Abstract
Polycomb repressive complex 1 (PRC1) and PRC2 are the major complexes composed of polycomb-group (PcG) proteins in plants. PRC2 catalyzes trimethylation of lysine 27 on histone 3 to silence target genes. Like Heterochromatin Protein 1/Terminal Flower 2 (LHP1/TFL2) recognizes and binds to H3K27me3 generated by PRC2 activities and enrolls PRC1 complex to further silence the chromatin through depositing monoubiquitylation of lysine 119 on H2A. Mutations in PcG genes display diverse developmental defects during shoot apical meristem (SAM) maintenance and differentiation, seed development and germination, floral transition, and so on so forth. PcG proteins play essential roles in regulating plant development through repressing gene expression. In this review, we are focusing on recent discovery about the regulatory roles of PcG proteins in SAM maintenance, root development, embryo development to seedling phase transition, and vegetative to reproductive phase transition.
1. Introduction
Higher eukaryotes have evolved mechanisms to temporally and specially control gene expression. Polycomb-group (PcG) proteins have been initially identified as transcriptional repressors of Homeobox (Hox) genes in Drosophila (Drosophila melanogaster) to control embryonic development, stem cell differentiation, and tissue homeostasis development [1,2]. PcG proteins usually form multiple complexes, such as polycomb repressive complex 1 (PRC1) and PRC2, to inactivate gene expression and maintain the silencing state of the target chromatin through covalent histone modifications [3]. PRC1 complexes are E3 ubiquitin ligases that modify the chromatin via depositing monoubiquitylation of lysine 119 mark on histone 2A (H2AK119ub). PRC2s are methyltransferases that target lysine 27 on histone 3 for trimethylation (H3K27me3) and generates H3K27me3 epigenetic mark on the target loci [4,5]. Most of the components in PRC1 and PRC2 complexes are conserved from Drosophila to mammals and plants, homologs of which have been characterized recently [6,7].
In Drosophila, PRC2 is composed of four core components, including the SET domain-containing histone methyltransferase enhancer of zeste [E(z)], Suppressor of Zeste 12 [Su(z)12], Extra sex combs (Esc), and Nucleosome remodeling factor 55 kDa subunit (Nurf55) or p55 [8] (Table 1).

Table 1.
Components of polycomb repressive complex 1 (PRC1) and polycomb repressive complex 2 (PRC2) complexes discovered in Drosophila, Arabidopsis, and rice.
In Arabidopsis (Arabidopsis thaliana), members of these core subunits are present in multigene families. CURLY LEAF (CLF), MEDEA (MEA) (also termed Fertilization Independent Seed 1, FIS1), and SWINGER (SWN) are three homologs of E(z) which have methyltransferase activity to trimethylate H3K27 of the target chromatin [9,10,11]. Embryonic Flower 2 (EMF2), VERNALIZATION 2 (VRN2), and Fertilization Independent Seed (FIS2) are the three homologs of Su(z)12, which promote the activity of H3K27 methyltransferase and confer target specificity to the corresponding PRC2s [11,12,13]. Multiple Suppressor of IRA 1 to 5 (MSI1 to MSI5) are the five homologs of Nurf55/p55, but only the functions of MSI1 and MSI4 have been characterized [14,15,16,17]. Fertilization Independent Endosperm (FIE) is the homolog of Esc in Arabidopsis [18,19,20,21]. Three different PRC2s, including EMF-PRC2, VRN-PRC2, and FIS-PRC2 complexes, have been characterized in Arabidopsis based on the homologs of Su(z)12 in the complexes [22]. Two components, FIE and MSI1, are present in all three PRC2 complexes in Arabidopsis [17,23] (Table 1).
In the rice (Oryza sativa) genome, two E(z)-like proteins in the rice genome have been identified termed OsCLF and OsSET1/OsiEZ1, respectively, because of their high similarity to Arabidopsis CLF and SET1 [24,25,26]. There are only two Su(z)12-like proteins, designated as OsEMF2a and OsEMF2b, which are the Arabidopsis homologs of EMF2. In contrast to the single copy of an Esc homolog, FIE in Arabidopsis, there are two Esc homologs in rice, OsFIE1 and OsFIE2. The two genes are located close to each other on chromosome 8 separated by one gene. The amino acid sequences of OsFIE1 and OsFIE2 are 72% identical [24]. One homolog of MSI1 in rice termed OsMSI1 has also been reported [27] (Table 1).
The PRC1 complex in Drosophila is composed of dRing/Sex comb extra (Sce), Posterior sex combs (Psc), and Polyhomeotic (Ph) sununits [7,28]. In Arabidopsis, AtRING1A/B and AtRING1B are the homologs of dRing/Sce [29]. AtBMI1A, AtBMI1B, and AtBMI1C are the homologs of Psc [29,30,31]. AtRING1A, AtRING1B, AtBMI1A, and AtBMI1B possess the E3 ligase activities to monoubiquitylate H2AK119. Like Heterochromatin Protein 1/Terminal Flower 2 (LHP1/TFL2), performing as the reader of histone modification, recognizes and binds to H3K27me3 generated by PRC2 activities [32,33]. In addition, plant-specific proteins, including Embryonic Flower 1 (EMF1), VERNALIZATION 1 (VRN1), and VP1/ABI3-Like 1/2/3 (VAL1/2/3), are involved in PRC1 function [34,35,36]. However, no functional homolog of Ph has been identified in Arabidopsis genome. LHP1 integrates into PRC1 complex via interacting with both AtBMI1 and AtRING1 [29,37,38]. In addition to the involvement in PRC1 complex, LHP1 is also associated with PRC2 through interacting with MSI1 and EMF2 to deposit H3K27me3 mark on the chromatin [17,39]. In rice, a homolog of EMF1, AtRING1A/1B, and LHP1 in rice designated as Curved Chimeric Palea 1 (CCP1) or OsEMF1, OsRING1, and OsLHP1, respectively, have been reported [27,40].
During the life cycle, plants undergo several important phase transitions, from gametophytic to sporophytic stage; from embryogenesis to germination, from juvenile to adult vegetative phase, and from vegetative to reproductive phase [41]. PcG proteins play pivotal roles in controlling these processes. Whole genome data have shown that about 20% of Arabidopsis genes are marked by H3K27me3 [32,42,43]. In this review, we are focusing on recent findings on roles of PcG proteins in SAM maintenance, root development, seed to germination phase transition, and floral transition.
2. Roles of PcG Proteins in SAM Maintenance and Differentiation
Generation of new organs during life cycle of plants is dependent on the maintenance and differentiation of stem cells [42,44,45]. Root apical meristem (RAM) is involved in generation of underground architecture. Shoot apical meristem (SAM), axillary meristems (AMs), inflorescence meristem (IM), and floral meristem (FM) generate aboveground organs [46].
In Arabidopsis, SAMs are regulated by a negative feedback loop comprising the stem cell-promoting transcription factor WUSCHEL (WUS) and differentiation-promoting peptide CLAVATA 3 (CLV3) [47,48,49]. WUS, a homeodomain transcription factor, is required for maintenance of stem cell identity [50,51,52,53]. WUS activates CLV3 expression [50,54,55,56]. CLV1 (a receptor kinase), CLV2 (receptor like protein), and CLV3 (a ligand of CLV1) form a receptor kinase-signaling cascade to repress WUS expression, maintain appropriate meristem size, and promote meristem cells toward organ initiation [54,57,58,59,60].
These regulatory mechanisms are conserved in rice [61]. The WUS ortholog in rice termed OsWUS is expressed mainly in leaf margins but could not be always detected in SAM [62,63]. The functions of Oryza sativa WUSCHEL-Related Homeobox 4 (OsWOX4) is similar to WUS in Arabidopsis. RNAi transgenic lines of OsWOX4 reduce the size of SAM and flatten SAM [64]. Rice Floral Organ Number 1 (FON1) and FON4 (also termed FON2) are homologs of CLV1 and CLV3, which play important roles in maintaining activities of the stem cells within the FM [65,66,67,68].
2.1. PRC2 Represses the Expression of WUS Directly or Indirectly
The expression of WUS is transcriptionally regulated by PRC2 complex [69]. The chromatin of WUS is marked by H3K27me3 [70]. The Polycomb Response Elements (PREs) from WUS locus, termed CArG boxes, have been characterized and are shown to be essential for establishing and spreading of H3K27me3 through PRC2 activity [70,71,72]. AGAMOUS (AG), a MADS domain-containing protein, binds directly to the CArG boxes located in the WUS coding region, which in turn recruits PRC2 complex to catalyze trimethylation of H3K27 and repress WUS expression [70]. Mutations in the CArG boxes of WUS disrupt the binding of AG to WUS and activate the expression of WUS (Figure 1a). Mutation in AG results in the decreased H3K27me3 level at the WUS locus [70].
