Collagen 24 α1 Is Increased in Insulin-Resistant Skeletal Muscle and Adipose Tissue
Abstract
1. Introduction
2. Results
2.1. In Vivo Quantitative Proteomics of the ECM Compartment
2.2. Col24α1 mRNA Was Increased in Skeletal Muscle of HF-Fed Mice
2.3. Col24α1 mRNA Was Increased in White Adipose Tissue, but Not in Liver of HF-Fed Mice
2.4. Col24α1 mRNA Was Increased in Visceral Adipose Tissue of Obese Diabetic Human Subjects
3. Discussion
4. Materials and Methods
4.1. Animal Models
4.2. ECM-Specific Mass Spectrometry-Based Proteomics
4.2.1. Isolation of the ECM
4.2.2. iTRAQ Labelling Peptide Analysis by LC-MS/MS
4.3. Gene-Annotation Enrichment Analysis and KEGG Pathway Mapping
4.4. String Network Analysis
4.5. Mouse Tissue Collection and Preparation
4.6. Human Sample Collection and Preparation
4.7. Measurement of Gene Expression by Real-Time PCR
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Akt | Protein kinase B |
| BCA | Bicinchoninic acid |
| ECM | Extracellular matrix |
| HF | High fat |
| iTRAQ | Isobaric tags for relative and absolute quantitation |
| KEGG | Kyoto Encyclopaedia of Genes and Genomes |
| LC-MS | Liquid chromatography-mass spectrometry |
| MMPs | Matrix metalloproteinases |
| MMP9 | Matrix metalloproteinases 9 |
| MSPI3K | Mass spectrometry |
| PI3K | Phosphoinositide 3-kinase |
| SAT | Subcutaneous adipose tissue |
| SDS | Sodium dodecyl sulfate |
| 2-DE | Two-dimensional gel electrophoresis |
| VAT | Visceral adipose tissue |
| WAT | White adipose tissue |
References
- Williams, A.S.; Kang, L.; Wasserman, D.H. The extracellular matrix and insulin resistance. Trends Endocrinol. Metab. 2015, 26, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, K.; Choi, I.; Lee, Y.H. Implications of Skeletal Muscle Extracellular Matrix Remodeling in Metabolic Disorders: Diabetes Perspective. Int. J. Mol. Sci. 2020, 21, 3845. [Google Scholar] [CrossRef] [PubMed]
