SUMOylation-Mediated Response to Mitochondrial Stress
Abstract
1. Introduction
1.1. Mitochondrial Stress Response
1.2. SUMOylation is a Stress-Induced Process
2. Roles of SUMOylation in Mitochondrial Biogenesis
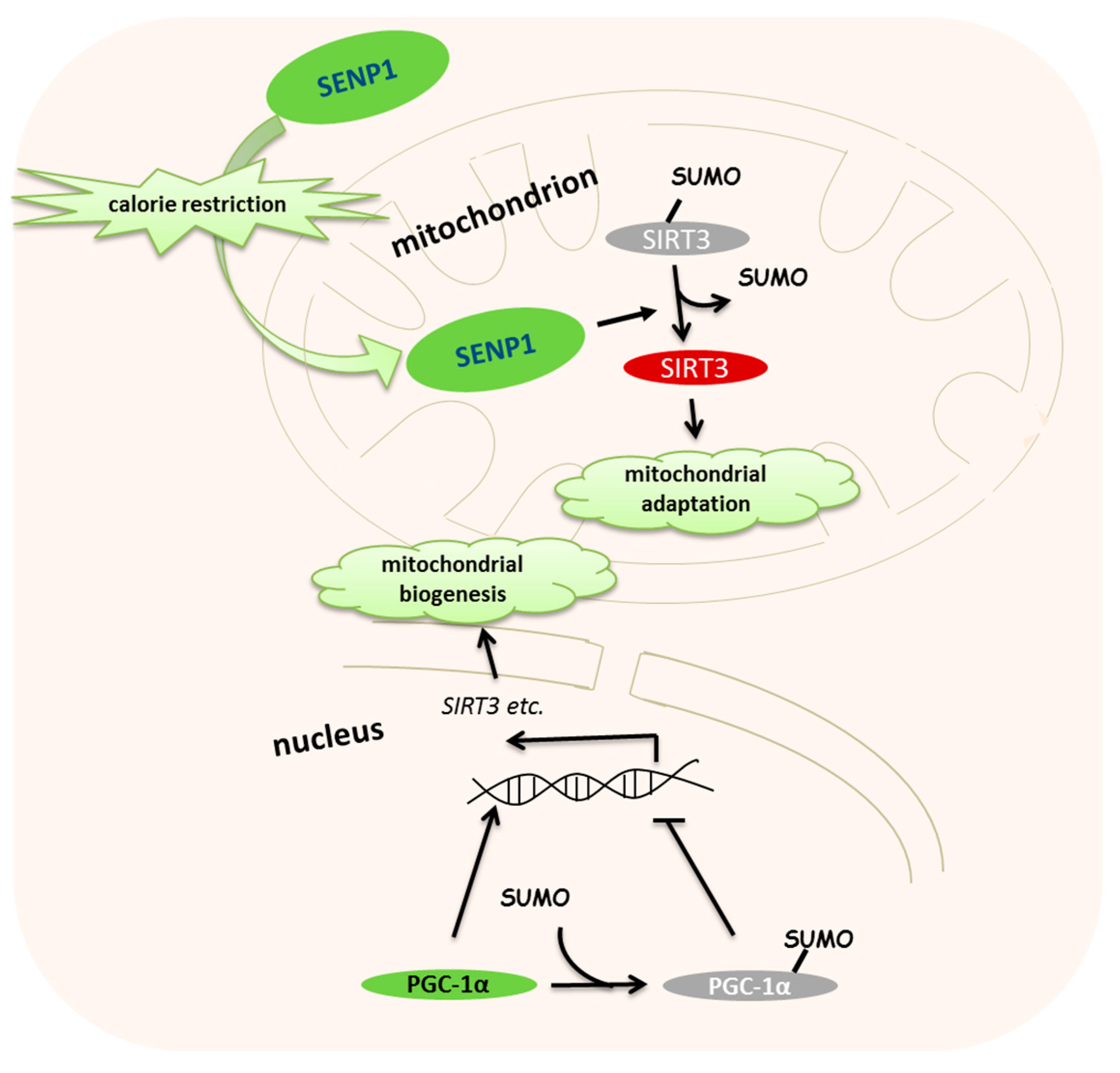
3. Roles of SUMOylation in Calorie Restriction-Induced Mitochondrial Metabolism
4. Roles of SUMOylation in Oxygen-Induced Mitochondrial Dysfunction
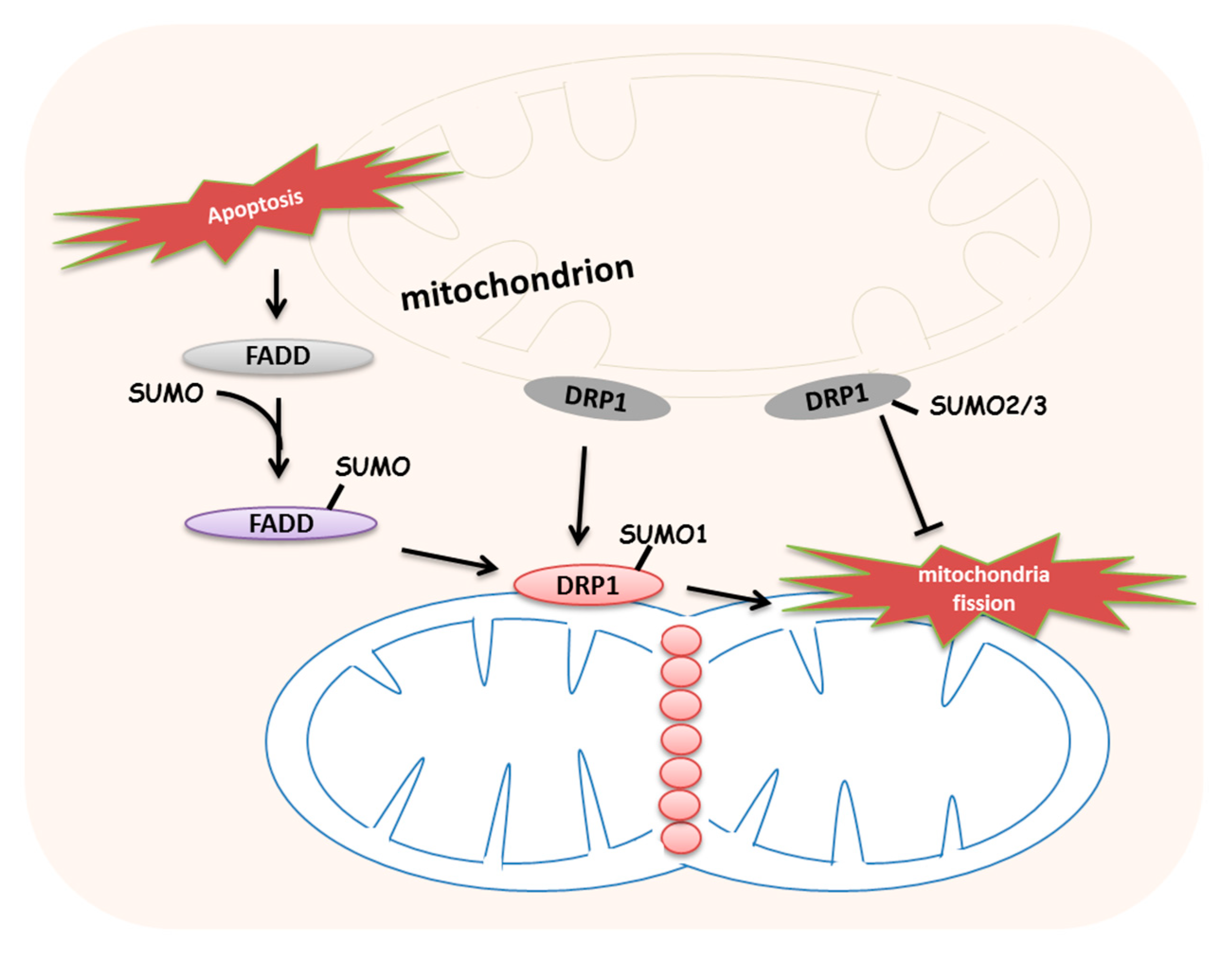
5. Roles of SUMOylation in Stress-Induced Mitochondrial Dynamics and Mitophagy
6. Roles of SUMOylation in Mitochondrial Stress-Induced UPRmt
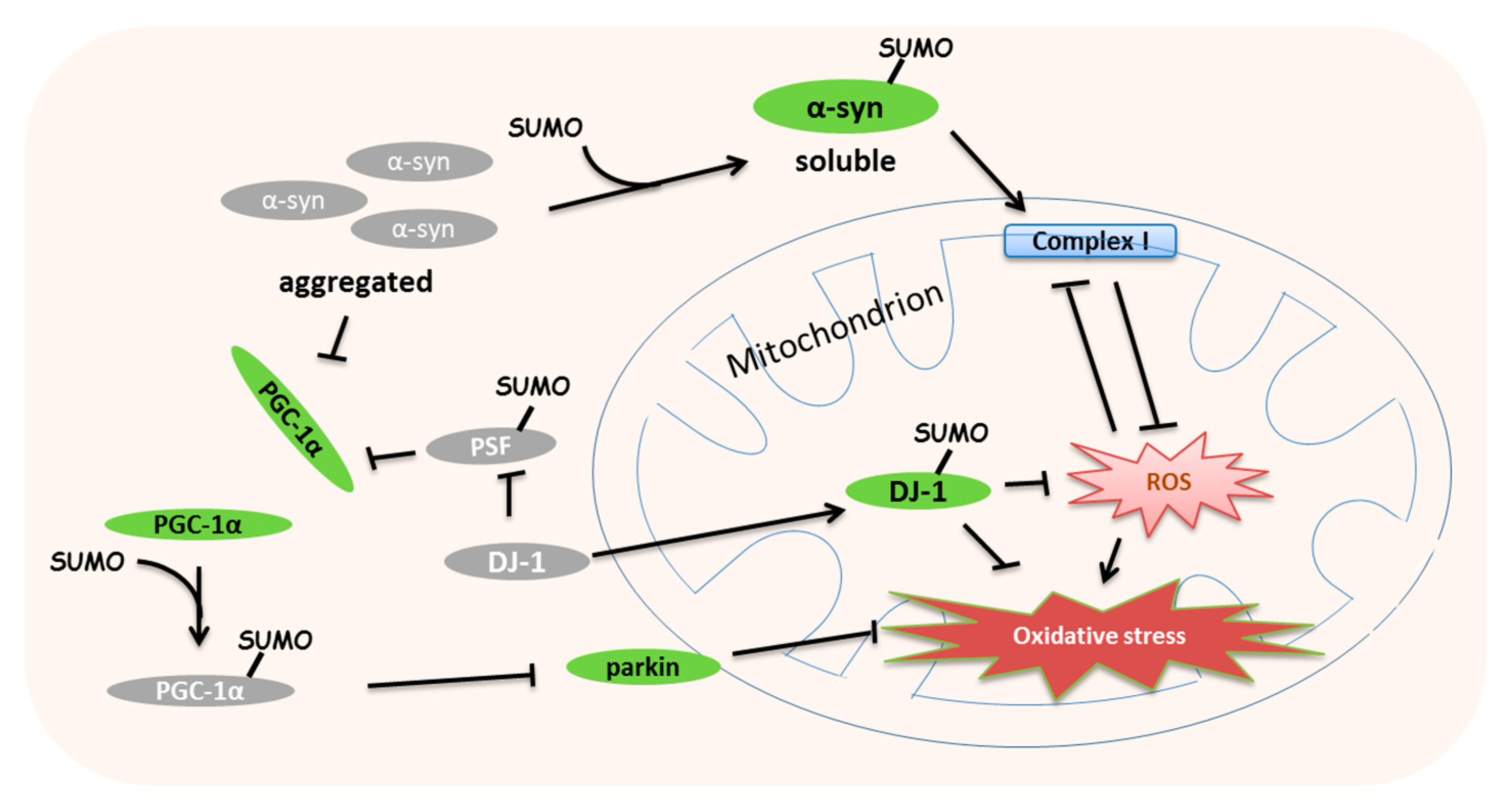
7. Roles of SUMOylation in Mitochondrial Stress-Induced Diseases
8. Conclusions and Further Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Fedorenko, A.; Lishko, P.V.; Kirichok, Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 2012, 151, 400–413. [Google Scholar] [CrossRef] [PubMed]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Bonkowski, M.S.; Sinclair, D.A. Slowing ageing by design: The rise of NAD(+) and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 2016, 17, 679–690. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Detappe, A.; Cai, K.; Keys, H.R.; Brune, Z.; Ying, W.; Thiru, P.; Reidy, M.; Kugener, G.; Rossen, J.; et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat. Chem. Biol. 2019, 15, 681–689. [Google Scholar] [CrossRef]
- Forsstrom, S.; Jackson, C.B.; Carroll, C.J.; Kuronen, M.; Pirinen, E.; Pradhan, S.; Marmyleva, A.; Auranen, M.; Kleine, I.M.; Khan, N.A.; et al. Fibroblast Growth Factor 21 Drives Dynamics of Local and Systemic Stress Responses in Mitochondrial Myopathy with mtDNA Deletions. Cell Metab. 2019, 30, 1040–1054. [Google Scholar] [CrossRef]
- Pla-Martin, D.; Schatton, D.; Wiederstein, J.L.; Marx, M.C.; Khiati, S.; Kruger, M.; Rugarli, E.I. CLUH granules coordinate translation of mitochondrial proteins with mTORC1 signaling and mitophagy. Embo. J. 2020, 39, e102731. [Google Scholar]
- Jain, I.H.; Calvo, S.E.; Markhard, A.L.; Skinner, O.S.; To, T.L.; Ast, T.; Mootha, V.K. Genetic Screen for Cell Fitness in High or Low Oxygen Highlights Mitochondrial and Lipid Metabolism. Cell 2020, 181, 716–727. [Google Scholar]
- Liesa, M.; Shirihai, O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013, 17, 491–506. [Google Scholar]
- Goodpaster, B.H.; Sparks, L.M. Metabolic Flexibility in Health and Disease. Cell Metab. 2017, 25, 1027–1036. [Google Scholar]
- Meluh, P.B.; Koshland, D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 1995, 6, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Wang, C.L.; Hsiao, P.J.; Lu, Y.C.; Chen, S.Y.; Lin, K.D.; Hsin, S.C.; Hsieh, M.C.; Shin, S.J. SUMO4 M55V variant is associated with diabetic nephropathy in type 2 diabetes. Diabetes 2007, 56, 1177–1180. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Tatham, M.H.; Jaffray, E.; Vaughan, O.A.; Desterro, J.M.; Botting, C.H.; Naismith, J.H.; Hay, R.T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001, 276, 35368–35374. [Google Scholar] [CrossRef]
- Saitoh, H.; Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000, 275, 6252–6258. [Google Scholar] [CrossRef]
- Zhou, W.; Ryan, J.J.; Zhou, H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 2004, 279, 32262–32268. [Google Scholar] [CrossRef]
- Castro, P.H.; Tavares, R.M.; Bejarano, E.R.; Azevedo, H. SUMO, a heavyweight player in plant abiotic stress responses. Cell Mol. Life Sci. 2012, 69, 3269–3283. [Google Scholar] [CrossRef]
- Enserink, J.M. Sumo and the cellular stress response. Cell Div. 2015, 10, 4. [Google Scholar] [CrossRef]