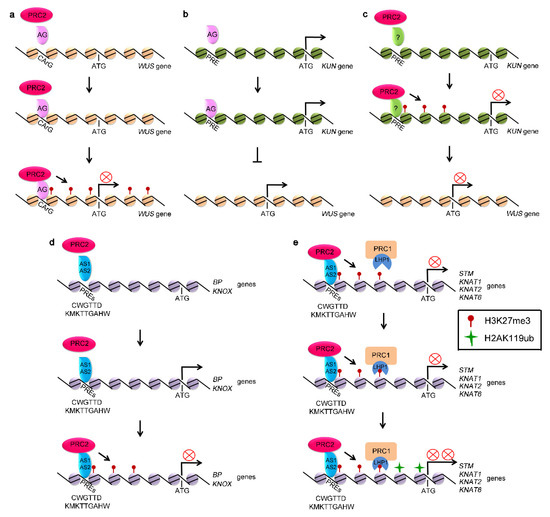
Figure 1.
Polycomb-group (PcG) proteins repressed the expression of stem cell fate related genes. (a) polycomb repressive complex 2 (PRC2) silences the expression of WUSCHEL (WUS) through binding to the Polycomb Response Elements (PREs), such as CArG located in the promoter of WUS and generating H3K27me3 at WUS locus. (b) AG represses the expression of WUS through activating KUN, a suppressor of WUS. (c) PRC2 activates the expression of WUS through silencing KUN, competing with AG. (d) PRC2 represses the expression of BP and KNOX genes through binding to PREs, such as CWGTTD and KMKTTGAHW. (e) KNOX genes, including STM, KNAT1, KNAT2, and KNAT6 genes, marked with H3K27me3 by PRC2.
In addition to directly repressing WUS, AG inactivates WUS to regulate stem cell activity through activating KNUCKLES (KNU) [72,73,74]. KNU, a C2H2-type zinc finger protein, represses the expression of WUS [72,73]. The expression of KNU is repressed by PRC2 [75,76]. There are three AG binding sites, termed KNU-PREs, in the promoter region of KNU (Figure 1b). KNU activation by AG is associated with the loss of H3K27me3 [71,72]. AG binds to the KNU-PREs by competing with PRC2, leading to the loss of H3K27me3 at the coding region and activation of KNU, further suppressing the expression of WUS (Figure 1c) [72,74,77,78,79].
2.2. PRC2 Inactivates KNOX Genes
The Knotted 1-like homeobox (KNOX) genes, including Shoot Meristemless (STM), BREVIPEDICELLUS (BP), Knotted 1-Like from Arabidopsis thaliana 1 (KNAT1), KNAT2, and KNAT6 are important to maintain the meristem activity of SAM [80,81,82]. They are localized at the shoot apex and function redundantly with STM [83]. The KNOX genes are expressed in the meristems. The Asymmetric leaves 1 (AS1) and AS2 genes, encoding MYB-domain and LOB-domain containing transcription factors, respectively, are expressed in the organ primordial [83]. The KNOX genes restrict the expression of AS1 and AS2 genes to organ primordia, thus preventing ectopic organ initiation. AS1 and AS2 can, in turn, repress KNOX gene expression [84,85]. These genes are crucial for distinguishing stem cells and organ founder cells.
It has been recently reported that H3K27 trimethylation of these cell fate-determination genes depends on FIE [86]. The expression of the genes, including STM, WUS, KNATs, and WOX1, are upregulated by fie mutation, along with the loss of H3K27me3 [86,87]. Mechanisms on PRC2-mediated H3K27me3 modification of KNOX genes have been recently reported [88,89]. AS1 physically associates with AS2, forming AS1-AS2 complex [85,90,91]. AS1-AS2 complex has DNA-binding activity and directly binds to the CWGTTD and KMKTTGAHW PREs localized in the promoters of BP and KNAT2 to recruit PRC2, thus triggering H3K27me3 modification to silence their expression [88,89]. AS1 integrates into PRC2 through physically interacting with CLF and FIE, whereas AS2 is directly associated with EMF2 and recruit PRC2 complex to BP and KNAT2 regions to modify the chromatin with H3K27me3 mark and silence SAM-specific homeobox domain genes in leaf tissue (Figure 1d) [88].
In addition, AS1-AS2 complex also interacts with LHP1 and recruits PRC2 to cis-acting element in the promoter regions of BP and KNAT2 [92]. Mutations in AS1 or AS2 lead to reduction in H3K27me3 levels at BP and KNAT2 loci. Furthermore, mutations in AS1-AS2 binding sites also result in decrease of H3K27me3 accumulation and the derepression of BP and KNAT2 genes [88,92].
2.3. PRC1 Regulates Stem Cell Fates
PRC1 complex also plays roles in maintenance of SAM. Mutations of AtRING1A and AtRING1B lead to ectopic-meristem formation in cotyledons and leaves. The atring1a/atring1b double mutant plants display an enlarged SAM [29]. The class I KNOX genes, including STM, KNAT1, KNAT2, and KNAT6, are direct targets of AtRING1s [29,38]. H2AK119ub modifications in KNOX genes are significantly reduced in atring1a/atring1b double mutants, suggesting that AtRING1A and AtRING1B control stem cell fate by generating H2AK119ub and repressing the expression of target genes (Figure 1e). However, H3K27me3 deposition on the chromatin of STM, KNAT2, and KNAT6 is undisturbed in atring1a/atring1b mutant plants [29,37]. Similarly, disruption of AtBMI1A and AtBMI1B activity decreases the level of H2AK119ub in vivo, but did not alter H3K27me3 deposition at target genes (Figure 1e) [29,31,36,37]. These results indicate that AtRING1s and AtBMI1s are required for H2AK119ub, but that they are not required for the trimethylation of H3K27 by PRC2 [29,37].
3. Roles of PcG Proteins in Root Development
3.1. PRC2 Represses ABI4 to Control Root Development
Involvement of PRC2 in root development has been characterized recently. Abscisic acid Insensitive 4 (ABI4), an AP2/ERF domain-containing transcription factor, regulates root development [93]. Disruption of ABI4 gene increases the number of lateral roots, whereas overexpression of ABI4 decreases lateral root density. The effect of ABI4 on lateral root development is mediated through repression of PIN-FORMED 1 (PIN1), which encodes an auxin-efflux carrier and regulates auxin distribution [93]. Basic Pentacysteine (BPC) proteins have been identified as plant-specific transcription factors [94,95]. BPCs proteins contain a conserved DNA-binding domain at the C-terminus and can bind to the GAGA-cis elements of their target genes [96,97]. BPCs are able to recruit PRC2 through interacting with SWN to the promoter of ABI4 to repress the expression of ABI4 by catalyzing H3K27me3 and regulate root development [98].
3.2. PcG Proteins Control Root Development through NRT2.1 under Lower Conditions
Nitrogen (N) is a limiting factor for plant development. Low levels of nitrate stimulate lateral root formation, whereas high levels suppress lateral root formation. In addition, different N forms such as nitrate (), ammonium (), or some amino acids are signaling molecules modulating plant development [99,100]. In Arabidopsis, Nitrate Transporter 2.1 (NRT2.1) encodes a key high-affinity root nitrate () transporter, crucial for uptake and thus for N nutrition during lateral root development and plant growth [101]. The nrt2.1 mutants show a dramatic reduction of growth under low and limiting availability. It has recently been observed that NRT2.1 is modified by H3K27me3 [33,102]. PRC2 directly targets and downregulates NRT2.1 in the nutritional conditions where this gene is one of the most highly expressed genes [103]. The mutation of CLF leads to the loss of H3K27me3 at NRT2.1 and results in the activation of NRT2.1 expression.
3.3. APOLO lncRNA Recruits PRC1 to Control Root Development
APOLO (AUXIN-REGULATED PROMOTER LOOP), a long noncoding RNAs (lncRNAs), is involved in lateral root development in Arabidopsis [104]. APOLO recognizes its targets by sequence complementarity to form DNA-RNA duplexes (R-loops). The recognition of APOLO to the target DNA recruits PRC1 through LHP1, which regulates the local chromatin three-dimensional conformation changes. APOLO lncRNA modulates the expression of distal unrelated auxin-responsive genes during lateral root development [104]. The findings are consistent with the report in Drosophila. R-loops are able to be formed at many PREs in Drosophila embryos and are correlated with repressive states of the target genes. PRC1 and PRC2 can recognize R-loops and open DNA bubbles. In addition, PRC2 activity drives formation of RNA-DNA hybrids, the key component of R-loops, from RNA and dsDNA [105].
4. Roles of PcG Proteins in Embryo to Seedling Phase Transition
The repressive activities of PcG proteins are required for the embryo to seedling phase transition [86,87,106]. Plants produce and store food in the seed, especially in endosperm to support germination and seedling growth. During germination, genes specifying embryogenesis, seed dormancy, and accumulation of seed storage compounds must be repressed to allow embryo to grow and differentiate into plants [107].