- Aumailley, M.; Gayraud, B. Structure and biological activity of the extracellular matrix. J. Mol. Med. 1998, 76, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Ayala, J.E.; Lee-Young, R.S.; Zhang, Z.; James, F.D.; Neufer, P.D.; Pozzi, A.; Zutter, M.M.; Wasserman, D.H. Diet-induced muscle insulin resistance is associated with extracellular matrix remodeling and interaction with integrin alpha2beta1 in mice. Diabetes 2011, 60, 416–426. [Google Scholar] [CrossRef]
- Kang, L.; Lantier, L.; Kennedy, A.; Bonner, J.S.; Mayes, W.H.; Bracy, D.P.; Bookbinder, L.H.; Hasty, A.H.; Thompson, C.B.; Wasserman, D.H. Hyaluronan accumulates with high-fat feeding and contributes to insulin resistance. Diabetes 2013, 62, 1888–1896. [Google Scholar] [CrossRef]
- Hasib, A.; Hennayake, C.K.; Bracy, D.P.; Bugler-Lamb, A.R.; Lantier, L.; Khan, F.; Ashford, M.L.J.; McCrimmon, R.J.; Wasserman, D.H.; Kang, L. CD44 contributes to hyaluronan-mediated insulin resistance in skeletal muscle of high-fat-fed C57BL/6 mice. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E973–E983. [Google Scholar] [CrossRef]
- Pasarica, M.; Gowronska-Kozak, B.; Burk, D.; Remedios, I.; Hymel, D.; Gimble, J.; Ravussin, E.; Bray, G.A.; Smith, S.R. Adipose tissue collagen VI in obesity. J. Clin. Endocrinol. Metab. 2009, 94, 5155–5162. [Google Scholar] [CrossRef]
- Nomiyama, T.; Perez-Tilve, D.; Ogawa, D.; Gizard, F.; Zhao, Y.; Heywood, E.B.; Jones, K.L.; Kawamori, R.; Cassis, L.A.; Tschöp, M.H.; et al. Osteopontin mediates obesity-induced adipose tissue macrophage infiltration and insulin resistance in mice. J. Clin. Investig. 2007, 117, 2877–2888. [Google Scholar] [CrossRef]
- Williams, A.S.; Trefts, E.; Lantier, L.; Grueter, C.A.; Bracy, D.P.; James, F.D.; Pozzi, A.; Zent, R.; Wasserman, D.H. Integrin-Linked Kinase Is Necessary for the Development of Diet-Induced Hepatic Insulin Resistance. Diabetes 2017, 66, 325–334. [Google Scholar] [CrossRef]
- Schmid, G.M.; Converset, V.; Walter, N.; Sennitt, M.V.; Leung, K.Y.; Byers, H.; Ward, M.; Hochstrasser, D.F.; Cawthorne, M.A.; Sanchez, J.C. Effect of high-fat diet on the expression of proteins in muscle, adipose tissues, and liver of C57BL/6 mice. Proteomics 2004, 4, 2270–2282. [Google Scholar] [CrossRef]