- Hickey, C.M.; Wilson, N.R.; Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012, 13, 755–766. [Google Scholar] [CrossRef]
- Braschi, E.; Zunino, R.; McBride, H.M. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. Embo. Rep. 2009, 10, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Prudent, J.; Zunino, R.; Sugiura, A.; Mattie, S.; Shore, G.C.; McBride, H.M. MAPL SUMOylation of Drp1 Stabilizes an ER/Mitochondrial Platform Required for Cell Death. Mol. Cell 2015, 59, 941–955. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.T. SUMO: A history of modification. Mol. Cell 2005, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Cao, Y.; Zheng, Q.; Tu, J.; Zhou, W.; He, J.; Zhong, J.; Chen, Y.; Wang, J.; Cai, R.; et al. SENP1-Sirt3 Signaling Controls Mitochondrial Protein Acetylation and Metabolism. Mol. Cell 2019, 75, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tian, J.; Huang, C.; Ma, J.; Hu, G.; Chen, Y.; Wang, T.; Cai, R.; Zuo, Y.; Tan, H.; et al. P53 suppresses SENP3 phosphorylation to mediate G2 checkpoint. Cell Discov. 2020, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kang, X.; Zhang, S.; Yeh, E.T. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 2007, 131, 584–595. [Google Scholar] [CrossRef]
- Zheng, Q.; Cao, Y.; Chen, Y.; Wang, J.; Fan, Q.; Huang, X.; Wang, Y.; Wang, T.; Wang, X.; Ma, J.; et al. Senp2 regulates adipose lipid storage by de-SUMOylation of Setdb1. J. Mol. Cell Biol. 2018, 10, 258–266. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, K.; Yang, J.; Lao, Y.; Deng, L.; Deng, G.; Yi, J.; Sun, X.; Wang, Q. SENP3 Suppresses Osteoclastogenesis by De-conjugating SUMO2/3 from IRF8 in Bone Marrow-Derived Monocytes. Cell Rep. 2020, 30, 1951–1963. [Google Scholar] [CrossRef]
- Shalizi, A.; Gaudilliere, B.; Yuan, Z.; Stegmuller, J.; Shirogane, T.; Ge, Q.; Tan, Y.; Schulman, B.; Harper, J.W.; Bonni, A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 2006, 311, 1012–1017. [Google Scholar] [CrossRef]
- Zhang, L.; Li, F.; Dimayuga, E.; Craddock, J.; Keller, J.N. Effects of aging and dietary restriction on ubiquitination, sumoylation, and the proteasome in the spleen. FEBS Lett. 2007, 581, 5543–5547. [Google Scholar] [CrossRef]
- Comerford, K.M.; Leonard, M.O.; Karhausen, J.; Carey, R.; Colgan, S.P.; Taylor, C.T. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc. Natl. Acad. Sci. USA 2003, 100, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Feige, J.N.; Auwerx, J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007, 17, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Gleyzer, N.; Scarpulla, R.C. PGC-1-related coactivator (PRC), a sensor of metabolic stress, orchestrates a redox-sensitive program of inflammatory gene expression. J. Biol. Chem. 2011, 286, 39715–39725. [Google Scholar] [CrossRef]
- Soriano, F.X.; Liesa, M.; Bach, D.; Chan, D.C.; Palacin, M.; Zorzano, A. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1 alpha, estrogen-related receptor-alpha, and mitofusin 2. Diabetes 2006, 55, 1783–1791. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, S.N.; Emter, R.; Hock, M.B.; Knutti, D.; Cardenas, J.; Podvinec, M.; Oakeley, E.J.; Kralli, A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 6472–6477. [Google Scholar] [CrossRef]
- Rytinki, M.M.; Palvimo, J.J. SUMOylation attenuates the function of PGC-1alpha. J. Biol. Chem. 2009, 284, 26184–26193. [Google Scholar] [CrossRef]
- Cai, R.; Gu, J.; Sun, H.; Liu, X.; Mei, W.; Qi, Y.; Xue, S.; Ren, S.; Rabinowitz, J.E.; Wang, Y.; et al. Induction of SENP1 in myocardium contributes to abnormities of mitochondria and cardiomyopathy. J. Mol. Cell. Cardiol. 2015, 79, 115–122. [Google Scholar] [CrossRef]
