Suppression of SHROOM1 Improves In Vitro and In Vivo Gene Integration by Promoting Homology-Directed Repair
Abstract
1. Introduction
2. Results
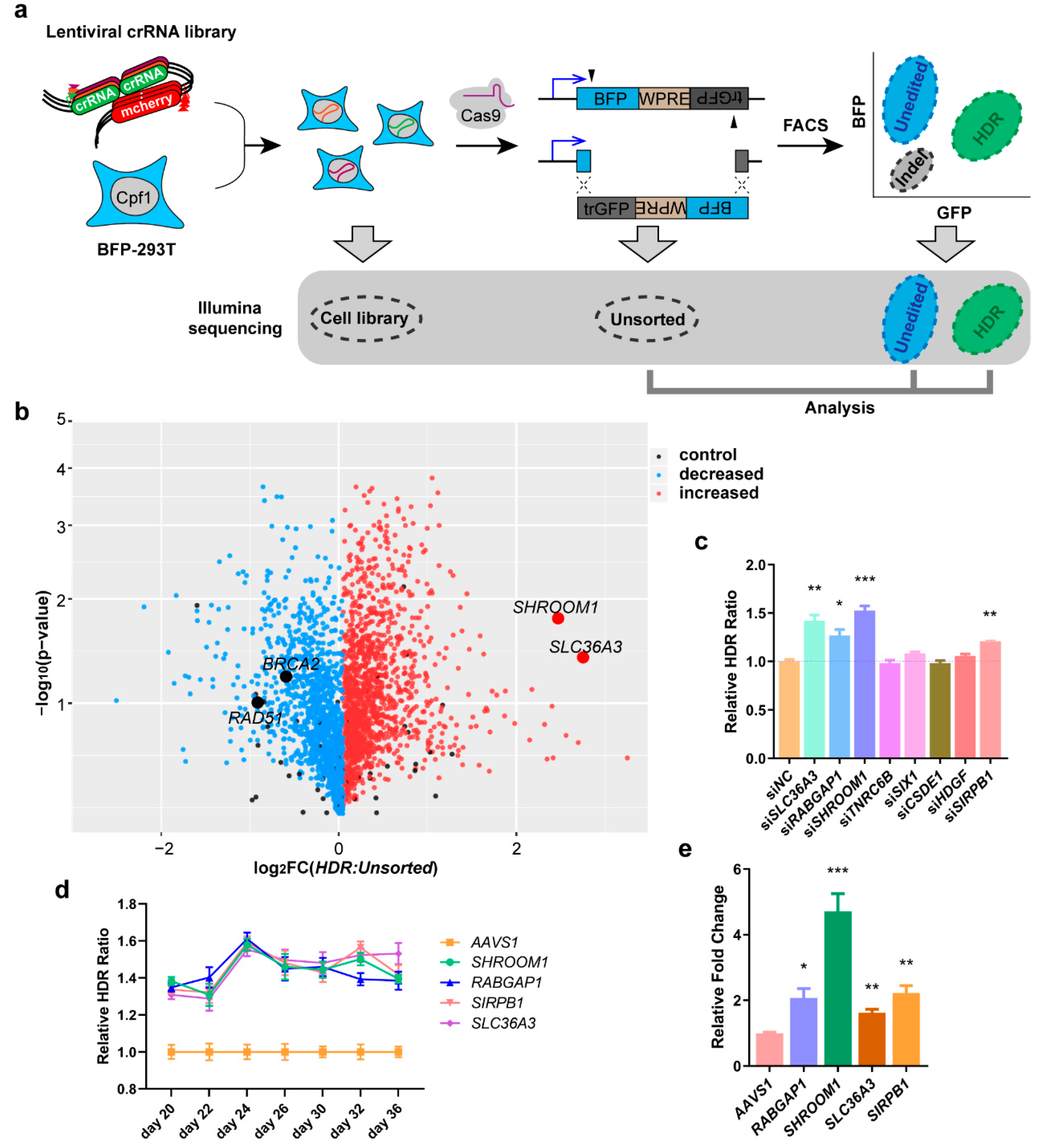
2.1. Genome-Wide Screening is Conducted to Search for Genes that Modulate HDR
2.2. Suppressing SHROOM1 Promotes HDR in BFP-293T Cells
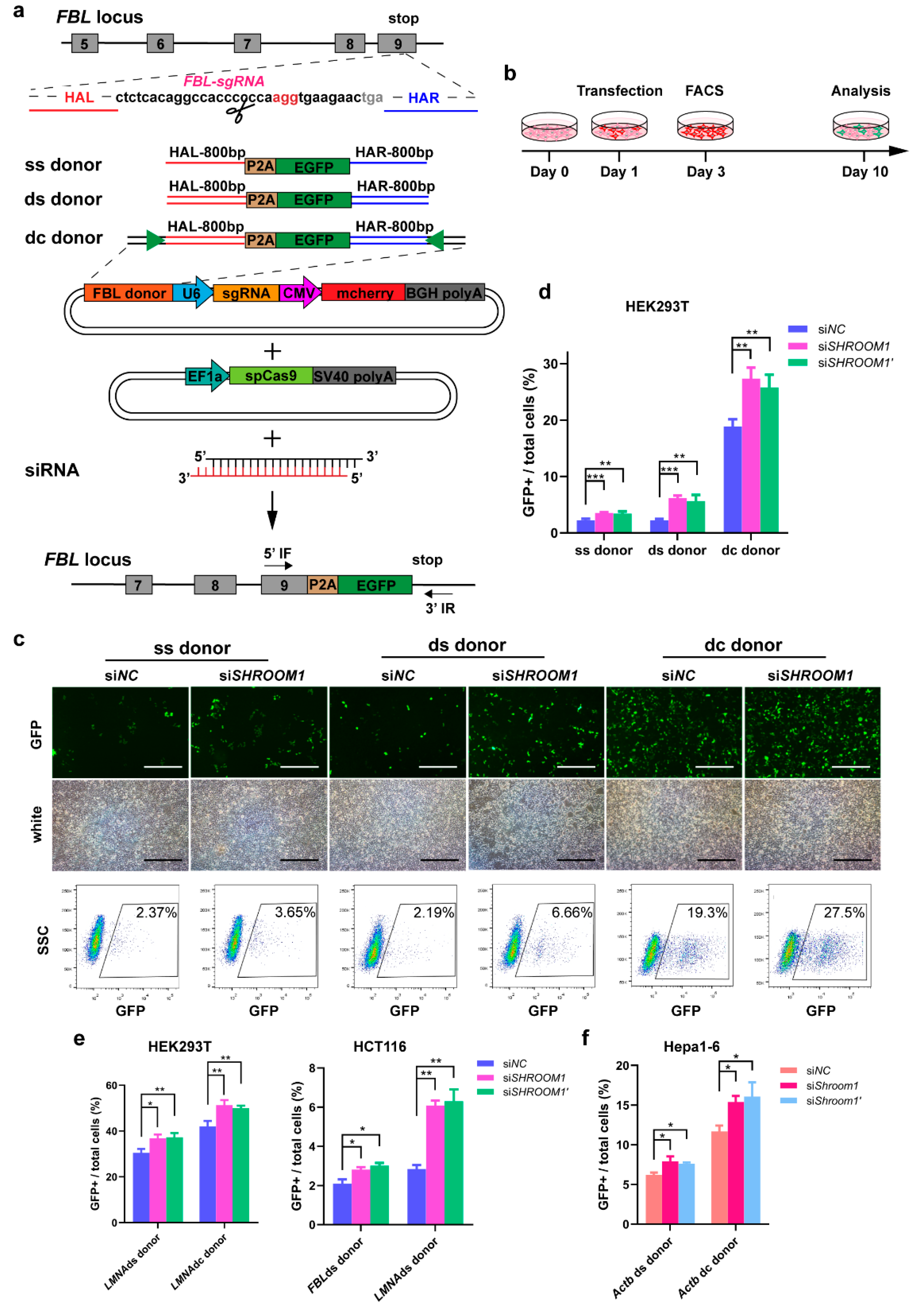
2.3. Knockdown of SHROOM1 Can Promote In Vitro Genome Editing Efficiency
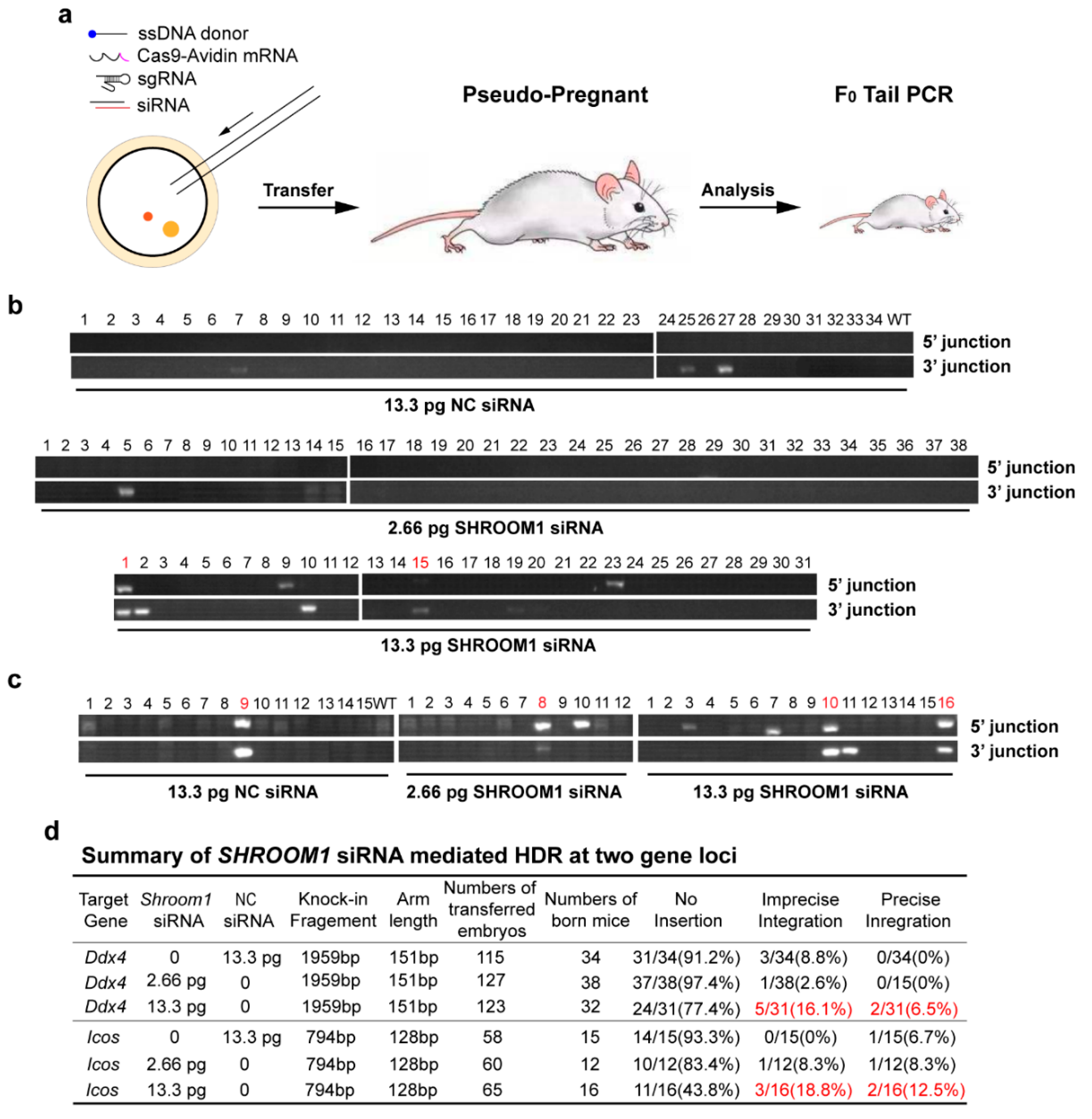
2.4. SHROOM1 Knockdown Can Promote Gene Integration in Mouse Embryos
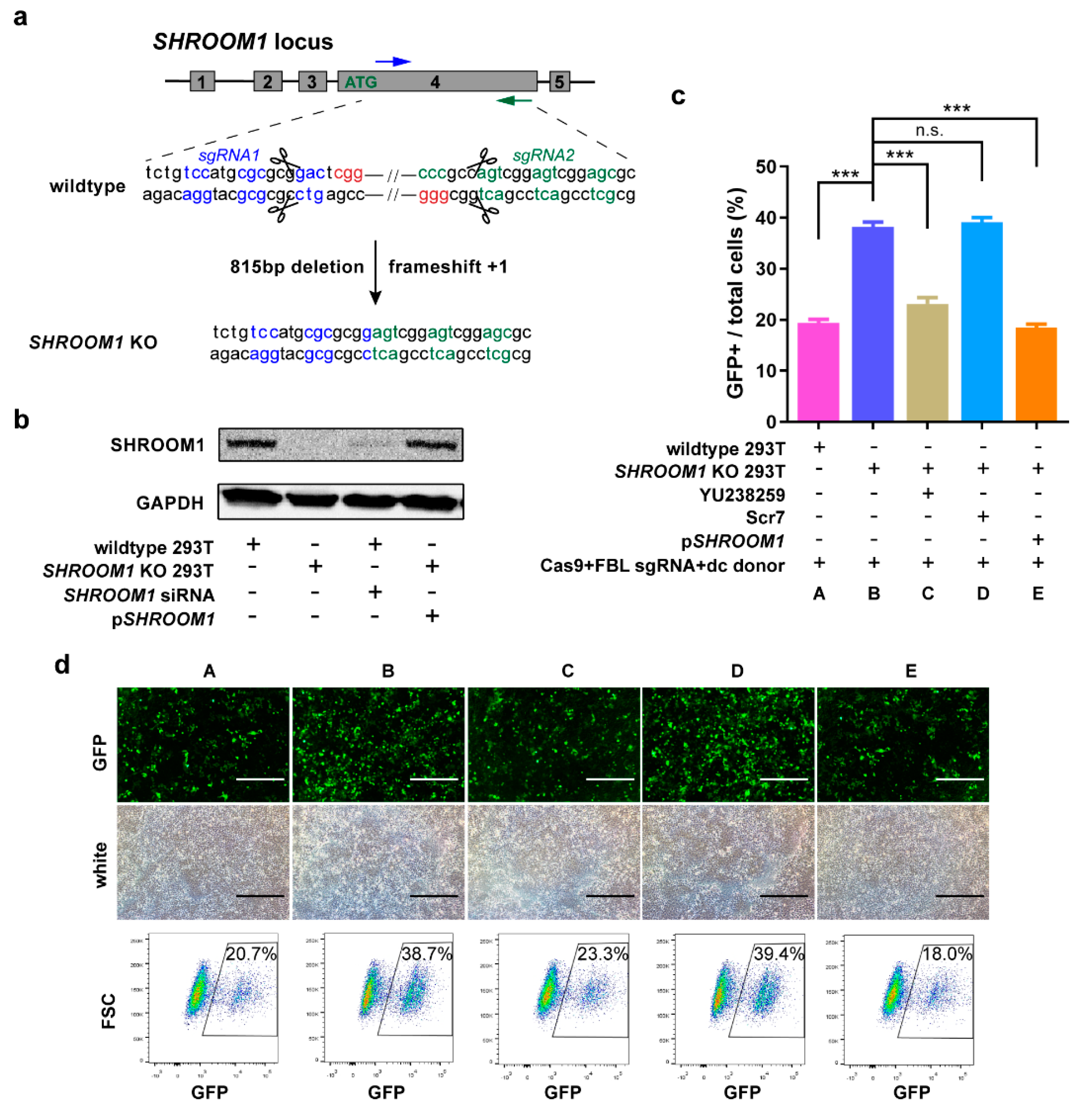
2.5. SHROOM1 Is a Potent Suppressor of HDR in Cells
3. Discussion
4. Methods
4.1. Construction of Plasmids
4.2. Cell Culture and Cell Lines
4.3. Genome-Wide Screening
4.4. Genomic DNA Extraction and PCR Amplification for Editing Region
4.5. Next-Generation Sequencing (NGS) Library Generation and Sequencing
4.6. Pooled Screen
4.7. Pooled Screen Analysis
4.8. qPCR
4.9. siRNA Interference and HDR Assays with Dual-Cut BFP Reporter
4.10. In Vitro Gene Knock-in
4.11. Western Blotting
4.12. Micro-Injection and Genotyping
4.13. Cell Cycle Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| siRNA | Small interfering RNA |
| BRCA2 | Breast Cancer Type 2 Susceptibility Protein |
| BRCA1 | Breast Cancer Type 1 Susceptibility Protein |
| RAD51 | DNA Repair Protein RAD51 Homolog 1 |
| SSTR3 | Somatostatin Receptor Type 3 |
| SLC36A3 | Proton-coupled Amino Acid Transporter 3 |
| CtIP | DNA endonuclease RBBP8 |
| SHROOM1 | Protein Shroom1 |
| UPF1 | Regulator of nonsense transcripts 1 |
