678. [Google Scholar] [CrossRef]
- Zhou, J.G.; Feng, X.; Ban, B.; Liu, J.X.; Wang, Z.; Xiao, W.H. Elongation factor ell (eleven-nineteen lysine-rich leukemia) acts as a transcription factor for direct thrombospondin-1 regulation. J. Biol. Chem. 2009, 284, 19142–19152. [Google Scholar] [CrossRef]
- Conti, M. Specificity of the cyclic adenosine 3’,5’-monophosphate signal in granulosa cell function. Biol. Reprod. 2002, 67, 1653–1661. [Google Scholar] [CrossRef]
- Menon, K.M.; Menon, B. Structure, function and regulation of gonadotropin receptors—A perspective. Mol. Cell. Endocrinol. 2012, 356, 88–97. [Google Scholar] [CrossRef]
- Zhang, P.; Louhio, H.; Tuuri, T.; Sjöberg, J.; Hreinsson, J.; Telfer, E.E.; Hovatta, O. In vitro effect of cyclic adenosine 3’, 5’-monophosphate (cAMP) on early human ovarian follicles. J. Assist. Reprod. Genet. 2004, 21, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Balazi, A.; Chrenek, P. The cAMP analogue, dbcAMP, affects rabbit ovarian cell proliferation, apoptosis, release of steroids and response to hormones. Folia Biol.-Prague 2014, 62, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Ben, O.A.; Tandlmajerová, A.; Lauková, M.; Vaší Ek, D.; Laurin Ik, J.; Kornhauser, J.; Alwasel, S.; Harrath, A.H. cAMP response element-binding protein 1 controls porcine ovarian cell proliferation, apoptosis, and FSH and insulin-like growth factor 1 response. Reprod. Fertil. Dev. 2018, 30, 1145–1153. [Google Scholar] [CrossRef]
- Ionta, M.; Rosa, M.C.; Almeida, R.B.; Freitas, V.M.; Rezende-Teixeira, P.; Machado-Santelli, G.M. Retinoic acid and cAMP inhibit rat hepatocellular carcinoma cell proliferation and enhance cell differentiation. Braz. J. Med. Biol. Res. 2012, 45, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.N.; Yang, H.H.; Li, M.H.; Shi, H.J.; Zhang, X.B.; Wang, D.S. Gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 2016, 83, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Xiao, H.S.; Jie, M.M.; Dai, S.F.; Wu, X.; Li, M.H.; Wang, D.S. Amh regulate female folliculogenesis and fertility in a dose-dependent manner through Amhr2 in Nile tilapia. Mol. Cell. Endocrinol. 2020, 499, 110593. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Nagahama, Y. Induction of XY sex reversal by estrogen involves altered gene expression in a teleost, tilapia. Cytogenet. Genome Res. 2003, 101, 289–294. [Google Scholar] [CrossRef]
- Sun, L.N.; Jiang, X.L.; Xie, Q.P.; Yuan, J.; Huang, B.F.; Tao, W.J.; Zhou, L.Y.; Nagahama, Y.; Wang, D.S. Transdifferentiation of differentiated ovary into functional testis by long-term treatment of aromatase inhibitor in Nile tilapia. Endocrinology 2014, 155, 1476–1488. [Google Scholar] [CrossRef]
- Zhang, X.B.; Li, M.R.; Ma, H.; Liu, X.Y.; Shi, H.J.; Li, M.H.; Wang, D.S. Mutation of foxl2 or cyp19a1a results in female to male sex reversal in XX Nile tilapia. Endocrinology 2017, 158, 2634–2647. [Google Scholar] [CrossRef]
- Li, M.H.; Wu, F.R.; Gu, Y.; Wang, T.R.; Wang, H.; Yang, S.J.; Sun, Y.L.; Zhou, L.Y.; Huang, X.G.; Jiao, B.W.; et al. Insulin-Like growth factor 3 regulates expression of genes encoding steroidogenic enzymes and key transcription factors in the Nile tilapia gonad1. Biol. Reprod. 2012, 86, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, M.H.; Liu, X.Y.; Dai, S.F.; Xiao, H.S.; Qi, S.S.; Li, Y.B.; Zheng, Q.Y.; Jie, M.M.; Cheng, C.H.K.; Wang, D.S. Regulation of spermatogenesis and reproductive capacity by Igf3 in tilapia. Cell. Mol. Life Sci. 2020, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Coward, K.; Bromage, N.R. Histological classification of oocyte growth and the dynamics of ovarian recrudescence in Tilapia zillii. J. Fish Biol. 1998, 53, 285–302. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jie, M.; Ma, H.; Zhou, L.; Wu, J.; Li, M.; Liu, X.; Wang, D. Regulation of Female Folliculogenesis by Tsp1a in Nile Tilapia (Oreochromis niloticus). Int. J. Mol. Sci. 2020, 21, 5893. https://doi.org/10.3390/ijms21165893
Jie M, Ma H, Zhou L, Wu J, Li M, Liu X, Wang D. Regulation of Female Folliculogenesis by Tsp1a in Nile Tilapia (Oreochromis niloticus). International Journal of Molecular Sciences. 2020; 21(16):5893. https://doi.org/10.3390/ijms21165893
Chicago/Turabian StyleJie, Mimi, He Ma, Li Zhou, Jiahong Wu, Minghui Li, Xingyong Liu, and Deshou Wang. 2020. "Regulation of Female Folliculogenesis by Tsp1a in Nile Tilapia (Oreochromis niloticus)" International Journal of Molecular Sciences 21, no. 16: 5893. https://doi.org/10.3390/ijms21165893
APA StyleJie, M., Ma, H., Zhou, L., Wu, J., Li, M., Liu, X., & Wang, D. (2020). Regulation of Female Folliculogenesis by Tsp1a in Nile Tilapia (Oreochromis niloticus). International Journal of Molecular Sciences, 21(16), 5893. https://doi.org/10.3390/ijms21165893