- Szalowska, E.; Dijkstra, M.; Elferink, M.G.; Weening, D.; de Vries, M.; Bruinenberg, M.; Hoek, A.; Roelofsen, H.; Groothuis, G.M.; Vonk, R.J. Comparative analysis of the human hepatic and adipose tissue transcriptomes during LPS-induced inflammation leads to the identification of differential biological pathways and candidate biomarkers. BMC Med. Genom. 2011, 4, 71. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Eraslan, B.; Wieland, T.; Hallström, B.; Hopf, T.; Zolg, D.P.; Zecha, J.; Asplund, A.; Li, L.H.; Meng, C.; et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 2019, 15, e8503. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.H.; Batchelder, C.A.; Lee, C.I.; Tarantal, A.F. Decellularized rhesus monkey kidney as a three-dimensional scaffold for renal tissue engineering. Tissue Eng. Part A 2010, 16, 2207–2216. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Chun, T.H.; Kang, L. Adipose extracellular matrix remodelling in obesity and insulin resistance. Biochem. Pharmacol. 2016, 119, 8–16. [Google Scholar] [CrossRef]
- Wang, W.; Olson, D.; Liang, G.; Franceschi, R.T.; Li, C.; Wang, B.; Wang, S.S.; Yang, S. Collagen XXIV (Col24α1) promotes osteoblastic differentiation and mineralization through TGF-β/Smads signaling pathway. Int. J. Biol. Sci. 2012, 8, 1310–1322. [Google Scholar] [CrossRef]
- Matsuo, N.; Tanaka, S.; Yoshioka, H.; Koch, M.; Gordon, M.K.; Ramirez, F. Collagen XXIV (Col24a1) gene expression is a specific marker of osteoblast differentiation and bone formation. Connect. Tissue Res. 2008, 49, 68–75. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Xiao, Z.; Blonder, J.; Zhou, M.; Veenstra, T.D. Proteomic analysis of extracellular matrix and vesicles. J. Proteom. 2009, 72, 34–45. [Google Scholar] [CrossRef]
- Inoue, M.; Jiang, Y.; Barnes, R.H., 2nd; Tokunaga, M.; Martinez-Santibañez, G.; Geletka, L.; Lumeng, C.N.; Buchner, D.A.; Chun, T.H. Thrombospondin 1 mediates high-fat diet-induced muscle fibrosis and insulin resistance in male mice. Endocrinology 2013, 154, 4548–4559. [Google Scholar] [CrossRef]