| AAVS1 | Adeno-associated Virus Integration Site 1 |
| FBL | rRNA 2′-O-methyltransferase Fibrillarin |
| LMNA | Prelamin-A/C |
| Actin | Actin, cytoplasmic 1 |
| Ddx4 | ATP-dependent RNA Helicase DDX4 |
| Icos | Inducible T-cell Costimulator |
| GAPDH | Glyceraldehyde 3-Phosphate Dehydrogenase |
References
- Hockemeyer, D.; Soldner, F.; Beard, C.; Gao, Q.; Mitalipova, M.; DeKelver, R.C.; Katibah, G.E.; Amora, R.; Boydston, E.A.; Zeitler, B.; et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009, 27, 851–857. [Google Scholar] [CrossRef]
- Hockemeyer, D.; Wang, H.; Kiani, S.; Lai, C.S.; Gao, Q.; Cassady, J.P.; Cost, G.J.; Zhang, L.; Santiago, Y.; Miller, J.C.; et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011, 29, 731–734. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef]
- Porteus, M.H.; Baltimore, D. Chimeric nucleases stimulate gene targeting in human cells. Science 2003, 300, 763. [Google Scholar] [CrossRef]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.H.; Kim, J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Maresca, M.; Lin, V.G.; Guo, N.; Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): Custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013, 23, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Tsunekawa, Y.; Hernandez-Benitez, R.; Wu, J.; Zhu, J.; Kim, E.J.; Hatanaka, F.; Yamamoto, M.; Araoka, T.; Li, Z.; et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016, 540, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Nakade, S.; Tsubota, T.; Sakane, Y.; Kume, S.; Sakamoto, N.; Obara, M.; Daimon, T.; Sezutsu, H.; Yamamoto, T.; Sakuma, T.; et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014, 5, 5560. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.; Suresh, S.; Guo, D.; Daman, K.; Chen, J.C.J.; Liu, P.; Zieger, M.; Luk, K.; Roscoe, B.P.; Mueller, C.; et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 2019, 568, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.; Platt, R.J.; Zhang, F. Therapeutic genome editing: Prospects and challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef]
- Maruyama, T.; Dougan, S.K.; Truttmann, M.C.; Bilate, A.M.; Ingram, J.R.; Ploegh, H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015, 33, 538–542. [Google Scholar] [CrossRef]
- Gutierrez-Triana, J.A.; Tavhelidse, T.; Thumberger, T.; Thomas, I.; Wittbrodt, B.; Kellner, T.; Anlas, K.; Tsingos, E.; Wittbrodt, J. Efficient single-copy HDR by 5′ modified long dsDNA donors. eLife 2018, 7, e39468. [Google Scholar] [CrossRef]
- Yu, Y.; Guo, Y.; Tian, Q.; Lan, Y.; Yeh, H.; Zhang, M.; Tasan, I.; Jain, S.; Zhao, H. An efficient gene knock-in strategy using 5′-modified double-stranded DNA donors with short homology arms. Nat. Chem. Biol. 2020, 16, 387–390. [Google Scholar] [CrossRef]
- Ma, M.; Zhuang, F.; Hu, X.; Wang, B.; Wen, X.Z.; Ji, J.F.; Xi, J.J. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res. 2017, 27, 578–581. [Google Scholar] [CrossRef]
- Yao, X.; Wang, X.; Hu, X.; Liu, Z.; Liu, J.; Zhou, H.; Shen, X.; Wei, Y.; Huang, Z.; Ying, W.; et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017, 27, 801–814. [Google Scholar] [CrossRef]