4.1. PRC2 Is Required for Transition from Embryogenesis to Seed Maturation
In Arabidopsis, as in most of the Angiosperms species, the female gametophyte (embryo sac) contains a haploid egg cell and a diploid central cell. After fertilization, the central cell forms the triploid endosperm and the egg cell generates the diploid zygote [108]. In the absence of fertilization, the autonomous endosperm development is repressed by FIS-PRC2, while seed coat development is suppressed by EMF-PRC2 and VRN-PRC2 [109,110,111]. Mutations in FIS-PRC2 or impairment of auxin synthesis and signaling affect the development of embryo, endosperm, and seed coat causing seed abortion [111,112,113].
SEEDSTICK (STK), a MADS-box gene, is specifically expressed during ovule and seed development and functions in these tissues [114,115]. During carpel development, the expression of STK is confined to placental tissues and ovule primordial. In mature ovules, STK is expressed strongly in the funiculus and integuments, which form the seed coat later [115]. Class I BPCs and the MADS-domain factors, including Short Vegetative Phase (SVP), APELATA 1 (AP1), and AGAMOUS-Like 24 (AGL24), are the key regulators of STK. Class I BPCs, including BPC1, BPC2, and BPC3, form homodimers and heterodimers and bind to the C-boxes in the promoter of STK [95,116]. SVP acts redundantly as AP1 and AGL24 to repress STK expression during early stages of flower development, through binding to its promoter [117,118]. Class I BPCs physically interact with SVP to repress STK expression in the floral meristem [117]. Class I and class II BPCs control the expression of STK, by depositing and/or maintaining H3K27me3 marks through interacting with LHP1 [119,120].
It has been shown that a protein homologous to HEME Activator Protein 3 (HAP3) subunits of the CAAT Box binding Factors, termed Leafy Cotyledon 1 (LEC1), and three B3-domain transcription factors, including ABA Insensitive 3 (ABI3), FUSCA 3 (FUS3), and LEC2, termed LAFL proteins, are the major factors activating the seed maturation program in a complex network [121,122,123]. The fus3-3 mutant exhibits ovule and seed abortion [124]. The repression of LAFL genes during early embryogenesis has been reported [125]. LAFL expression is tightly controlled in specific tissues of seeds. FUS3 is expressed during ovule development. Before fertilization, the expression of FUS3 is restricted to the chalaza and funiculus of mature ovules. After fertilization, FUS3 is confined in the funiculus, seed coat, and chalaza [126]. BPC1 and 2 promote the transition from ovule to seed development by repressing FUS3 in ovule integuments. Class I BPCs interact with FIS2 and MEA in FIS-PRC2 and bind to the GA/CT cis-element located in FUS3 promoter to represses the expression of FUS3 in the integuments of mature ovules and the endosperm of developing seeds. These results suggest that BPCs-mediated repression of FUS3 in the endosperm is necessary to coordinate endosperm and embryo growth.
4.2. PRC2 Regulates Seed Maturation to Germination Transition
The LAFL genes are sequentially expressed during seed development, but silenced during and after seed germination [127,128,129]. PRC2 is involved in the repression of these genes in vegetative tissue [106,125]. In fie mutant, genes functioning in late seed development, including AGAMOUS-Like 15 (AGL15), ABI3, FUS3, and LEC2, are upregulated. The expression of genes encoding storage compounds in seeds, including CRUCIFERIN 3 (CRU3), CRUCIFERINA 1 (CRA1), Seed Storage Albumin 1 (SESA1), Seed Storage Albumin 2, (SESA2), Late Embryogenesis Abundant proteins (LEAs), and OLEOSINs (OLEOs), are also upregulated by fie mutation [87,106]. It has also been shown that BPC1 interacts with EMF2 and recruit EMF-PRC2 to the promoter of FUS3 to repress its expression during vegetative development [126].
In plants, hormones are required for regulating seed development and germination. Abscisic acid (ABA) promotes seed development, maturation, and dormancy, whereas gibberellic acid (GA) facilitates germination and seedling growth [107]. PcG proteins regulate seedling development through multiple hormone signaling pathways [86,106]. During germination, FIE and EMF1 bind to genes encoding positive regulators in ABA signaling pathway, such as ABI3 and ABI4, to genes encoding negative regulators of GA signaling, e.g., RGA-Like 3 (RGL3) and to genes encoding major regulatory factors that promote embryo development and maturation, such as LEC2, to repress their expression [86,87,106]. In fie mutant, positive regulator genes of ABA signaling and negative regulator genes of GA signaling, such as ABI4, Delay of Germination 1 (DOG1), CHOTTO 1/AINTEGUMENTA-LIKE 5 (CHO1/AIL5), and SOMNUS (SOM), and SQUAMOSA Promoter Binding Protein-Like 8 (SPL8), are activated because of the loss of H3K27me3 modification [86,106].
4.3. PRC1 Represses Seed Development Genes to Allow Germination
PRC1 is involved in seed development and seedling growth. Genes required for seed development are activated by emf1 mutation. EMF1 binds to major seed regulated genes, such as FUS3, ABI3, and the downstream seed maturation genes, including LEC2, LEA, OLEO2, Lipid Transfer Protein 3 (LTP3), and seed storage protein genes, which are modified by H3K27me3 [130,131]. The VAL (VP1/ABI3-LIKE) 1/2/3 proteins suppress the expression of ABI3, FUS3, and LEC2 to initiate germination and vegetative development [121]. VAL proteins interact with AtBMI1 proteins to initiate repression of seed maturation genes. The H2AK119ub levels of the seed development genes are lower in atbmi1a/b than in WT seedlings. In val1/2 mutants, the levels of H2AK119ub are also reduced at seed maturation genes, but are not affected at WUS, indicating that VAL proteins are specifically required for generating H2AK119ub mark at seed maturation genes [36]. The regulation of seed maturation genes by AtBMI1s does not follow the classic hierarchical model proposed for animal PcG-mediated repression [132]. The PRC1 activity is required for the H3K27me3 modification of these genes to maintain the suppression status. Whether VALs is responsible for recruiting PRC1 to the targets remain unclear.
5. Roles of PcG Proteins in Floral Transition
The timing of floral transition is essential for the life cycle of plants. In Arabidopsis, a facultative long-day plant, the autonomous, vernalization, photoperiod, PAF1 complexes, FRIGIDA (FRI)/FRIGIDA-Like (FRL), GA, and aging pathways form a complicated network to regulate floral transition [133,134]. Till date, key flowering time-related genes in the network have been characterized and their regulatory mechanisms have been intensively investigated. Flowering Locus C (FLC), a MADS box protein, is a central suppressor of flowering through repressing the expression of two flowering time integrators, Flowering locus T (FT) and Suppressor of COSTANS 1 (SOC1) [135,136,137]. The expression of FLC is activated by FRI/FRL and PAF1 complexes, but repressed by vernalization and the autonomous-pathway proteins [138]. Two PRC2 complexes, VRN-PRC2 and EMF-PRC2, are involved in the inactivation of FLC through generating H3K27me3 modification on its chromatin [6,139,140].
5.1. VRN-PRC2 in Vernalization Pathway
Vernalization is the process in which flowering is accelerated by prolonged cold [141,142]. It involves PcG protein-mediated gene silencing through marking H3K27me3 modification at FLC chromatin [143,144,145]. The VRN2-PRC2 complex is required for vernalization-induced flowering through repressing FLC [13]. Vernalization Insensitive 3 (VIN3) and VRN5/Vernalization Insensitive 3-Like 1 (VIL1), two plant homeodomain (PHD) proteins, associated with PRC2 suppress the expression of FLC [13,140,146,147]. During vernalization, VIN3 is induced and physically interacts with VRN5/VIL1. This two PHD protein-formed heterodimer is associated with VRN-PRC2 at promoter region of FLC to form PHD–PRC2 complex. The PHD–PRC2 complexes deposit H3K27me3 mark at FLC locus during cold exposure [13,23,140,144,145,146,147,148]. After transferring to the warm condition, the PHD–PRC2 complex without VIN3 spreads across the gene resulting in high H3K27me3 across FLC locus, necessary for epigenetic stability through the rest of development [23,145,148]. Maintenance of this silenced state requires LHP1, suggesting the cross-talk between PRC2 and PRC1 in flowering time control [144,149].
COLDAIR, a sense noncoding RNA (ncRNA), is also induced after cold exposure. FLC is downregulated concomitantly with increased COLDAIR transcripts [150]. The cold-induced COLDAIR is involved in PRC2 recruitment [151].