- Pincu, Y.; Linden, M.A.; Zou, K.; Baynard, T.; Boppart, M.D. The effects of high fat diet and moderate exercise on TGFβ1 and collagen deposition in mouse skeletal muscle. Cytokine 2015, 73, 23–29. [Google Scholar] [CrossRef]
- Sabatelli, P.; Gualandi, F.; Gara, S.K.; Grumati, P.; Zamparelli, A.; Martoni, E.; Pellegrini, C.; Merlini, L.; Ferlini, A.; Bonaldo, P.; et al. Expression of collagen VI α5 and α6 chains in human muscle and in Duchenne muscular dystrophy-related muscle fibrosis. Matrix. Biol. 2012, 31, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Berria, R.; Wang, L.; Richardson, D.K.; Finlayson, J.; Belfort, R.; Pratipanawatr, T.; De Filippis, E.A.; Kashyap, S.; Mandarino, L.J. Increased collagen content in insulin-resistant skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E560–E565. [Google Scholar] [CrossRef] [PubMed]
- Arriazu, E.; Ruiz de Galarreta, M.; Cubero, F.J.; Varela-Rey, M.; Pérez de Obanos, M.P.; Leung, T.M.; Lopategi, A.; Benedicto, A.; Abraham-Enachescu, I.; Nieto, N. Extracellular matrix and liver disease. Antioxid. Redox Signal. 2014, 21, 1078–1097. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.K.; Kashyap, S.; Bajaj, M.; Cusi, K.; Mandarino, S.J.; Finlayson, J.; DeFronzo, R.A.; Jenkinson, C.P.; Mandarino, L.J. Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases the expression of extracellular matrix genes in human skeletal muscle. J. Biol. Chem. 2005, 280, 10290–10297. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Mayes, W.H.; James, F.D.; Bracy, D.P.; Wasserman, D.H. Matrix metalloproteinase 9 opposes diet-induced muscle insulin resistance in mice. Diabetologia 2014, 57, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.S.; Kim, Y.J.; Kwon, E.Y.; Ryoo, J.Y.; Kim, S.R.; Jung, U.J. High-fat diet decreases energy expenditure and expression of genes controlling lipid metabolism, mitochondrial function and skeletal system development in the adipose tissue, along with increased expression of extracellular matrix remodelling- and inflammation-related genes. Br. J. Nutr. 2015, 113, 867–877. [Google Scholar] [PubMed]