- Zhang, J.P.; Li, X.L.; Li, G.H.; Chen, W.; Arakaki, C.; Botimer, G.D.; Baylink, D.; Zhang, L.; Wen, W.; Fu, Y.W.; et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017, 18, 35. [Google Scholar] [CrossRef] [PubMed]
- Vartak, S.V.; Raghavan, S.C. Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing. FEBS J. 2015, 282, 4289–4294. [Google Scholar] [CrossRef] [PubMed]
- Chu, V.T.; Weber, T.; Wefers, B.; Wurst, W.; Sander, S.; Rajewsky, K.; Kuhn, R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015, 33, 543–548. [Google Scholar] [CrossRef]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 2014, 3, e04766. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Ren, C.; Liu, Z.; Bai, Y.; Chen, Z.; Wei, Z.; Wang, X.; Zhang, Z.; Xu, K. Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int. J. Biochem. Cell Biol. 2017, 92, 43–52. [Google Scholar] [CrossRef]
- Di Primio, C.; Galli, A.; Cervelli, T.; Zoppe, M.; Rainaldi, G. Potentiation of gene targeting in human cells by expression of Saccharomyces cerevisiae Rad52. Nucleic Acids Res. 2005, 33, 4639–4648. [Google Scholar] [CrossRef]
- Nambiar, T.S.; Billon, P.; Diedenhofen, G.; Hayward, S.B.; Taglialatela, A.; Cai, K.; Huang, J.W.; Leuzzi, G.; Cuella-Martin, R.; Palacios, A.; et al. Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat. Commun. 2019, 10, 3395. [Google Scholar] [CrossRef]
- Charpentier, M.; Khedher, A.H.Y.; Menoret, S.; Brion, A.; Lamribet, K.; Dardillac, E.; Boix, C.; Perrouault, L.; Tesson, L.; Geny, S.; et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 2018, 9, 1133. [Google Scholar] [CrossRef]