- Cai, R.; Yu, T.; Huang, C.; Xia, X.; Liu, X.; Gu, J.; Xue, S.; Yeh, E.T.; Cheng, J. SUMO-specific protease 1 regulates mitochondrial biogenesis through PGC-1alpha. J. Biol. Chem. 2012, 287, 44464–44470. [Google Scholar]
- Abid, H.; Cartier, D.; Hamieh, A.; Francois-Bellan, A.M.; Bucharles, C.; Pothion, H.; Manecka, D.L.; Leprince, J.; Adriouch, S.; Boyer, O.; et al. AMPK Activation of PGC-1alpha/NRF-1-Dependent SELENOT Gene Transcription Promotes PACAP-Induced Neuroendocrine Cell Differentiation Through Tolerance to Oxidative Stress. Mol. Neurobiol. 2019, 56, 4086–4101. [Google Scholar] [CrossRef]
- Vernier, M.; Dufour, C.R.; McGuirk, S.; Scholtes, C.; Li, X.; Bourmeau, G.; Kuasne, H.; Park, M.; St-Pierre, J.; Audet-Walsh, E.; et al. Estrogen-related receptors are targetable ROS sensors. Genes Dev. 2020, 34, 544–559. [Google Scholar] [CrossRef]
- Sekulic-Jablanovic, M.; Petkovic, V.; Wright, M.B.; Kucharava, K.; Huerzeler, N.; Levano, S.; Brand, Y.; Leitmeyer, K.; Glutz, A.; Bausch, A.; et al. Effects of peroxisome proliferator activated receptors (PPAR)-gamma and -alpha agonists on cochlear protection from oxidative stress. PLoS ONE 2017, 12, e0188596. [Google Scholar] [CrossRef] [PubMed]
- Guerra de Souza, A.C.; Prediger, R.D.; Cimarosti, H. SUMO-regulated mitochondrial function in Parkinson’s disease. J. Neurochem 2016, 137, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Ahn, B.H.; Kim, H.S.; Song, S.; Lee, I.H.; Liu, J.; Vassilopoulos, A.; Deng, C.X.; Finkel, T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. USA 2008, 105, 14447–14452. [Google Scholar] [CrossRef] [PubMed]
- Hirschey, M.D.; Shimazu, T.; Goetzman, E.; Jing, E.; Schwer, B.; Lombard, D.B.; Grueter, C.A.; Harris, C.; Biddinger, S.; Ilkayeva, O.R.; et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 2010, 464, 121–125. [Google Scholar] [CrossRef]
- Qiu, X.; Brown, K.; Hirschey, M.D.; Verdin, E.; Chen, D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010, 12, 662–667. [Google Scholar] [CrossRef]
- Schlicker, C.; Gertz, M.; Papatheodorou, P.; Kachholz, B.; Becker, C.F.; Steegborn, C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J. Mol. Biol. 2008, 382, 790–801. [Google Scholar] [CrossRef]
- Someya, S.; Yu, W.; Hallows, W.C.; Xu, J.; Vann, J.M.; Leeuwenburgh, C.; Tanokura, M.; Denu, J.M.; Prolla, T.A. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 2010, 143, 802–812. [Google Scholar] [CrossRef]
- Kim, H.S.; Patel, K.; Muldoon-Jacobs, K.; Bisht, K.S.; Aykin-Burns, N.; Pennington, J.D.; van der Meer, R.; Nguyen, P.; Savage, J.; Owens, K.M.; et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010, 17, 41–52. [Google Scholar] [CrossRef]
- Finley, L.W.; Carracedo, A.; Lee, J.; Souza, A.; Egia, A.; Zhang, J.; Teruya-Feldstein, J.; Moreira, P.I.; Cardoso, S.M.; Clish, C.B.; et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 2011, 19, 416–428. [Google Scholar] [CrossRef]
- Hallows, W.C.; Yu, W.; Smith, B.C.; Devries, M.K.; Ellinger, J.J.; Someya, S.; Shortreed, M.R.; Prolla, T.; Markley, J.L.; Smith, L.M.; et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell 2011, 41, 139–149. [Google Scholar] [CrossRef]