- Spencer, M.; Yao-Borengasser, A.; Unal, R.; Rasouli, N.; Gurley, C.M.; Zhu, B.; Peterson, C.A.; Kern, P.A. Adipose tissue macrophages in insulin-resistant subjects are associated with collagen VI and fibrosis and demonstrate alternative activation. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E1016–E1027. [Google Scholar] [CrossRef]
- Petäistö, T.; Vicente, D.; Mäkelä, K.A.; Finnilä, M.A.; Miinalainen, I.; Koivunen, J.; Izzi, V.; Aikio, M.; Karppinen, S.M.; Devarajan, R.; et al. Lack of collagen XVIII leads to lipodystrophy and perturbs hepatic glucose and lipid homeostasis. J. Physiol. 2020. [Google Scholar] [CrossRef]
- Khan, T.; Muise, E.S.; Iyengar, P.; Wang, Z.V.; Chandalia, M.; Abate, N.; Zhang, B.B.; Bonaldo, P.; Chua, S.; Scherer, P.E. Metabolic dysregulation and adipose tissue fibrosis: Role of collagen VI. Mol. Cell Biol. 2009, 29, 1575–1591. [Google Scholar] [CrossRef]
- Mori, S.; Kiuchi, S.; Ouchi, A.; Hase, T.; Murase, T. Characteristic expression of extracellular matrix in subcutaneous adipose tissue development and adipogenesis; comparison with visceral adipose tissue. Int. J. Biol. Sci. 2014, 10, 825–833. [Google Scholar] [CrossRef]
- Jeffery, E.; Wing, A.; Holtrup, B.; Sebo, Z.; Kaplan, J.L.; Saavedra-Peña, R.; Church, C.D.; Colman, L.; Berry, R.; Rodeheffer, M.S. The Adipose Tissue Microenvironment Regulates Depot-Specific Adipogenesis in Obesity. Cell Metab. 2016, 24, 142–150. [Google Scholar] [CrossRef] [PubMed]
- White, U.A.; Tchoukalova, Y.D. Sex dimorphism and depot differences in adipose tissue function. Biochim. Biophys. Acta 2014, 1842, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Strieder-Barboza, C.; Baker, N.A.; Flesher, C.G.; Karmakar, M.; Patel, A.; Lumeng, C.N.; O’Rourke, R.W. Depot-specific adipocyte-extracellular matrix metabolic crosstalk in murine obesity. Adipocyte 2020, 9, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.K.; Hahn, R.A. Collagens. Cell Tissue Res. 2010, 339, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Askenazi, M.; Jiang, J.; Luckey, C.J.; Griffin, J.D.; Marto, J.A. A robust error model for iTRAQ quantification reveals divergent signaling between oncogenic FLT3 mutants in acute myeloid leukemia. Mol. Cell Proteom. 2010, 9, 780–790. [Google Scholar] [CrossRef]