5.2. EMF-PRC2 in Floral Transition
The EMF-PRC2 plays important roles in repressing reproductive development during vegetative growth [9,12,18,152]. Mutants of EMF2 gene skip vegetative growth and flower upon germination [153]. EMF-PRC2 complex is attributed to the ectopic expression of flower organ identity or flower MADS box genes, such as AG, APETALA1 (AP1), AP3, and PISTILATA (PI), to delay flowering [130,131,154]. In addition, EMF-PRC2 inhibits the expression of FLC in a vernalization independent manner, which may function in concert with Flowering Locus D (FLD) [155]. FLD encodes a plant ortholog of the human protein Lys-Specific Demethylase 1 (LSD1) that is involved in H3K4 demethylation [155,156]. Loss of FLD function causes a reduction in H3K27me3, and, conversely, loss of EMF-PRC2 activity leads to an increase in H3K4me3 [157,158]. This suggests that EMF-PRC2 and the FLD complex function coordinately to demethylate H3K4, deacetylate histones, and deposit H3K27me3 on FLC chromatin, leading to the establishment of a repressive chromatin environment to repress FLC expression. EMF-PRC2-mediated repression is an endogenous mechanism that is independent of environmental signals [159].
5.3. PRC1 in Floral Transition
AtBMI1C, a component of PRC1 in Arabidopsis, has been identified as flowering time accelerator. Overexpression of AtBMI1C promotes flowering in Arabidopsis [31]. LHP1 is a component of PRC1 through interacting with AtRING1a and acts as a reader component of PRC1, similar to Drosophila Pc. LHP1 displays the binding specificity to H3K27me3 through its chromodomain [29,160]. A genome-wide analysis of LHP1 showed that LHP1 is highly enriched with H3K27me3-modified loci [32,33]. The lhp1 mutant exhibits early flowering because of the activation of the expression of flowering time-related genes FT and floral organ identity genes, such as AG, AP3, PI, and SEPALLATA 3 (SEP3) [32,149,161]. In addition, LHP1 is required to maintain the epigenetically repressed state of FLC upon return to warm conditions [162]. To maintain SEP3 chromatin in a silenced state, Short Vegetative Phase (SVP) interacts with LHP1/TFL2 to modulate H3K27me3 [163].
EMF1, another Arabidopsis gene required for vegetative development, encodes a plant-specific protein containing sequence motifs found in transcriptional regulators [164]. The emf1 mutants have similar phenotypes to emf2 mutants, flowering early upon germination [18,106,152,153]. Tissue-specific removal of EMF1 activity from leaf primordia allows vegetative growth, but leads to early flowering plants with curly leaves similar to clf mutants [165]. The expression of floral organ identity such as AG, AP1, AP3, and PI are upregulated by emf1, which confers its early flowering phenotype [130,131,154]. EMF1 binds to the chromatin regions of the floral organ identity genes such as AG, AP1, AP3, and PI and displays characteristics similar to PRC1 component, Psc, in Drosophila [35]. EMF1 is also required for Arabidopsis RING-finger protein-mediated H2AK119 monoubiquitination [37].
It has also been revealed that EMF1, LHP1, and a trimethyl histone H3 lysine-4 (H3K4) demethylase form a PcG complex, termed EMF1 complex (EMF1c) to mediate FT silencing in Arabidopsis. In leaf veins, EMF1c directly represses FT expression before the end of long days (LDs) and at night, and that in response to inductive LDs, CONSTANS (CO) accumulation at dusk antagonizes EMF1c binding to FT chromatin to promote FT transcriptional activation and thus the onset of flowering [159].
5.4. PcG Proteins in Flowering Time Control of Rice
Photoperiodic regulation is a major mechanism on flowering time control of rice. Short day (SD) promotes rice flowering. The genes involved in the photoperiod pathway are conserved between Arabidopsis and rice. OsGIGANTEA (OsGI), Heading date 1 (Hd1), Heading date 3a (Hd3a), and Rice Flowering locus T 1 (RFT1) are the ortholog of Arabidopsis GI, CO, and FT in rice. Hd3a and RFT1 proteins are the florigens in rice [166,167,168]. OsGI, Hd1, and Hd3a form OsGI-Hd1-Hd3a module to control rice flowering. OsGI activates the expression of Hd1 to promote synthesis of Hd3a and RFT1 to accelerate flowering [169,170,171,172].
Early heading date 1 (Ehd1) encodes a plant specific B-type response regulator, which is activated by OsMADS51. Ehd1 activates the expression of Hd3a and RFT1 to promote rice flowering [173]. Ghd7, OsMADS56, OsLEC1 and FUSCA-Like 1 (OsLFL1), and DTH8/Ghd8 repress the expression of Ehd1 to delay flowering time of rice [174,175,176,177,178]. Ehd2, Ehd3, and Ehd4 activate the expression of Ehd1. Activated Ehd1 enhances the expression of Hd3a to promote flowering [179,180,181]. Interaction Protein 1 (SIP1), a C2H2 zinc finger domain-containing protein, binds to the promoter of Ehd1 and recruits Trithorax-like protein, OsTrx1, to modify the chromatin with H3K4me3 mark and activates the expression of Ehd1 to promote flowering of rice [182].
PRC2 is involved in regulation of flowering time. OsVIN3-Like (OsVIL) proteins are PHD domain containing protein, a homolog of Arabidopsis VIN3. There are four VIN3-like proteins, PHD proteins, in rice, termed OsVIL1 to OsVIL4 [183,184]. OsVIL2 and LC2/OsVIL3 have been reported to associate with PRC2 to regulate flowering time in rice [183,184,185]. The osvil2 mutant and OsVIL2–RNAi lines display late flowering phenotype under both LD and SD conditions [184,185]. The expression of OsLFL1 is increased, but the expression of Ehd1, Hd3a, and RFT1 is reduced by osvil2 mutation [185]. OsLFL1 chromatin is modified by H3K27me3 mark. OsVIL2 physically interacts with OsEMF2b, a component of PRC2 in rice. OsVIL2 binds directly to OsLFL1 chromatin to recruit PRC2 through interacting with OsEMF2b and catalyze H3K27 trimethylation on the locus of OsLFL1. H3K27me3 enrichment is diminished in the osvil2 mutants. The osemf2b mutant flowers late by increasing OsLFL1 expression and decreasing Ehd1 expression [185]. These results suggest that OsVIL2, together with PRC2, represses OsLFL1 expression and increases the expression of Ehd1 to induce flowering [185].
LC2/OsVIL3 promote flowering of rice. The lc2 mutation delays heading date by reducing the expression of Hd1 and Hd3a under SD conditions. LC2 physically interacts with OsVIL2, but not with OsMSI1, OsCLF, OsEMF2a, OsEMF2b, OdFIE1, and OsFIE2. LC2 binds to the promoter region of a floral repressor OsLF and represses the OsLF expression via H3K27 trimethylation modification. OsLF directly regulates the Hd1 expression through binding to Hd1 promoter. OsLF, a bHLH transcription factor, interacts with OsPIL13 and OsPIL15 to repress rice flowering [186]. These results suggest that LC2 may bind to the promoter region of target genes to recruit PRC2 and modify its target genes [184].
6. Concluding Remarks
PRC2 and PRC1 play critical roles in maintaining the stable inactivated states of developmental genes by catalyzing H3K27me3 and H2AK119ub histone modifications, respectively. To ensure proper development, PcG proteins are required to precisely program gene expression in temporal- and spatial-dependent manners. The major components of PRC1 and PRC2 and their functions have extensively been explored in Arabidopsis and rice [6,7]. So far, the evidence suggests that both PRC1 and PRC2 are involved in the maintenance of stem cell activity and root development. After germination, multiple forms of PRC2s repress the expression of seed development genes, and PRC1 represses seed development and flower genes. EMF-PRC2, VRN-PRC2, and PRC1 control flowering time or floral transition by repressing the relative gene expression.
Though recent progresses have advanced our understanding of the PcG mechanisms on regulating major phase transitions in Arabidopsis and rice, more questions remain. It has been known that about 20% of the genes in Arabidopsis are modified by H3K27 tri-methylation [32,42,43]. Only small part of modified gene function is uncovered. Discovering the functions of modified genes is the challenges we are facing. For most of these identified targets, it is still not known precisely how and in which cells, the PcG system regulates gene expression. Because PcG proteins lack DNA-binding activity, the recruitment of PRC1 and PRC2 to their target chromatin is one of the major open questions in PcG-mediated gene silencing. Up to date, PRE cis-elements, transcription factors, DNA-binding proteins, and lncRNAs are proposed to contribute to the recruitment [6,187]. It is expected that various mechanisms exist, and further identifications of PcG-interacting factors would extend our understanding and allow us to extract fundamental principles underlying epigenetic regulation of eukaryote developmental programs.
Addressing these questions will further elucidate the functions of PcG proteins in silencing gene expression and regulating plant development.
Author Contributions
X.W., B.Y., Y.L., and C.Z. wrote the article. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program from Ministry of Science and Technology of China (grant number 2016YFD0100406 and 2017YFD0300107).
Acknowledgments
We apologize to colleagues whose work was not cited due to space limitations.
Conflicts of Interest
There is no any conflicts of interest being declared.