- Hirschey, M.D.; Shimazu, T.; Capra, J.A.; Pollard, K.S.; Verdin, E. SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging (Albany NY) 2011, 3, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Schumacker, P.T. SIRT3 controls cancer metabolic reprogramming by regulating ROS and HIF. Cancer Cell 2011, 19, 299–300. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitch, R.C.; Samborska, B.; Faubert, B.; Ma, E.H.; Gravel, S.P.; Andrzejewski, S.; Raissi, T.C.; Pause, A.; St-Pierre, J.; Jones, R.G. AMPK Maintains Cellular Metabolic Homeostasis through Regulation of Mitochondrial Reactive Oxygen Species. Cell Rep. 2017, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.P.; Wong, C.C.; Kai, A.K.; Ho, D.W.; Lau, E.Y.; Tsui, Y.M.; Chan, L.K.; Cheung, T.T.; Chok, K.S.; Chan, A.C.Y.; et al. SENP1 promotes hypoxia-induced cancer stemness by HIF-1alpha deSUMOylation and SENP1/HIF-1alpha positive feedback loop. Gut 2017, 66, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Rubio, T.; Vernia, S.; Sanz, P. Sumoylation of AMPKbeta2 subunit enhances AMP-activated protein kinase activity. Mol. Biol. Cell. 2013, 24, 1801–1811. [Google Scholar] [CrossRef]
- Yan, Y.; Ollila, S.; Wong, I.P.L.; Vallenius, T.; Palvimo, J.J.; Vaahtomeri, K.; Makela, T.P. SUMOylation of AMPKalpha1 by PIAS4 specifically regulates mTORC1 signalling. Nat. Commun. 2015, 6, 8979. [Google Scholar] [CrossRef]
- Sack, M.N. Type 2 diabetes, mitochondrial biology and the heart. J. Mol. Cell. Cardiol. 2009, 46, 842–849. [Google Scholar] [CrossRef]
- Carvalho, C.; Correia, S.C.; Santos, R.X.; Cardoso, S.; Moreira, P.I.; Clark, T.A.; Zhu, X.; Smith, M.A.; Perry, G. Role of mitochondrial-mediated signaling pathways in Alzheimer disease and hypoxia. J. Bioenerg. Biomembr. 2009, 41, 433–440. [Google Scholar] [CrossRef]
- Zhang, P.; Ke, J.; Li, Y.; Huang, L.; Chen, Z.; Huang, X.; Zhang, L.; Xiao, D. Long-term exposure to high altitude hypoxia during pregnancy increases fetal heart susceptibility to ischemia/reperfusion injury and cardiac dysfunction. Int J. Cardiol. 2019, 274, 7–15. [Google Scholar] [CrossRef]
- Krikun, G.; Schatz, F.; Finlay, T.; Kadner, S.; Mesia, A.; Gerrets, R.; Lockwood, C.J. Expression of angiopoietin-2 by human endometrial endothelial cells: Regulation by hypoxia and inflammation. Biochem. Biophys. Res. Commun. 2000, 275, 159–163. [Google Scholar] [CrossRef]
- Hung, P.F.; Hong, T.M.; Chang, C.C.; Hung, C.L.; Hsu, Y.L.; Chang, Y.L.; Wu, C.T.; Chang, G.C.; Chan, N.L.; Yu, S.L.; et al. Hypoxia-induced Slug SUMOylation enhances lung cancer metastasis. J. Exp. Clin. Cancer Res. 2019, 38, 5. [Google Scholar] [CrossRef] [PubMed]
- Quintana, D.D.; Garcia, J.A.; Sarkar, S.N.; Jun, S.; Engler-Chiurazzi, E.B.; Russell, A.E.; Cavendish, J.Z.; Simpkins, J.W. Hypoxia-reoxygenation of primary astrocytes results in a redistribution of mitochondrial size and mitophagy. Mitochondrion 2019, 47, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.W.; Kim, J.H.; Ahn, Y.H.; Seo, J.; Ko, A.; Jeong, M.; Kim, S.J.; Ro, J.Y.; Park, K.M.; Lee, H.W.; et al. Ubiquitination and degradation of the FADD adaptor protein regulate death receptor-mediated apoptosis and necroptosis. Nat. Commun. 2012, 3, 978. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.G.; Kim, H.; Jeong, E.I.; Lee, H.J.; Park, S.; Lee, S.Y.; Lee, H.J.; Lee, S.W.; Chung, C.H.; Jung, Y.K. SUMO-Modified FADD Recruits Cytosolic Drp1 and Caspase-10 to Mitochondria for Regulated Necrosis. Mol. Cell. Biol. 2017, 37, e00254-16. [Google Scholar] [PubMed]
- Li, C.; Wang, L.; Zhang, J.; Huang, M.; Wong, F.; Liu, X.; Liu, F.; Cui, X.; Yang, G.; Chen, J.; et al. CERKL interacts with mitochondrial TRX2 and protects retinal cells from oxidative stress-induced apoptosis. Biochim. Biophys. Acta 2014, 1842, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, K.; Zhang, H.; Zhou, H.J.; Chen, Y.; Min, W. A Unique SUMO-Interacting Motif of Trx2 Is Critical for Its Mitochondrial Presequence Processing and Anti-oxidant Activity. Front. Physiol. 2019, 10, 1089. [Google Scholar] [CrossRef]