- Pierce, A.J.; Johnson, R.D.; Thompson, L.H.; Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999, 13, 2633–2638. [Google Scholar] [CrossRef]
- Buchholz, D.R. Tet-On binary systems for tissue-specific and inducible transgene expression. Methods Mol. Biol. 2012, 917, 265–275. [Google Scholar] [CrossRef]
- Willis, N.A.; Chandramouly, G.; Huang, B.; Kwok, A.; Follonier, C.; Deng, C.; Scully, R. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature 2014, 510, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Steinfeld, J.B.; Liang, F.; Chen, X.; Maranon, D.G.; Jian Ma, C.; Kwon, Y.; Rao, T.; Wang, W.; Sheng, C.; et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 2017, 550, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.; Beach, A.; Li, K.; Haber, J. Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature 2017, 544, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Sartori, A.A.; Lukas, C.; Coates, J.; Mistrik, M.; Fu, S.; Bartek, J.; Baer, R.; Lukas, J.; Jackson, S.P. Human CtIP promotes DNA end resection. Nature 2007, 450, 509–514. [Google Scholar] [CrossRef]
- Yun, M.H.; Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 2009, 459, 460–463. [Google Scholar] [CrossRef]
- Richardson, C.D.; Kazane, K.R.; Feng, S.J.; Zelin, E.; Bray, N.L.; Schafer, A.J.; Floor, S.N.; Corn, J.E. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 2018, 50, 1132–1139. [Google Scholar] [CrossRef]
- Zhu, S.; Li, W.; Liu, J.; Chen, C.H.; Liao, Q.; Xu, P.; Xu, H.; Xiao, T.; Cao, Z.; Peng, J.; et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol. 2016, 34, 1279–1286. [Google Scholar] [CrossRef]