References
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Struhl, G. A gene product required for correct initiation of segmental determination in Drosophila. Nature 1981, 293, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Mozgova, I.; Hennig, L. The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 2015, 66, 269–296. [Google Scholar] [CrossRef] [PubMed]
- Piunti, A.; Shilatifard, A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016, 352, 6290. [Google Scholar] [CrossRef] [PubMed]
- Entrevan, M.; Schuettengruber, B.; Cavalli, G. Regulation of Genome Architecture and Function by Polycomb Proteins. Trends Cell Biol. 2016, 26, 511–525. [Google Scholar] [CrossRef]
- Huo, Y.Q.; Yan, Z.Q.; Zhang, B.S.; Wang, X. Recruitment of Polycomb Repressive Complex 2 is Essential to Suppress the Target Chromatin in Arabidopsis. Crit. Rev. Plant Sci. 2016, 35, 131–145. [Google Scholar] [CrossRef]
- Yang, X.; Tong, A.; Yan, B.; Wang, X. Governing the Silencing State of Chromatin: The Roles of Polycomb Repressive Complex 1 in Arabidopsis. Plant Cell Physiol. 2017, 58, 198–206. [Google Scholar] [CrossRef]
- Hugues, A.; Jacobs, C.S.; Roudier, F. Mitotic Inheritance of PRC2-Mediated Silencing: Mechanistic Insights and Developmental Perspectives. Front. Plant Sci. 2020, 11, 262. [Google Scholar] [CrossRef]
- Goodrich, J.; Puangsomlee, P.; Martin, M.; Long, D.; Meyerowitz, E.M.; Coupland, G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 1997, 386, 44–51. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Vielle-Calzada, J.P.; Hoeppner, M.A.; Gagliano, W.B. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 1998, 280, 446–450. [Google Scholar] [CrossRef]
- Luo, M.; Bilodeau, P.; Koltunow, A.; Dennis, E.S.; Peacock, W.J.; Chaudhury, A.M. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1999, 96, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Yanai, Y.; Chen, L.; Kato, Y.; Hiratsuka, J.; Miwa, T.; Sung, Z.R.; Takahashi, S. EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 2001, 13, 2471–2481. [Google Scholar] [CrossRef] [PubMed]
- Gendall, A.R.; Levy, Y.Y.; Wilson, A.; Dean, C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 2001, 107, 525–535. [Google Scholar] [CrossRef]
- Ach, R.A.; Taranto, P.; Gruissem, W. A conserved family of WD-40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell 1997, 9, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Kenzior, A.L.; Folk, W.R. AtMSI4 and RbAp48 WD-40 repeat proteins bind metal ions. FEBS Lett. 1998, 440, 425–429. [Google Scholar] [CrossRef]
- Kohler, C.; Hennig, L.; Bouveret, R.; Gheyselinck, J.; Grossniklaus, U.; Gruissem, W. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 2003, 22, 4804–4814. [Google Scholar] [CrossRef]
- Derkacheva, M.; Steinbach, Y.; Wildhaber, T.; Mozgova, I.; Mahrez, W.; Nanni, P.; Bischof, S.; Gruissem, W.; Hennig, L. Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 2013, 32, 2073–2085. [Google Scholar] [CrossRef]
- Kinoshita, T.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. USA 2001, 98, 14156–14161. [Google Scholar] [CrossRef]
- Ohad, N.; Yadegari, R.; Margossian, L.; Hannon, M.; Michaeli, D.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 1999, 11, 407–416. [Google Scholar] [CrossRef]
- Spillane, C.; MacDougall, C.; Stock, C.; Kohler, C.; Vielle-Calzada, J.P.; Nunes, S.M.; Grossniklaus, U.; Goodrich, J. Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr. Biol. 2000, 10, 1535–1538. [Google Scholar] [CrossRef]
- Yadegari, R.; Kinoshita, T.; Lotan, O.; Cohen, G.; Katz, A.; Choi, Y.; Nakashima, K.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L.; et al. Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 2000, 12, 2367–2382. [Google Scholar] [CrossRef]
- Bemer, M.; Grossniklaus, U. Dynamic regulation of Polycomb group activity during plant development. Curr. Opin. Plant Biol. 2012, 15, 523–529. [Google Scholar] [CrossRef] [PubMed]
- De Lucia, F.; Crevillen, P.; Jones, A.M.; Greb, T.; Dean, C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. USA 2008, 105, 16831–16836. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Platten, D.; Chaudhury, A.; Peacock, W.J.; Dennis, E.S. Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol. Plant 2009, 2, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.K.; Wang, Y.; Zhang, Y.; Li, S.G.; Lu, X.C.; Li, H.; Zou, C.; Xu, Z.H.; Bai, S.N. OsSET1, a novel SET-domain-containing gene from rice. J. Exp. Bot. 2003, 54, 1995–1996. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thakur, J.K.; Malik, M.R.; Bhatt, V.; Reddy, M.K.; Sopory, S.K.; Tyagi, A.K.; Khurana, J.P. A POLYCOMB group gene of rice (Oryza sativa L. subspecies indica), OsiEZ1, codes for a nuclear-localized protein expressed preferentially in young seedlings and during reproductive development. Gene 2003, 314, 1–13. [Google Scholar] [CrossRef]
- Yan, D.; Zhang, X.; Zhang, L.; Ye, S.; Zeng, L.; Liu, J.; Li, Q.; He, Z. Curved chimeric palea 1 encoding an EMF1-like protein maintains epigenetic repression of OsMADS58 in rice palea development. Plant J. 2015, 82, 12–24. [Google Scholar] [CrossRef]
- Molitor, A.; Shen, W.H. The polycomb complex PRC1: Composition and function in plants. J. Genet. Genom. 2013, 40, 231–238. [Google Scholar] [CrossRef]
- Xu, L.; Shen, W.H. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 2008, 18, 1966–1971. [Google Scholar] [CrossRef]
- Sanchez-Pulido, L.; Devos, D.; Sung, Z.R.; Calonje, M. RAWUL: A new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genom. 2008, 9, 308. [Google Scholar] [CrossRef]
- Li, W.; Wang, Z.; Li, J.; Yang, H.; Cui, S.; Wang, X.; Ma, L. Overexpression of AtBMI1C, a polycomb group protein gene, accelerates flowering in Arabidopsis. PLoS ONE 2011, 6, e21364. [Google Scholar] [CrossRef] [PubMed]
- Turck, F.; Roudier, F.; Farrona, S.; Martin-Magniette, M.L.; Guillaume, E.; Buisine, N.; Gagnot, S.; Martienssen, R.A.; Coupland, G.; Colot, V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007, 3, e86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Germann, S.; Blus, B.J.; Khorasanizadeh, S.; Gaudin, V.; Jacobsen, S.E. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 2007, 14, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Beh, L.Y.; Colwell, L.J.; Francis, N.J. A core subunit of Polycomb repressive complex 1 is broadly conserved in function but not primary sequence. Proc. Natl. Acad. Sci. USA 2012, 109, E1063–E1071. [Google Scholar] [CrossRef]
- Calonje, M.; Sanchez, R.; Chen, L.; Sung, Z.R. EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 2008, 20, 277–291. [Google Scholar] [CrossRef]
- Yang, C.; Bratzel, F.; Hohmann, N.; Koch, M.; Turck, F.; Calonje, M. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 2013, 23, 1324–1329. [Google Scholar] [CrossRef]
- Bratzel, F.; Lopez-Torrejon, G.; Koch, M.; Del Pozo, J.C.; Calonje, M. Keeping cell identity in arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 2010, 20, 1853–1859. [Google Scholar] [CrossRef]
- Chen, D.; Molitor, A.; Liu, C.; Shen, W.H. The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 2010, 20, 1332–1344. [Google Scholar] [CrossRef]
- Wang, H.; Liu, C.; Cheng, J.; Liu, J.; Zhang, L.; He, C.; Shen, W.H.; Jin, H.; Xu, L.; Zhang, Y. Arabidopsis Flower and Embryo Developmental Genes are Repressed in Seedlings by Different Combinations of Polycomb Group Proteins in Association with Distinct Sets of Cis-regulatory Elements. PLoS Genet. 2016, 12, e1005771. [Google Scholar] [CrossRef]
- Guan, H.; Zheng, Z.; Grey, P.H.; Li, Y.; Oppenheimer, D.G. Conservation and divergence of plant LHP1 protein sequences and expression patterns in angiosperms and gymnosperms. Mol. Genet. Genom. 2011, 285, 357–373. [Google Scholar] [CrossRef]