- Zemirli, N.; Morel, E.; Molino, D. Mitochondrial Dynamics in Basal and Stressful Conditions. Int. J. Mol. Sci. 2018, 19, 564. [Google Scholar]
- Harder, Z.; Zunino, R.; McBride, H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr. Biol. 2004, 14, 340–345. [Google Scholar] [CrossRef]
- Figueroa-Romero, C.; Iniguez-Lluhi, J.A.; Stadler, J.; Chang, C.R.; Arnoult, D.; Keller, P.J.; Hong, Y.; Blackstone, C.; Feldman, E.L. SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle. FASEB J. 2009, 23, 3917–3927. [Google Scholar] [CrossRef]
- Zunino, R.; Braschi, E.; Xu, L.; McBride, H.M. Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J. Biol. Chem. 2009, 284, 17783–17795. [Google Scholar]
- Guo, C.; Hildick, K.L.; Luo, J.; Dearden, L.; Wilkinson, K.A.; Henley, J.M. SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia. Embo. J. 2013, 32, 1514–1528. [Google Scholar] [CrossRef] [PubMed]
- Wasiak, S.; Zunino, R.; McBride, H.M. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J. Cell Biol. 2007, 177, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Deng, X.; Lim, G.G.Y.; Xie, S.; Zhou, Z.D.; Lim, K.L.; Tan, E.K. Superoxide drives progression of Parkin/PINK1-dependent mitophagy following translocation of Parkin to mitochondria. Cell Death Dis. 2017, 8, e3097. [Google Scholar] [CrossRef] [PubMed]
- Um, J.W.; Chung, K.C. Functional modulation of parkin through physical interaction with SUMO-1. J. Neurosci. Res. 2006, 84, 1543–1554. [Google Scholar] [CrossRef]
- Ashrafi, G.; Schlehe, J.S.; LaVoie, M.J.; Schwarz, T.L. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J. Cell Biol. 2014, 206, 655–670. [Google Scholar] [CrossRef]
- Ding, D.; Ao, X.; Liu, Y.; Wang, Y.Y.; Fa, H.G.; Wang, M.Y.; He, Y.Q.; Wang, J.X. Post-translational modification of Parkin and its research progress in cancer. Cancer Commun. 2019, 39, 77. [Google Scholar] [CrossRef]
- Nishida, T.; Yamada, Y. RNF4-mediated SUMO-targeted ubiquitination relieves PARIS/ZNF746-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 2020, 526, 110–116. [Google Scholar] [CrossRef]
- Benedetti, C.; Haynes, C.M.; Yang, Y.; Harding, H.P.; Ron, D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 2006, 174, 229–239. [Google Scholar]
- Houtkooper, R.H.; Mouchiroud, L.; Ryu, D.; Moullan, N.; Katsyuba, E.; Knott, G.; Williams, R.W.; Auwerx, J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 2013, 497, 451–457. [Google Scholar] [CrossRef]
- Nargund, A.M.; Fiorese, C.J.; Pellegrino, M.W.; Deng, P.; Haynes, C.M. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol. Cell 2015, 58, 123–133. [Google Scholar] [CrossRef]
- Pellegrino, M.W.; Nargund, A.M.; Kirienko, N.V.; Gillis, R.; Fiorese, C.J.; Haynes, C.M. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 2014, 516, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Merkwirth, C.; Jovaisaite, V.; Durieux, J.; Matilainen, O.; Jordan, S.D.; Quiros, P.M.; Steffen, K.K.; Williams, E.G.; Mouchiroud, L.; Tronnes, S.U.; et al. Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell 2016, 165, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Nargund, A.M.; Pellegrino, M.W.; Fiorese, C.J.; Baker, B.M.; Haynes, C.M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 2012, 337, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Garcia, G.; Bian, Q.; Steffen, K.K.; Joe, L.; Wolff, S.; Meyer, B.J.; Dillin, A. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt). Cell 2016, 165, 1197–1208. [Google Scholar] [CrossRef]