- Kim, H.J.; Lin, D.; Lee, H.J.; Li, M.; Liebler, D.C. Quantitative Profiling of Protein Tyrosine Kinases in Human Cancer Cell Lines by Multiplexed Parallel Reaction Monitoring Assays. Mol. Cell Proteom. 2016, 15, 682–691. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- da Huang, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
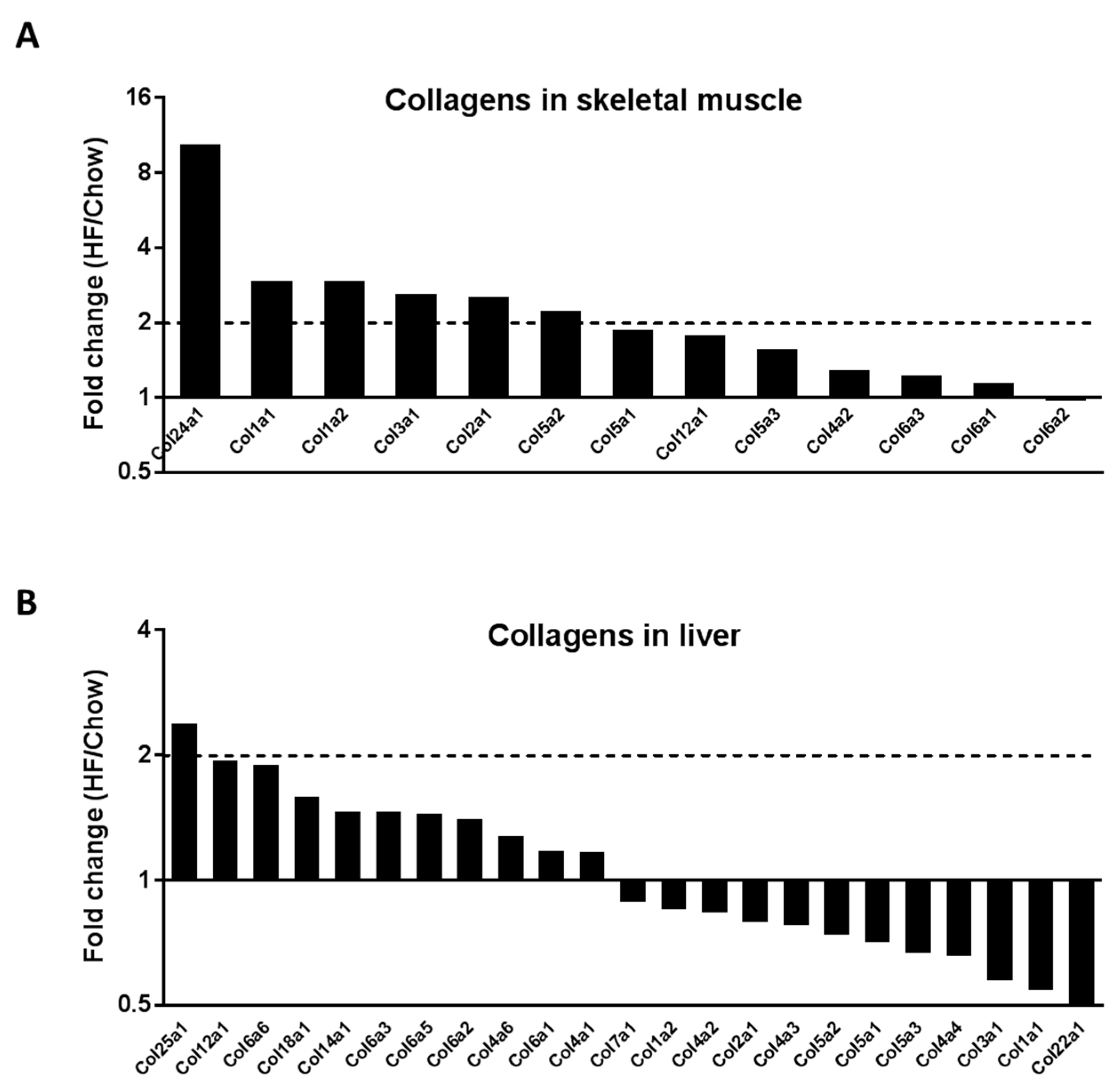
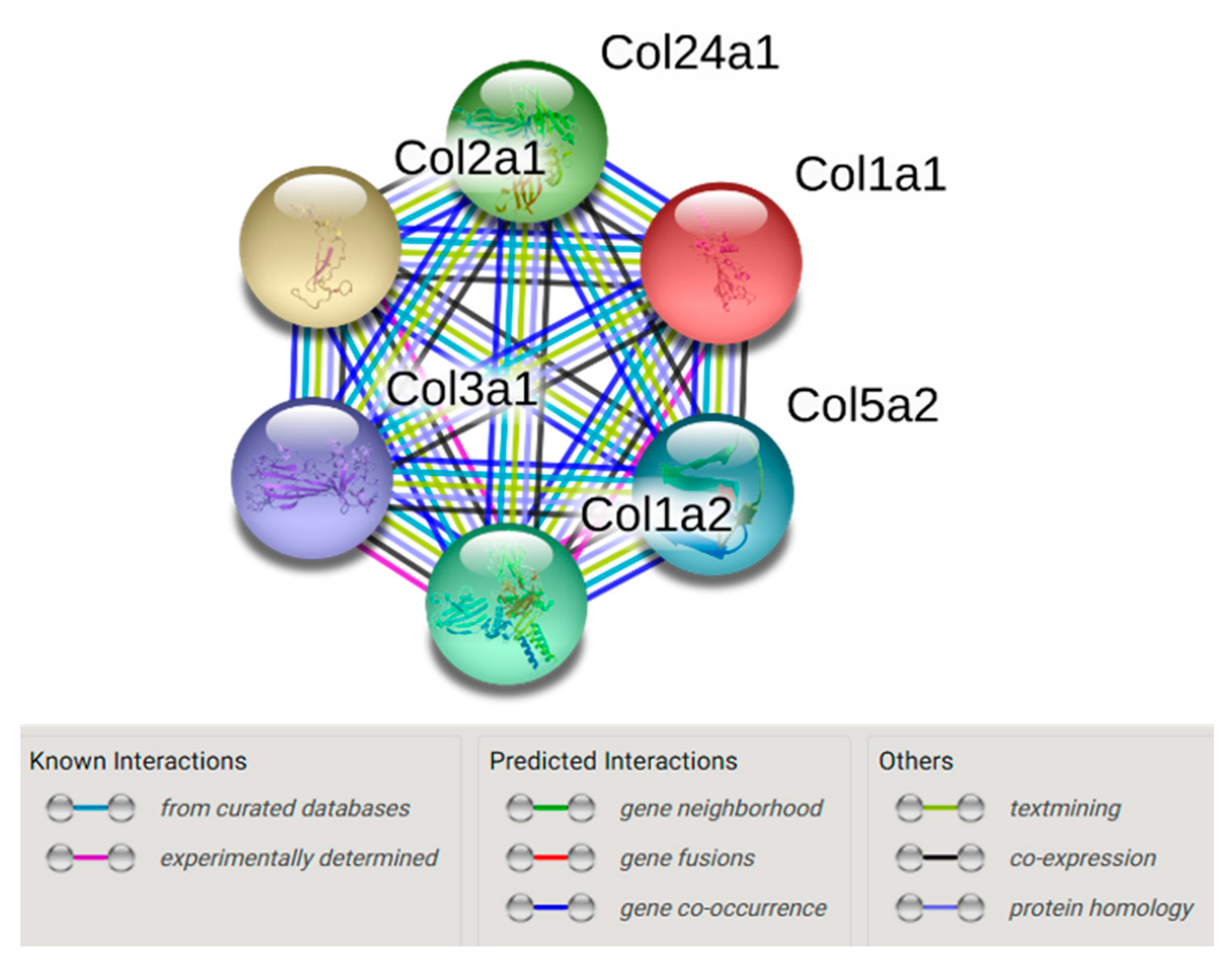

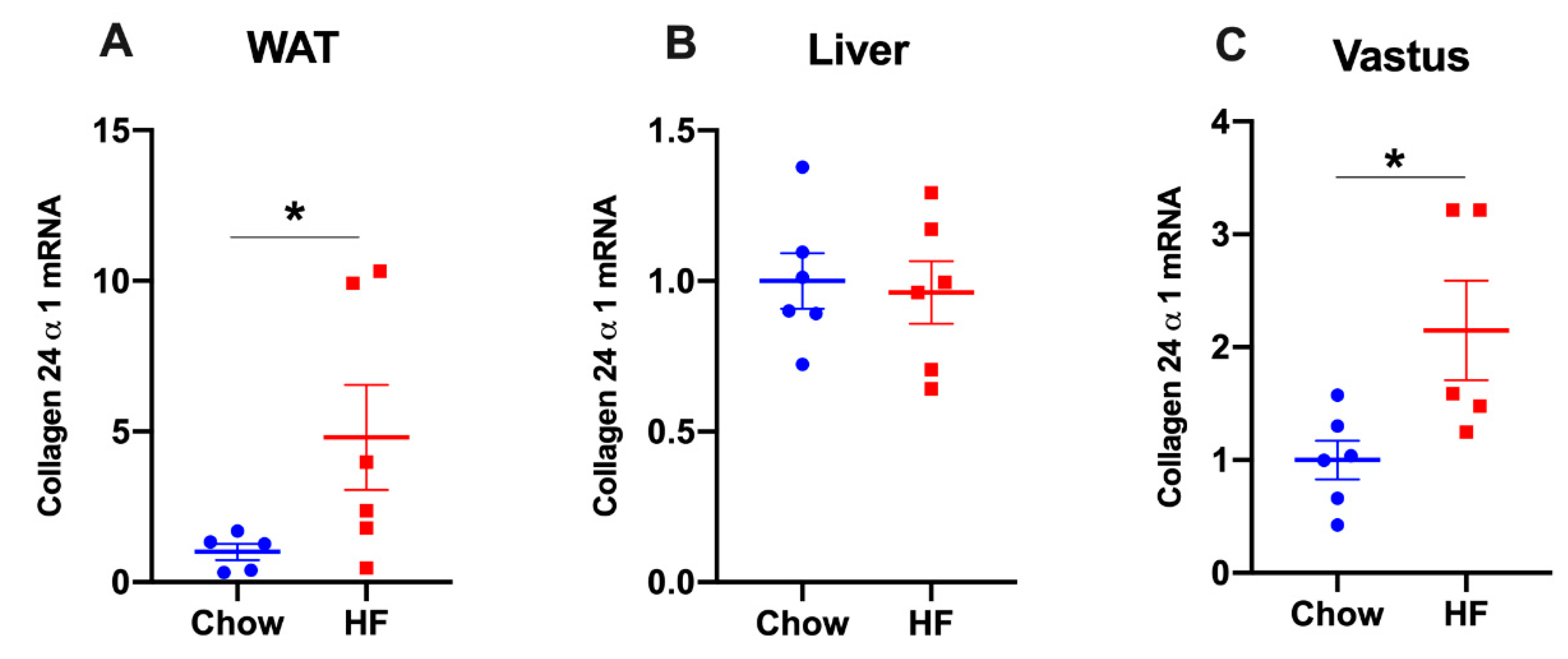

| Skeletal Muscle | ||||||
| Category | Term | Count | % | Genes | pValue | Benjamini |
| KEGG_PATHWAY | Protein digestion and absorption | 6 | 54.5 | COL3A1, COL1A2, COL2A1, COL1A1, COL24A1, COL5A2 | 1.75 × 10−10 | 1.22 × 10−9 |
| KEGG_PATHWAY | ECM–receptor interaction | 6 | 54.5 | COL3A1, COL1A2, COL2A1, COL1A1, COL24A1, COL5A2 | 1.75 × 10−10 | 1.22 × 10−9 |
| KEGG_PATHWAY | Amoebiasis | 6 | 54.5 | COL3A1, COL1A2, COL2A1, COL1A1, COL24A1, COL5A2 | 7.48 × 10−10 | 2.62 × 10−9 |
| KEGG_PATHWAY | Platelet activation | 6 | 54.5 | COL3A1, COL1A2, COL2A1, COL1A1, COL24A1, COL5A2 | 1.33 × 10−9 | 3.10 × 10−9 |
| KEGG_PATHWAY | Focal adhesion | 6 | 54.5 | COL3A1, COL1A2, COL2A1, COL1A1, COL24A1, COL5A2 | 1.35 × 10−8 | 2.36 × 10−8 |
| KEGG_PATHWAY | PI3K-Akt signalling pathway | 6 | 54.5 | COL3A1, COL1A2, COL2A1, COL1A1, COL24A1, COL5A2 | 1.93 × 10−7 | 2.70 × 10−7 |
| Liver | ||||||
| Category | Term | Count | % | Genes | pValue | Benjamini |
| KEGG_PATHWAY | African trypanosomias-is | 4 | 7.0 | HBA-A1, HBA-A2, APOA1, HBB-B2 | 7.60 × 10−4 | 0.058 |
| KEGG_PATHWAY | Biosynthesis of antibiotics | 6 | 10.5 | LDHA, ALDOB, FBP1, ALDH2, HADH, ACAT1 | 0.005 | 0.171 |
| KEGG_PATHWAY | Glycolysis/Gluconeogenesis | 4 | 7.0 | LDHA, ALDOB, FBP1, ALDH2 | 0.005 | 0.118 |
| KEGG_PATHWAY | Butanoate metabolism | 3 | 5.3 | HADH, ACAT1, BDH1 | 0.009 | 0.156 |