- Baurle, I.; Dean, C. The timing of developmental transitions in plants. Cell 2006, 125, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Lafos, M.; Kroll, P.; Hohenstatt, M.L.; Thorpe, F.L.; Clarenz, O.; Schubert, D. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 2011, 7, e1002040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Clarenz, O.; Cokus, S.; Bernatavichute, Y.V.; Pellegrini, M.; Goodrich, J.; Jacobsen, S.E. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007, 5, e129. [Google Scholar] [CrossRef] [PubMed]
- Aloia, L.; Di Stefano, B.; Di Croce, L. Polycomb complexes in stem cells and embryonic development. Development 2013, 140, 2525–2534. [Google Scholar] [CrossRef]
- Xue, Z.; Liu, L.; Zhang, C. Regulation of Shoot Apical Meristem and Axillary Meristem Development in Plants. Int. J. Mol. Sci. 2020, 21, 2917. [Google Scholar] [CrossRef]
- Kaufmann, K.; Pajoro, A.; Angenent, G.C. Regulation of transcription in plants: Mechanisms controlling developmental switches. Nat. Rev. Genet. 2010, 11, 830–842. [Google Scholar] [CrossRef]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jurgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Ha, C.M.; Jun, J.H.; Fletcher, J.C. Current Topics in Developmental Biology; Shoot apical meristem form and function; Academic Press: Cambridge, MA, USA, 2010; Volume 91, pp. 103–140. [Google Scholar] [CrossRef]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jurgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Yadav, R.K.; Perales, M.; Gruel, J.; Ohno, C.; Heisler, M.; Girke, T.; Jonsson, H.; Reddy, G.V. Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 2013, 9, 654. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.; Berger, J.; Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [PubMed]
- Aichinger, E.; Kornet, N.; Friedrich, T.; Laux, T. Plant stem cell niches. Annu. Rev. Plant Biol. 2012, 63, 615–636. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.C.; Brand, U.; Running, M.P.; Simon, R.; Meyerowitz, E.M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 1999, 283, 1911–1914. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jonsson, H.; Reddy, G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, A.; Han, H.; Li, T.; Geng, Y.; Liu, X.; Meyerowitz, E.M. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science 2018, 361, 502–506. [Google Scholar] [CrossRef]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 1993, 119, 397–418. [Google Scholar]
- Kayes, J.M.; Clark, S.E. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 1998, 125, 3843–3851. [Google Scholar]
- Clark, S.E.; Williams, R.W.; Meyerowitz, E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 1997, 89, 575–585. [Google Scholar] [CrossRef]
- Jeong, S.; Trotochaud, A.E.; Clark, S.E. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 1999, 11, 1925–1934. [Google Scholar] [CrossRef]
- Somssich, M.; Je, B.I.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef]
- Nardmann, J.; Werr, W. The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 2006, 23, 2492–2504. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, W.; Ohmori, Y.; Ushijima, T.; Matsusaka, H.; Matsushita, T.; Kumamaru, T.; Kawano, S.; Hirano, H.Y. Axillary Meristem Formation in Rice Requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 2015, 27, 1173–1184. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, Y.; Tanaka, W.; Kojima, M.; Sakakibara, H.; Hirano, H.Y. WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell 2013, 25, 229–241. [Google Scholar] [CrossRef]
- Suzaki, T.; Sato, M.; Ashikari, M.; Miyoshi, M.; Nagato, Y.; Hirano, H.Y. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 2004, 131, 5649–5657. [Google Scholar] [CrossRef]
- Suzaki, T.; Toriba, T.; Fujimoto, M.; Tsutsumi, N.; Kitano, H.; Hirano, H.Y. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 2006, 47, 1591–1602. [Google Scholar] [CrossRef]
- Chu, H.; Qian, Q.; Liang, W.; Yin, C.; Tan, H.; Yao, X.; Yuan, Z.; Yang, J.; Huang, H.; Luo, D.; et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006, 142, 1039–1052. [Google Scholar] [CrossRef]
- Xu, W.; Tao, J.; Chen, M.; Dreni, L.; Luo, Z.; Hu, Y.; Liang, W.; Zhang, D. Interactions between FLORAL ORGAN NUMBER4 and floral homeotic genes in regulating rice flower development. J. Exp. Bot. 2017, 68, 483–498. [Google Scholar] [CrossRef]
- Barton, M.K. Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 2010, 341, 95–113. [Google Scholar] [CrossRef]
- Liu, X.; Kim, Y.J.; Muller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef]
- Sun, B.; Looi, L.S.; Guo, S.; He, Z.; Gan, E.S.; Huang, J.; Xu, Y.; Wee, W.Y.; Ito, T. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 2014, 343, 1248559. [Google Scholar] [CrossRef]
- Sun, B.; Xu, Y.; Ng, K.H.; Ito, T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009, 23, 1791–1804. [Google Scholar] [CrossRef]
- Payne, T.; Johnson, S.D.; Koltunow, A.M. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 2004, 131, 3737–3749. [Google Scholar] [CrossRef] [PubMed]
- Urbanus, S.L.; de Folter, S.; Shchennikova, A.V.; Kaufmann, K.; Immink, R.G.; Angenent, G.C. In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol. 2009, 9, 5. [Google Scholar] [CrossRef]
- Sawarkar, R.; Paro, R. Interpretation of developmental signaling at chromatin: The Polycomb perspective. Dev. Cell 2010, 19, 651–661. [Google Scholar] [CrossRef]
- Simon, J.A.; Kingston, R.E. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 2013, 49, 808–824. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Lohmann, J.U.; Hong, R.L.; Hobe, M.; Busch, M.A.; Parcy, F.; Simon, R.; Weigel, D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 2001, 105, 793–803. [Google Scholar] [CrossRef]
- Lenhard, M.; Bohnert, A.; Jurgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef]
- Hay, A.; Tsiantis, M. KNOX genes: Versatile regulators of plant development and diversity. Development 2010, 137, 3153–3165. [Google Scholar] [CrossRef]
- Carles, C.C.; Fletcher, J.C. Shoot apical meristem maintenance: The art of a dynamic balance. Trends Plant Sci. 2003, 8, 394–401. [Google Scholar] [CrossRef]
- Byrne, M.E.; Simorowski, J.; Martienssen, R.A. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 2002, 129, 1957–1965. [Google Scholar]
- Semiarti, E.; Ueno, Y.; Tsukaya, H.; Iwakawa, H.; Machida, C.; Machida, Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 2001, 128, 1771–1783. [Google Scholar]
- Kim, S.Y.; Lee, J.; Eshed-Williams, L.; Zilberman, D.; Sung, Z.R. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 2012, 8, e1002512. [Google Scholar] [CrossRef]
- Deng, W.; Buzas, D.M.; Ying, H.; Robertson, M.; Taylor, J.; Peacock, W.J.; Dennis, E.S.; Helliwell, C. Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genom. 2013, 14, 593. [Google Scholar] [CrossRef]
- Lodha, M.; Marco, C.F.; Timmermans, M.C. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 2013, 27, 596–601. [Google Scholar] [CrossRef]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef]
- Lin, W.C.; Shuai, B.; Springer, P.S. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 2003, 15, 2241–2252. [Google Scholar] [CrossRef]
- Li, Z.; Li, B.; Liu, J.; Guo, Z.; Liu, Y.; Li, Y.; Shen, W.H.; Huang, Y.; Huang, H.; Zhang, Y.; et al. Transcription factors AS1 and AS2 interact with LHP1 to repress KNOX genes in Arabidopsis. J. Integr. Plant. Biol. 2016, 58, 959–970. [Google Scholar] [CrossRef]
- Shkolnik-Inbar, D.; Bar-Zvi, D. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 2010, 22, 3560–3573. [Google Scholar] [CrossRef]
- Monfared, M.M.; Simon, M.K.; Meister, R.J.; Roig-Villanova, I.; Kooiker, M.; Colombo, L.; Fletcher, J.C.; Gasser, C.S. Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis. Plant J. 2011, 66, 1020–1031. [Google Scholar] [CrossRef]
- Kooiker, M.; Airoldi, C.A.; Losa, A.; Manzotti, P.S.; Finzi, L.; Kater, M.M.; Colombo, L. BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 2005, 17, 722–729. [Google Scholar] [CrossRef]