- Gao, K.; Li, Y.; Hu, S.; Liu, Y. SUMO peptidase ULP-4 regulates mitochondrial UPR-mediated innate immunity and lifespan extension. Elife 2019, 8. [Google Scholar] [CrossRef]
- Dobreva, G.; Dambacher, J.; Grosschedl, R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003, 17, 3048–3061. [Google Scholar] [CrossRef]
- Tan, J.A.; Sun, Y.; Song, J.; Chen, Y.; Krontiris, T.G.; Durrin, L.K. SUMO conjugation to the matrix attachment region-binding protein, special AT-rich sequence-binding protein-1 (SATB1), targets SATB1 to promyelocytic nuclear bodies where it undergoes caspase cleavage. J. Biol. Chem. 2008, 283, 18124–18134. [Google Scholar] [CrossRef]
- Cai, S.; Han, H.J.; Kohwi-Shigematsu, T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 2003, 34, 42–51. [Google Scholar] [CrossRef]
- Fiorese, C.J.; Schulz, A.M.; Lin, Y.F.; Rosin, N.; Pellegrino, M.W.; Haynes, C.M. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol. 2016, 26, 2037–2043. [Google Scholar] [CrossRef]
- Henley, J.M.; Craig, T.J.; Wilkinson, K.A. Neuronal SUMOylation: Mechanisms, physiology, and roles in neuronal dysfunction. Physiol. Rev. 2014, 94, 1249–1285. [Google Scholar]
- Zarranz, J.J.; Alegre, J.; Gomez-Esteban, J.C.; Lezcano, E.; Ros, R.; Ampuero, I.; Vidal, L.; Hoenicka, J.; Rodriguez, O.; Atares, B.; et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 2004, 55, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Jang, W.H.; Quezado, M.M.; Oh, Y.; Chung, K.C.; Junn, E.; Mouradian, M.M. Proteasome inhibition induces alpha-synuclein SUMOylation and aggregate formation. J. Neurol. Sci. 2011, 307, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Krumova, P.; Meulmeester, E.; Garrido, M.; Tirard, M.; Hsiao, H.H.; Bossis, G.; Urlaub, H.; Zweckstetter, M.; Kugler, S.; Melchior, F.; et al. Sumoylation inhibits alpha-synuclein aggregation and toxicity. J. Cell Biol. 2011, 194, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Kim, Y.M.; Mouradian, M.M.; Chung, K.C. Human Polycomb protein 2 promotes alpha-synuclein aggregate formation through covalent SUMOylation. Brain Res. 2011, 1381, 78–89. [Google Scholar] [CrossRef]
- Dias, V.; Junn, E.; Mouradian, M.M. The role of oxidative stress in Parkinson’s disease. J. Parkinsons Dis. 2013, 3, 461–491. [Google Scholar]
- Devi, L.; Raghavendran, V.; Prabhu, B.M.; Avadhani, N.G.; Anandatheerthavarada, H.K. Mitochondrial import and accumulation of alpha-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J. Biol. Chem. 2008, 283, 9089–9100. [Google Scholar] [CrossRef]
- Shinbo, Y.; Niki, T.; Taira, T.; Ooe, H.; Takahashi-Niki, K.; Maita, C.; Seino, C.; Iguchi-Ariga, S.M.; Ariga, H. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 2006, 13, 96–108. [Google Scholar] [CrossRef]
- Takahashi, K.; Taira, T.; Niki, T.; Seino, C.; Iguchi-Ariga, S.M.; Ariga, H. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J. Biol. Chem. 2001, 276, 37556–37563. [Google Scholar] [CrossRef]
- Tao, X.; Tong, L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson’s disease. J. Biol. Chem. 2003, 278, 31372–31379. [Google Scholar]
- Fan, J.; Ren, H.; Fei, E.; Jia, N.; Ying, Z.; Jiang, P.; Wu, M.; Wang, G. Sumoylation is critical for DJ-1 to repress p53 transcriptional activity. FEBS Lett. 2008, 582, 1151–1156. [Google Scholar] [CrossRef]