- Lee, C.; Le, M.P.; Wallingford, J.B. The shroom family proteins play broad roles in the morphogenesis of thickened epithelial sheets. Dev. Dyn. 2009, 238, 1480–1491. [Google Scholar] [CrossRef]
- Lee, C.; Scherr, H.M.; Wallingford, J.B. Shroom family proteins regulate gamma-tubulin distribution and microtubule architecture during epithelial cell shape change. Development 2007, 134, 1431–1441. [Google Scholar] [CrossRef]
- Hildebrand, J.D.; Soriano, P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 1999, 99, 485–497. [Google Scholar] [CrossRef]
- Zalewski, J.K.; Mo, J.H.; Heber, S.; Heroux, A.; Gardner, R.G.; Hildebrand, J.D.; VanDemark, A.P. Structure of the Shroom-Rho Kinase Complex Reveals a Binding Interface with Monomeric Shroom That Regulates Cell Morphology and Stimulates Kinase Activity. J. Biol. Chem. 2016, 291, 25364–25374. [Google Scholar] [CrossRef] [PubMed]
- Thwaites, D.T.; Anderson, C.M. The SLC36 family of proton-coupled amino acid transporters and their potential role in drug transport. Br. J. Pharmacol. 2011, 164, 1802–1816. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, J.R., Jr.; Pennington, J. Organization and expression of the SLC36 cluster of amino acid transporter genes. Mamm. Genome 2004, 15, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Stachelek, G.C.; Peterson-Roth, E.; Liu, Y.; Fernandez, R.J., 3rd; Pike, L.R.; Qian, J.M.; Abriola, L.; Hoyer, D.; Hungerford, W.; Merkel, J.; et al. YU238259 Is a Novel Inhibitor of Homology-Dependent DNA Repair That Exhibits Synthetic Lethality and Radiosensitization in Repair-Deficient Tumors. Mol. Cancer Res. 