- Wanke, D.; Hohenstatt, M.L.; Dynowski, M.; Bloss, U.; Hecker, A.; Elgass, K.; Hummel, S.; Hahn, A.; Caesar, K.; Schleifenbaum, F.; et al. Alanine zipper-like coiled-coil domains are necessary for homotypic dimerization of plant GAGA-factors in the nucleus and nucleolus. PLoS ONE 2011, 6, e16070. [Google Scholar] [CrossRef]
- Meister, R.J.; Williams, L.A.; Monfared, M.M.; Gallagher, T.L.; Kraft, E.A.; Nelson, C.G.; Gasser, C.S. Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 2004, 37, 426–438. [Google Scholar] [CrossRef]
- Mu, Y.; Zou, M.; Sun, X.; He, B.; Xu, X.; Liu, Y.; Zhang, L.; Chi, W. BASIC PENTACYSTEINE Proteins Repress ABSCISIC ACID INSENSITIVE4 Expression via Direct Recruitment of the Polycomb-Repressive Complex 2 in Arabidopsis Root Development. Plant Cell Physiol. 2017, 58, 607–621. [Google Scholar] [CrossRef]
- Bellegarde, F.; Gojon, A.; Martin, A. Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2553–2565. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Leyser, O. A plant’s diet, surviving in a variable nutrient environment. Science 2020, 368. [Google Scholar] [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Girin, T.; Tillard, P.; Lepetit, M.; Gojon, A. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006, 140, 909–921. [Google Scholar] [CrossRef]
- Roudier, F.; Ahmed, I.; Berard, C.; Sarazin, A.; Mary-Huard, T.; Cortijo, S.; Bouyer, D.; Caillieux, E.; Duvernois-Berthet, E.; Al-Shikhley, L.; et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011, 30, 1928–1938. [Google Scholar] [CrossRef]
- Bellegarde, F.; Herbert, L.; Sere, D.; Caillieux, E.; Boucherez, J.; Fizames, C.; Roudier, F.; Gojon, A.; Martin, A. Polycomb Repressive Complex 2 attenuates the very high expression of the Arabidopsis gene NRT2.1. Sci. Rep. 2018, 8, 7905. [Google Scholar] [CrossRef]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e1054. [Google Scholar] [CrossRef]
- Alecki, C.; Chiwara, V.; Sanz, L.A.; Grau, D.; Arias Perez, O.; Boulier, E.L.; Armache, K.J.; Chedin, F.; Francis, N.J. RNA-DNA strand exchange by the Drosophila Polycomb complex PRC2. Nat. Commun. 2020, 11, 1781. [Google Scholar] [CrossRef]
- Bouyer, D.; Roudier, F.; Heese, M.; Andersen, E.D.; Gey, D.; Nowack, M.K.; Goodrich, J.; Renou, J.P.; Grini, P.E.; Colot, V.; et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011, 7, e1002014. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef]
- Gasser, C.S.; Skinner, D.J. Development and evolution of the unique ovules of flowering plants. Curr. Top. Dev. Biol. 2019, 131, 373–399. [Google Scholar] [CrossRef]
- Lau, S.; Slane, D.; Herud, O.; Kong, J.; Jurgens, G. Early embryogenesis in flowering plants: Setting up the basic body pattern. Annu. Rev. Plant Biol. 2012, 63, 483–506. [Google Scholar] [CrossRef]
- De Vries, S.C.; Weijers, D. Plant embryogenesis. Curr. Biol. 2017, 27, R870–R873. [Google Scholar] [CrossRef]
- Figueiredo, D.D.; Kohler, C. Auxin: A molecular trigger of seed development. Genes Dev. 2018, 32, 479–490. [Google Scholar] [CrossRef]
- Robert, H.S.; Park, C.; Gutierrez, C.L.; Wojcikowska, B.; Pencik, A.; Novak, O.; Chen, J.; Grunewald, W.; Dresselhaus, T.; Friml, J.; et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 2018, 4, 548–553. [Google Scholar] [CrossRef]
- Robert, H.S. Molecular Communication for Coordinated Seed and Fruit Development: What Can We Learn from Auxin and Sugars? Int. J. Mol. Sci. 2019, 20, 936. [Google Scholar] [CrossRef]
- Ezquer, I.; Mizzotti, C.; Nguema-Ona, E.; Gotte, M.; Beauzamy, L.; Viana, V.E.; Dubrulle, N.; Costa de Oliveira, A.; Caporali, E.; Koroney, A.S.; et al. The Developmental Regulator SEEDSTICK Controls Structural and Mechanical Properties of the Arabidopsis Seed Coat. Plant Cell 2016, 28, 2478–2492. [Google Scholar] [CrossRef]
- Mizzotti, C.; Ezquer, I.; Paolo, D.; Rueda-Romero, P.; Guerra, R.F.; Battaglia, R.; Rogachev, I.; Aharoni, A.; Kater, M.M.; Caporali, E.; et al. SEEDSTICK is a master regulator of development and metabolism in the Arabidopsis seed coat. PLoS Genet. 2014, 10, e1004856. [Google Scholar] [CrossRef]
- Simonini, S.; Kater, M.M. Class I BASIC PENTACYSTEINE factors regulate HOMEOBOX genes involved in meristem size maintenance. J. Exp. Bot. 2014, 65, 1455–1465. [Google Scholar] [CrossRef]
- Simonini, S.; Roig-Villanova, I.; Gregis, V.; Colombo, B.; Colombo, L.; Kater, M.M. Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. Plant Cell 2012, 24, 4163–4172. [Google Scholar] [CrossRef]
- Gregis, V.; Andres, F.; Sessa, A.; Guerra, R.F.; Simonini, S.; Mateos, J.L.; Torti, S.; Zambelli, F.; Prazzoli, G.M.; Bjerkan, K.N.; et al. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 2013, 14, R56. [Google Scholar] [CrossRef]
- Petrella, R.; Caselli, F.; Roig-Villanova, I.; Vignati, V.; Chiara, M.; Ezquer, I.; Tadini, L.; Kater, M.M.; Gregis, V. BPC transcription factors and a Polycomb Group protein confine the expression of the ovule identity gene SEEDSTICK in Arabidopsis. Plant J. 2020, 102, 582–599. [Google Scholar] [CrossRef]
- Hecker, A.; Brand, L.H.; Peter, S.; Simoncello, N.; Kilian, J.; Harter, K.; Gaudin, V.; Wanke, D. The Arabidopsis GAGA-Binding Factor BASIC PENTACYSTEINE6 Recruits the POLYCOMB-REPRESSIVE COMPLEX1 Component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA Motifs. Plant Physiol. 2015, 168, 1013–1024. [Google Scholar] [CrossRef]
- Suzuki, M.; Wang, H.H.; McCarty, D.R. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007, 143, 902–911. [Google Scholar] [CrossRef]
- Suzuki, M.; McCarty, D.R. Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 2008, 11, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Van Wuytswinkel, O.; Castelain, M.; Bellini, C. Combined networks regulating seed maturation. Trends Plant Sci. 2007, 12, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.; Carianopol, C.; Tsai, A.Y.; Varatharajah, K.; Chiu, R.S.; Gazzarrini, S. Corrigendum: SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J. Exp. Bot. 2017, 68, 5981. [Google Scholar] [CrossRef] [PubMed]
- Makarevich, G.; Leroy, O.; Akinci, U.; Schubert, D.; Clarenz, O.; Goodrich, J.; Grossniklaus, U.; Kohler, C. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006, 7, 947–952. [Google Scholar] [CrossRef]
- Wu, J.; Mohamed, D.; Dowhanik, S.; Petrella, R.; Gregis, V.; Li, J.; Wu, L.; Gazzarrini, S. Spatiotemporal Restriction of FUSCA3 Expression by Class I BPCs Promotes Ovule Development and Coordinates Embryo and Endosperm Growth. Plant Cell 2020, 32, 1886–1904. [Google Scholar] [CrossRef]
- Santos-Mendoza, M.; Dubreucq, B.; Baud, S.; Parcy, F.; Caboche, M.; Lepiniec, L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008, 54, 608–620. [Google Scholar] [CrossRef]
- Braybrook, S.A.; Harada, J.J. LECs go crazy in embryo development. Trends Plant Sci. 2008, 13, 624–630. [Google Scholar] [CrossRef]
- To, A.; Valon, C.; Savino, G.; Guilleminot, J.; Devic, M.; Giraudat, J.; Parcy, F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 2006, 18, 1642–1651. [Google Scholar] [CrossRef]
- Moon, Y.H.; Chen, L.; Pan, R.L.; Chang, H.S.; Zhu, T.; Maffeo, D.M.; Sung, Z.R. EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 2003, 15, 681–693. [Google Scholar] [CrossRef]
- Kim, S.Y.; Zhu, T.; Sung, Z.R. Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiol. 2010, 152, 516–528. [Google Scholar] [CrossRef]
- Wang, L.; Brown, J.L.; Cao, R.; Zhang, Y.; Kassis, J.A.; Jones, R.S. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 2004, 14, 637–646. [Google Scholar] [CrossRef]
- Amasino, R. Seasonal and developmental timing of flowering. Plant J. 2010, 61, 1001–1013. [Google Scholar] [CrossRef]
- Berry, S.; Dean, C. Environmental perception and epigenetic memory: Mechanistic insight through FLC. Plant J. 2015, 83, 133–148. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Burn, J.E.; Perez, P.P.; Metzger, J.; Edwards, J.A.; Peacock, W.J.; Dennis, E.S. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 1999, 11, 445–458. [Google Scholar] [CrossRef]