- Zhong, N.; Kim, C.Y.; Rizzu, P.; Geula, C.; Porter, D.R.; Pothos, E.N.; Squitieri, F.; Heutink, P.; Xu, J. DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor. J. Biol. Chem. 2006, 281, 20940–20948. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Ko, H.S.; Kang, H.; Lee, Y.; Lee, Y.I.; Pletinkova, O.; Troconso, J.C.; Dawson, V.L.; Dawson, T.M. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell 2011, 144, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Wareski, P.; Vaarmann, A.; Choubey, V.; Safiulina, D.; Liiv, J.; Kuum, M.; Kaasik, A. PGC-1{alpha} and PGC-1{beta} regulate mitochondrial density in neurons. J. Biol. Chem. 2009, 284, 21379–21385. [Google Scholar] [CrossRef] [PubMed]
- Zhong, N.; Xu, J. Synergistic activation of the human MnSOD promoter by DJ-1 and PGC-1alpha: Regulation by SUMOylation and oxidation. Hum. Mol. Genet. 2008, 17, 3357–3367. [Google Scholar] [CrossRef]
- Zanellati, M.C.; Monti, V.; Barzaghi, C.; Reale, C.; Nardocci, N.; Albanese, A.; Valente, E.M.; Ghezzi, D.; Garavaglia, B. Mitochondrial dysfunction in Parkinson disease: Evidence in mutant PARK2 fibroblasts. Front. Genet. 2015, 6, 78. [Google Scholar] [CrossRef]
- Vijayakumar, U.G.; Milla, V.; Cynthia Stafford, M.Y.; Bjourson, A.J.; Duddy, W.; Duguez, S.M. A Systematic Review of Suggested Molecular Strata, Biomarkers and Their Tissue Sources in ALS. Front. Neurol. 2019, 10, 400. [Google Scholar] [CrossRef]
- Kaur, S.J.; McKeown, S.R.; Rashid, S. Mutant SOD1 mediated pathogenesis of Amyotrophic Lateral Sclerosis. Gene 2016, 577, 109–118. [Google Scholar] [CrossRef]
- Niikura, T.; Kita, Y.; Abe, Y. SUMO3 modification accelerates the aggregation of ALS-linked SOD1 mutants. PLoS ONE 2014, 9, e101080. [Google Scholar] [CrossRef]
- Fei, E.; Jia, N.; Yan, M.; Ying, Z.; Sun, Q.; Wang, H.; Zhang, T.; Ma, X.; Ding, H.; Yao, X.; et al. SUMO-1 modification increases human SOD1 stability and aggregation. Biochem. Biophys. Res. Commun. 2006, 347, 406–412. [Google Scholar] [CrossRef]
- Wada, H.; Suzuki, D.; Niikura, T. Regulation of ALS-Associated SOD1 Mutant SUMOylation and Aggregation by SENP and PIAS Family Proteins. J. Mol. Neurosci. 2020. [Google Scholar] [CrossRef]
- De Vos, K.J.; Chapman, A.L.; Tennant, M.E.; Manser, C.; Tudor, E.L.; Lau, K.F.; Brownlees, J.; Ackerley, S.; Shaw, P.J.; McLoughlin, D.M.; et al. Familial amyotrophic lateral sclerosis-linked SOD1 mutants perturb fast axonal transport to reduce axonal mitochondria content. Hum. Mol. Genet. 2007, 16, 2720–2728. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Sarge, K.D. Sumoylation of amyloid precursor protein negatively regulates Abeta aggregate levels. Biochem. Biophys. Res. Commun. 2008, 374, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.B.; Xia, Y.Y.; Shu, X.J.; Liu, Z.C.; Feng, Y.; Liu, X.H.; Yu, G.; Yin, G.; Xiong, Y.S.; Zeng, K.; et al. SUMOylation at K340 inhibits tau degradation through deregulating its phosphorylation and ubiquitination. Proc. Natl. Acad. Sci. USA 2014, 111, 16586–16591. [Google Scholar] [CrossRef] [PubMed]
- Weidling, I.W.; Swerdlow, R.H. Mitochondria in Alzheimer’s disease and their potential role in Alzheimer’s proteostasis. Exp. Neurol. 2020, 330, 113321. [Google Scholar] [CrossRef]
- Ferdaoussi, M.; Dai, X.; Jensen, M.V.; Wang, R.; Peterson, B.S.; Huang, C.; Ilkayeva, O.; Smith, N.; Miller, N.; Hajmrle, C.; et al. Isocitrate-to-SENP1 signaling amplifies insulin secretion and rescues dysfunctional beta cells. J. Clin. Investig. 2015, 125, 3847–3860. [Google Scholar] [CrossRef]
- Galanty, Y.; Belotserkovskaya, R.; Coates, J.; Polo, S.; Miller, K.M.; Jackson, S.P. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009, 462, 935–939. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Vertegaal, A.C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016, 17, 581–595. [Google Scholar] [CrossRef]