2015, 13, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Beucher, A.; Birraux, J.; Tchouandong, L.; Barton, O.; Shibata, A.; Conrad, S.; Goodarzi, A.A.; Krempler, A.; Jeggo, P.A.; Lobrich, M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009, 28, 3413–3427. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Zhang, H.; Xiong, T.; Wang, J.; Yang, D.; Zhu, D.; Li, J.; Yang, Y.; Sun, C.; Zhao, Y.; et al. Suppression of SHROOM1 Improves In Vitro and In Vivo Gene Integration by Promoting Homology-Directed Repair. Int. J. Mol. Sci. 2020, 21, 5821. https://doi.org/10.3390/ijms21165821
Zhao Z, Zhang H, Xiong T, Wang J, Yang D, Zhu D, Li J, Yang Y, Sun C, Zhao Y, et al. Suppression of SHROOM1 Improves In Vitro and In Vivo Gene Integration by Promoting Homology-Directed Repair. International Journal of Molecular Sciences. 2020; 21(16):5821. https://doi.org/10.3390/ijms21165821
Chicago/Turabian StyleZhao, Zhihua, Hanshuo Zhang, Tuanlin Xiong, Junyi Wang, Di Yang, Dan Zhu, Juan Li, Ye Yang, Changhong Sun, Yuting Zhao, and et al. 2020. "Suppression of SHROOM1 Improves In Vitro and In Vivo Gene Integration by Promoting Homology-Directed Repair" International Journal of Molecular Sciences 21, no. 16: 5821. https://doi.org/10.3390/ijms21165821
APA StyleZhao, Z., Zhang, H., Xiong, T., Wang, J., Yang, D., Zhu, D., Li, J., Yang, Y., Sun, C., Zhao, Y., & Xi, J. J. (2020). Suppression of SHROOM1 Improves In Vitro and In Vivo Gene Integration by Promoting Homology-Directed Repair. International Journal of Molecular Sciences, 21(16), 5821. https://doi.org/10.3390/ijms21165821