- Song, J.; Irwin, J.; Dean, C. Remembering the Prolonged Cold of Winter. Curr. Biol. 2013, 23, R807–R811. [Google Scholar] [CrossRef]
- Kim, D.H.; Doyle, M.R.; Sung, S.; Amasino, R.M. Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 2009, 25, 277–299. [Google Scholar] [CrossRef]
- Bastow, R.; Mylne, J.S.; Lister, C.; Lippman, Z.; Martienssen, R.A.; Dean, C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 2004, 427, 164–167. [Google Scholar] [CrossRef]
- Sung, S.; Amasino, R.M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 2004, 427, 159–164. [Google Scholar] [CrossRef]
- Cui, Z.; Tong, A.; Huo, Y.; Yan, Z.; Yang, W.; Yang, X.; Wang, X.X. SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in Arabidopsis. BMC Biol. 2017, 15, 80. [Google Scholar] [CrossRef]
- Song, Y.H.; Ito, S.; Imaizumi, T. Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013, 18, 575–583. [Google Scholar] [CrossRef]
- Schubert, D.; Primavesi, L.; Bishopp, A.; Roberts, G.; Doonan, J.; Jenuwein, T.; Goodrich, J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006, 25, 4638–4649. [Google Scholar] [CrossRef]
- Sung, S.; Schmitz, R.J.; Amasino, R.M. A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev. 2006, 20, 3244–3248. [Google Scholar] [CrossRef]
- Finnegan, E.J.; Dennis, E.S. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr. Biol. 2007, 17, 1978–1983. [Google Scholar] [CrossRef]
- Wood, C.C.; Robertson, M.; Tanner, G.; Peacock, W.J.; Dennis, E.S.; Helliwell, C.A. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. USA 2006, 103, 14631–14636. [Google Scholar] [CrossRef]
- Greb, T.; Mylne, J.S.; Crevillen, P.; Geraldo, N.; An, H.; Gendall, A.R.; Dean, C. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 2007, 17, 73–78. [Google Scholar] [CrossRef]
- Angel, A.; Song, J.; Dean, C.; Howard, M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 2011, 476, 105–108. [Google Scholar] [CrossRef]
- Mylne, J.S.; Barrett, L.; Tessadori, F.; Mesnage, S.; Johnson, L.; Bernatavichute, Y.V.; Jacobsen, S.E.; Fransz, P.; Dean, C. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc. Natl. Acad. Sci. USA 2006, 103, 5012–5017. [Google Scholar] [CrossRef]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Yang, C.H.; Chen, L.J.; Sung, Z.R. Genetic regulation of shoot development in Arabidopsis: Role of the EMF genes. Dev. Biol. 1995, 169, 421–435. [Google Scholar] [CrossRef]
- Sung, Z.R.; Belachew, A.; Shunong, B.; Bertrand-Garcia, R. EMF, an Arabidopsis Gene Required for Vegetative Shoot Development. Science 1992, 258, 1645–1647. [Google Scholar] [CrossRef]
- Chen, L.; Cheng, J.C.; Castle, L.; Sung, Z.R. EMF genes regulate Arabidopsis inflorescence development. Plant Cell 1997, 9, 2011–2024. [Google Scholar] [CrossRef]
- Yu, C.W.; Liu, X.; Luo, M.; Chen, C.; Lin, X.; Tian, G.; Lu, Q.; Cui, Y.; Wu, K. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 2011, 156, 173–184. [Google Scholar] [CrossRef]
- Jiang, D.; Yang, W.; He, Y.; Amasino, R.M. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 2007, 19, 2975–2987. [Google Scholar] [CrossRef]
- Jiang, D.; Wang, Y.; He, Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 2008, 3, e3404. [Google Scholar] [CrossRef]
- Doyle, M.R.; Amasino, R.M. A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol. 2009, 151, 1688–1697. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, X.; Yuan, W.; Schmitz, R.J.; He, Y. Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev. Cell 2014, 28, 727–736. [Google Scholar] [CrossRef]
- Exner, V.; Aichinger, E.; Shu, H.; Wildhaber, T.; Alfarano, P.; Caflisch, A.; Gruissem, W.; Kohler, C.; Hennig, L. The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS ONE 2009, 4, e5335. [Google Scholar] [CrossRef]
- Kotake, T.; Takada, S.; Nakahigashi, K.; Ohto, M.; Goto, K. Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 2003, 44, 555–564. [Google Scholar] [CrossRef]
- Sung, S.; He, Y.; Eshoo, T.W.; Tamada, Y.; Johnson, L.; Nakahigashi, K.; Goto, K.; Jacobsen, S.E.; Amasino, R.M. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 2006, 38, 706–710. [Google Scholar] [CrossRef]
- Liu, C.; Xi, W.; Shen, L.; Tan, C.; Yu, H. Regulation of floral patterning by flowering time genes. Dev. Cell 2009, 16, 711–722. [Google Scholar] [CrossRef]
- Aubert, D.; Chen, L.; Moon, Y.H.; Martin, D.; Castle, L.A.; Yang, C.H.; Sung, Z.R. EMF1, a novel protein involved in the control of shoot architecture and flowering in Arabidopsis. Plant Cell 2001, 13, 1865–1875. [Google Scholar] [CrossRef]
- Sanchez, R.; Kim, M.Y.; Calonje, M.; Moon, Y.H.; Sung, Z.R. Temporal and spatial requirement of EMF1 activity for Arabidopsis vegetative and reproductive development. Mol. Plant 2009, 2, 643–653. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Itoh, H.; Nonoue, Y.; Yano, M.; Izawa, T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 2010, 42, 635–638. [Google Scholar] [CrossRef]
- Pasriga, R.; Yoon, J.; Cho, L.H.; An, G. Overexpression of RICE FLOWERING LOCUS T 1 (RFT1) Induces Extremely Early Flowering in Rice. Mol. Cells 2019, 42, 406–417. [Google Scholar] [CrossRef]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Ryu, C.H.; Lee, S.; Cho, L.H.; Kim, S.L.; Lee, Y.S.; Choi, S.C.; Jeong, H.J.; Yi, J.; Park, S.J.; Han, C.D.; et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009, 32, 1412–1427. [Google Scholar] [CrossRef]
- Peng, Y.; Lin, W.; Cai, W.; Arora, R. Overexpression of a Panax ginseng tonoplast aquaporin alters salt tolerance, drought tolerance and cold acclimation ability in transgenic Arabidopsis plants. Planta 2007, 226, 729–740. [Google Scholar] [CrossRef]
- Peng, L.T.; Shi, Z.Y.; Li, L.; Shen, G.Z.; Zhang, J.L. Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. J. Plant Physiol. 2008, 165, 876–885. [Google Scholar] [CrossRef]
- Yan, W.H.; Wang, P.; Chen, H.X.; Zhou, H.J.; Li, Q.P.; Wang, C.R.; Ding, Z.H.; Zhang, Y.S.; Yu, S.B.; Xing, Y.Z.; et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef]
- Coneva, V.; Zhu, T.; Colasanti, J. Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. J. Exp. Bot. 2007, 58, 3679–3693. [Google Scholar] [CrossRef]
- Matsubara, K.; Yamanouchi, U.; Nonoue, Y.; Sugimoto, K.; Wang, Z.X.; Minobe, Y.; Yano, M. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 2011, 66, 603–612. [Google Scholar] [CrossRef]
- Gao, H.; Zheng, X.M.; Fei, G.; Chen, J.; Jin, M.; Ren, Y.; Wu, W.; Zhou, K.; Sheng, P.; Zhou, F.; et al. Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet. 2013, 9, e1003281. [Google Scholar] [CrossRef]
- Jiang, P.; Wang, S.; Zheng, H.; Li, H.; Zhang, F.; Su, Y.; Xu, Z.; Lin, H.; Qian, Q.; Ding, Y. SIP1 participates in regulation of flowering time in rice by recruiting OsTrx1 to Ehd1. New Phytol. 2018, 219, 422–435. [Google Scholar] [CrossRef]
- Zhao, S.Q.; Hu, J.; Guo, L.B.; Qian, Q.; Xue, H.W. Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 2010, 20, 935–947. [Google Scholar] [CrossRef]
- Wang, J.; Hu, J.; Qian, Q.; Xue, H.-W. LC2 and OsVIL2 Promote Rice Flowering by Photoperoid-Induced Epigenetic Silencing of OsLF. Mol. Plant 2013, 6, 514–527. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.; Hang, R.; Kim, S.R.; Lee, Y.S.; Cao, X.; Amasino, R.; An, G. OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 2013, 73, 566–578. [Google Scholar] [CrossRef]
- Zhao, X.L.; Shi, Z.Y.; Peng, L.T.; Shen, G.Z.; Zhang, J.L. An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15. New Biotechnol. 2011, 28, 788–797. [Google Scholar] [CrossRef]
- Xiao, J.; Jin, R.; Yu, X.; Shen, M.; Wagner, J.D.; Pai, A.; Song, C.; Zhuang, M.; Klasfeld, S.; He, C.; et al. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 2017, 49, 1546–1552. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).