- He, Z.; Huang, T.; Ao, K.; Yan, X.; Huang, Y. Sumoylation, Phosphorylation, and Acetylation Fine-Tune the Turnover of Plant Immunity Components Mediated by Ubiquitination. Front. Plant. Sci. 2017, 8, 1682. [Google Scholar] [CrossRef]
- Tomanov, K.; Nukarinen, E.; Vicente, J.; Mendiondo, G.M.; Winter, N.; Nehlin, L.; Weckwerth, W.; Holdsworth, M.J.; Teige, M.; Bachmair, A. Sumoylation and phosphorylation: Hidden and overt links. J. Exp. Bot. 2018, 69, 4583–4590. [Google Scholar] [CrossRef]
- Tomasi, M.L.; Ramani, K. SUMOylation and phosphorylation cross-talk in hepatocellular carcinoma. Transl. Gastroenterol. Hepatol. 2018, 3, 20. [Google Scholar] [CrossRef]
- Xiao, Y.; Lucas, B.; Molcho, E.; Vigodner, M. Cross-talk between sumoylation and phosphorylation in mouse spermatocytes. Biochem. Biophys. Res. Commun. 2017, 487, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Seok, S.; Yau, P.; Li, X.; Kemper, B.; Kemper, J.K. Obesity and aging diminish sirtuin 1 (SIRT1)-mediated deacetylation of SIRT3, leading to hyperacetylation and decreased activity and stability of SIRT3. J. Biol. Chem. 2017, 292, 17312–17323. [Google Scholar] [CrossRef] [PubMed]
- Iwabu, M.; Yamauchi, T.; Okada-Iwabu, M.; Sato, K.; Nakagawa, T.; Funata, M.; Yamaguchi, M.; Namiki, S.; Nakayama, R.; Tabata, M.; et al. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature 2010, 464, 1313–1319. [Google Scholar] [CrossRef] [PubMed]

| Proteins | SUMOylation Site | SUMO-Specific Protease | Function of SUMOylation | References |
|---|---|---|---|---|
| PCG-1α | K183 (mouse) | SENP1, SENP2 | SUMOylation suppresses its function of transcriptional biogenesis | Rytinki and Palvimo, 2009 [36] |
| FADD | K120/K125/K149 (human) | - | SUMOylations promotes mitochondrial-associated necrosis | Choi et al., 2017 [64] |
| DRP1 | K532/K535/K558/ K568/K594/K597/ K606/K608 (human) | SENP3 | SUMO2/3 modification suppresses mitochondrial fragmentation | Figueroa-Romero et al., 2019 [69]; Zunino et al., 2009 [70]; Guo et al., 2013 [71] |
| SENP5 | SUMO1 modification promotes mitochondrial fragmentation | |||
| SNCA (α-synuclein) | K96/K102 (human) | - | SUMOylation inhibits α-syn accumulation to protect the mitochondrial complex I | Krumova et al., 2011 [93] |
| DJ-1 | K130 (human) | - | Abnormal SUMOylation lost the response to the mitochondrial stress | Shinbo et al., 2006 [97] |
| PSF | K338 (human) | SENP1 | SUMOylation promotes the interaction with PGC-1α to repress mitochondrial biogenesis | Zhong and Xu, 2008 [104] |
| SIRT3 | K228 (human) K223 (mouse) | SENP1 | SUMOylation suppresses its function as an NAD+-dependent deacetylase | Wang et al., 2019 [24] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Cheng, J.; Wang, T. SUMOylation-Mediated Response to Mitochondrial Stress. Int. J. Mol. Sci. 2020, 21, 5657. https://doi.org/10.3390/ijms21165657
He J, Cheng J, Wang T. SUMOylation-Mediated Response to Mitochondrial Stress. International Journal of Molecular Sciences. 2020; 21(16):5657. https://doi.org/10.3390/ijms21165657
Chicago/Turabian StyleHe, Jianli, Jinke Cheng, and Tianshi Wang. 2020. "SUMOylation-Mediated Response to Mitochondrial Stress" International Journal of Molecular Sciences 21, no. 16: 5657. https://doi.org/10.3390/ijms21165657
APA StyleHe, J., Cheng, J., & Wang, T. (2020). SUMOylation-Mediated Response to Mitochondrial Stress. International Journal of Molecular Sciences, 21(16), 5657. https://doi.org/10.3390/ijms21165657