| KEGG_PATHWAY | Metabolic pathways | 14 | 24.6 | LDHA, SORD, GLUD1, ALDOB, FBP1, ACAT1, LAP3, ALDH1A1, PRDX6, BHMT, ALDH2, ATP5A1, HADH, BDH1 | 0.010 | 0.141 |
| KEGG_PATHWAY | Gap junction | 4 | 7.0 | TUBB5, TUBA4A, TUBA1A, TUBA1C | 0.010 | 0.123 |
| KEGG_PATHWAY | Fructose and mannose metabolism | 3 | 5.3 | SORD, ALDOB, FBP1 | 0.013 | 0.140 |
| KEGG_PATHWAY | Pyruvate metabolism | 3 | 5.3 | LDHA, ALDH2, ACAT1 | 0.017 | 0.158 |
| KEGG_PATHWAY | Carbon metabolism | 4 | 7.0 | GLUD1, ALDOB, FBP1, ACAT1 | 0.022 | 0.178 |
| KEGG_PATHWAY | Tryptophan metabolism | 3 | 5.3 | ALDH2, HADH, ACAT1 | 0.025 | 0.179 |
| KEGG_PATHWAY | Malaria | 3 | 5.3 | HBA-A1, HBA-A2, HBB-B2 | 0.026 | 0.170 |
| KEGG_PATHWAY | Fatty acid degradation | 3 | 5.3 | ALDH2, HADH, ACAT1 | 0.027 | 0.163 |
| KEGG_PATHWAY | Lysine degradation | 3 | 5.3 | ALDH2, HADH, ACAT1 | 0.030 | 0.167 |
| KEGG_PATHWAY | Valine, leucine and isoleucine degradation | 3 | 5.3 | ALDH2, HADH, ACAT1 | 0.033 | 0.172 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weng, X.; Lin, D.; Huang, J.T.J.; Stimson, R.H.; Wasserman, D.H.; Kang, L. Collagen 24 α1 Is Increased in Insulin-Resistant Skeletal Muscle and Adipose Tissue. Int. J. Mol. Sci. 2020, 21, 5738. https://doi.org/10.3390/ijms21165738
Weng X, Lin D, Huang JTJ, Stimson RH, Wasserman DH, Kang L. Collagen 24 α1 Is Increased in Insulin-Resistant Skeletal Muscle and Adipose Tissue. International Journal of Molecular Sciences. 2020; 21(16):5738. https://doi.org/10.3390/ijms21165738
Chicago/Turabian StyleWeng, Xiong, De Lin, Jeffrey T. J. Huang, Roland H. Stimson, David H. Wasserman, and Li Kang. 2020. "Collagen 24 α1 Is Increased in Insulin-Resistant Skeletal Muscle and Adipose Tissue" International Journal of Molecular Sciences 21, no. 16: 5738. https://doi.org/10.3390/ijms21165738
APA StyleWeng, X., Lin, D., Huang, J. T. J., Stimson, R. H., Wasserman, D. H., & Kang, L. (2020). Collagen 24 α1 Is Increased in Insulin-Resistant Skeletal Muscle and Adipose Tissue. International Journal of Molecular Sciences, 21(16), 5738. https://doi.org/10.3390/ijms21165738





