Probing the Importance of the G-Quadruplex Grooves for the Activity of the Anti-HIV-Integrase Aptamer T30923
Abstract
1. Introduction
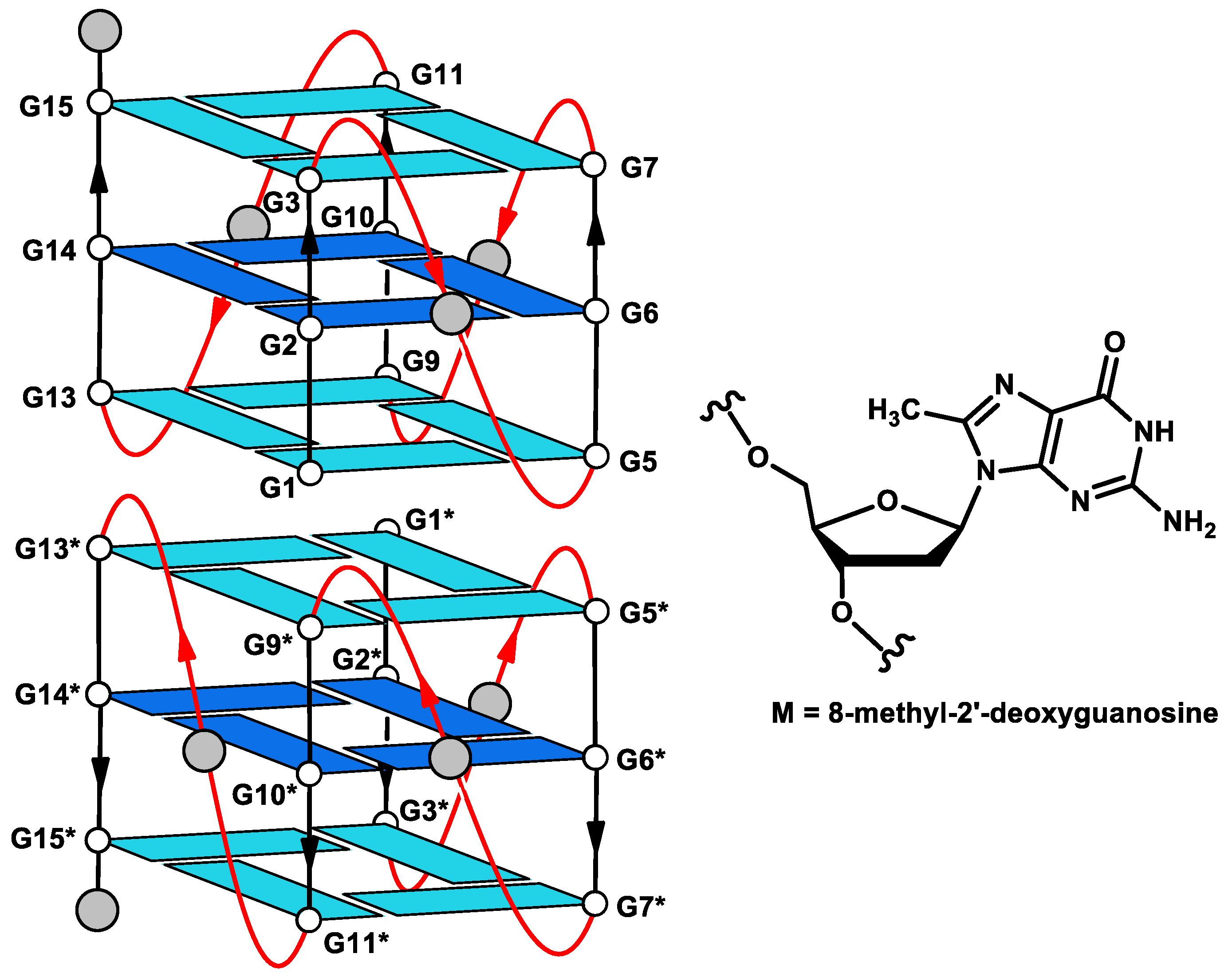
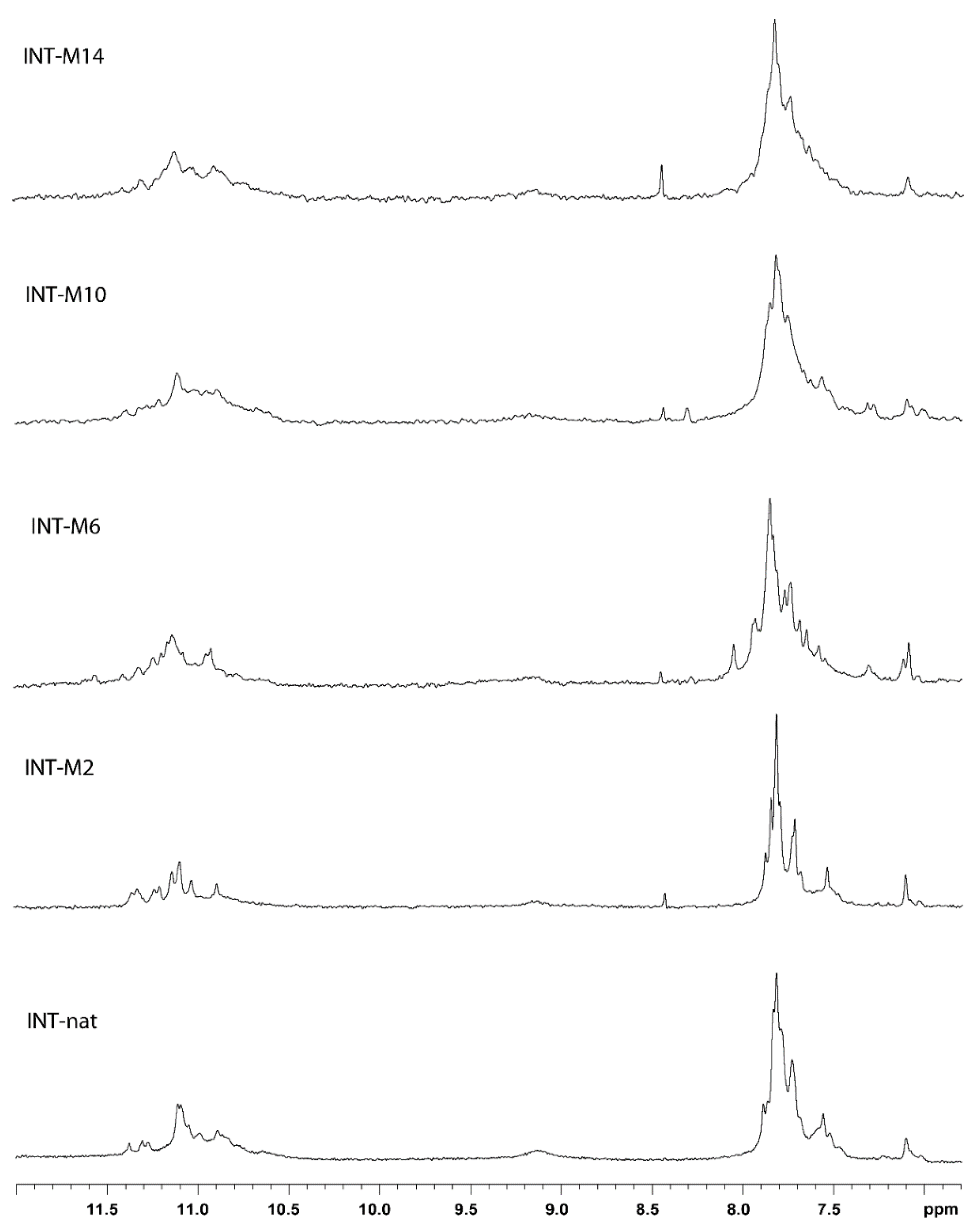
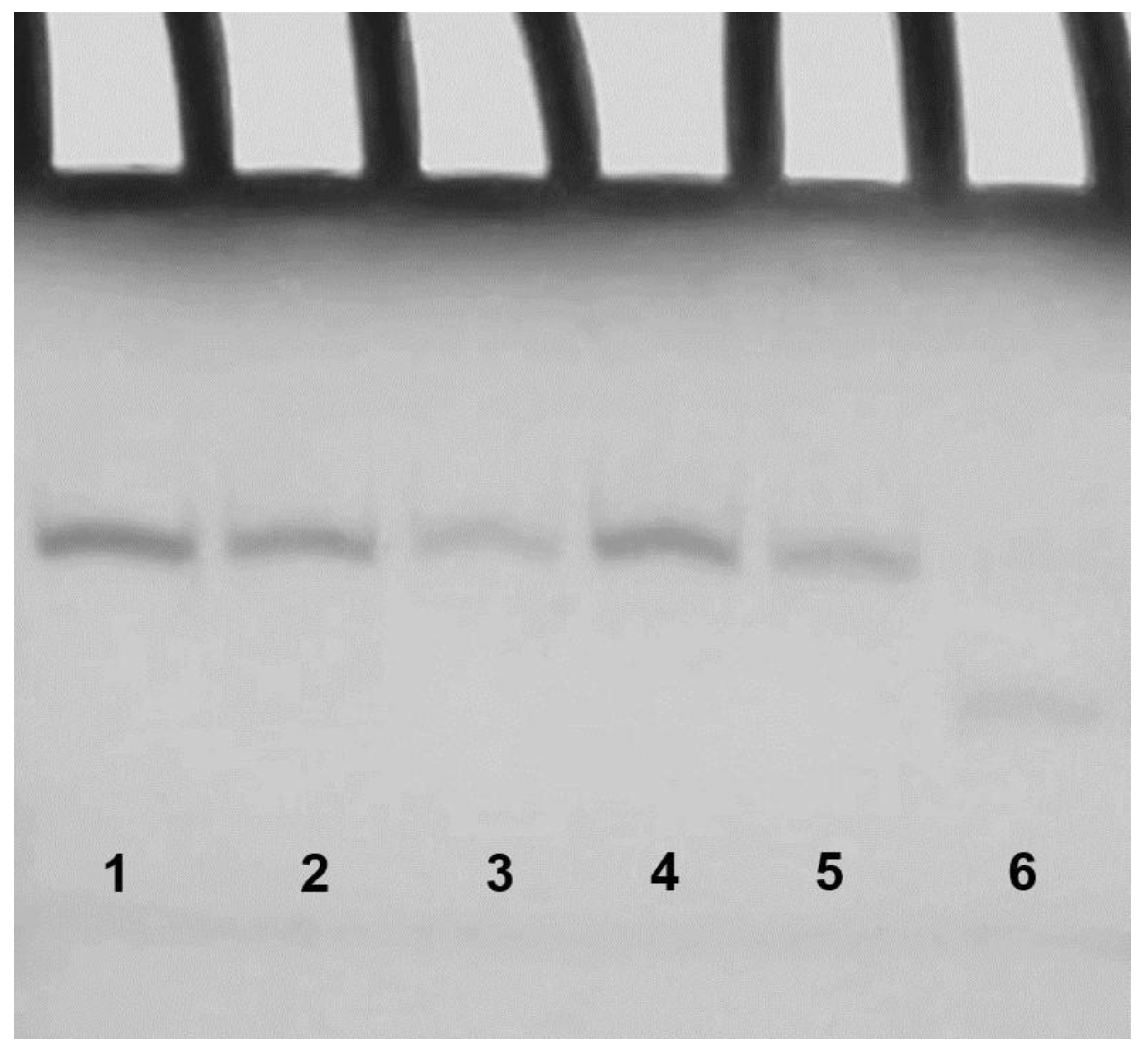
2. Results
2.1. NMR Spectroscopy
2.2. CD Spectroscopy
2.3. Polyacrylamide Gel Electrophoresis (PAGE)
2.4. Molecular Modeling (MM)
2.5. Effect of T30923 Derivatives on the HIV-1 IN LEDGF-Independent Activity
3. Discussion
4. Materials and Methods
4.1. Oligonucleotide Synthesis and Purification
4.2. NMR Spectroscopy
4.3. CD Spectroscopy
4.4. Gel Electrophoresis
4.5. Molecular Modeling
4.6. Expression and Purification of Recombinant HIV-1 IN
4.7. HTRF Integrase LEDGF-Independent Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shukla, E.; Chauhan, R. Host-HIV-1 Interactome: A Quest for Novel Therapeutic Intervention. Cells 2019, 8, 1155. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on Aptamer Research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef] [PubMed]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-Quadruplex-Forming Aptamers-Characteristics, Applications, and Perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [PubMed]
- Ecker, D.J.; Vickers, T.A.; Hanecak, R.; Diver, V.; Anderson, K. Rational screening of oligonucleotide combinatorial libraries for drug discovery. Nucleic Acids Res. 1993, 21, 1853–1856. [Google Scholar] [CrossRef]
- Romanucci, V.; Zarrelli, A.; Di Fabio, G. Hotoda’s Sequence and Anti-HIV Activity: Where Are We Now? Molecules 2019, 24, 1417. [Google Scholar] [CrossRef]
- Virgilio, A.; Esposito, V.; Citarella, G.; Mayol, L.; Galeone, A. Structural Investigations on the Anti-HIV G-Quadruplex-Forming Oligonucleotide TGGGAG and Its Analogues: Evidence for the Presence of an A-Tetrad. ChemBioChem 2012, 13, 2219–2224. [Google Scholar] [CrossRef]
- Romanucci, V.; Gaglione, M.; Messere, A.; Potenza, N.; Zarrelli, A.; Noppen, S.; Liekens, S.; Balzarini, J.; Di Fabio, G. Hairpin oligonucleotides forming G-quadruplexes: New aptamers with anti-HIV activity. Eur. J. Med. Chem. 2015, 89, 51–58. [Google Scholar] [CrossRef]
- Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Galeone, A.; Mayol, L.; Haider, S.; Olubiyi, O.; Hoorelbeke, B.; Balzarini, J.; et al. Tetra-end-linked oligonucleotides forming DNA G-quadruplexes: A new class of aptamers showing anti-HIV activity. Chem. Commun. (Camb) 2010, 46, 8971–8973. [Google Scholar] [CrossRef]
- Oliviero, G.; Borbone, N.; Amato, J.; D’Errico, S.; Galeone, A.; Piccialli, G.; Varra, M.; Mayol, L. Synthesis of quadruplex-forming tetra-end-linked oligonucleotides: Effects of the linker size on quadruplex topology and stability. Biopolymers 2009, 91, 466–477. [Google Scholar] [CrossRef]
- Andreola, M.L.; Pileur, F.; Calmels, C.; Ventura, M.; Tarrago-Litvak, L.; Toulme, J.J.; Litvak, S. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry 2001, 40, 10087–10094. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Ma, J.-B.; Faure, A.; Andréola, M.-L.; Patel, D.J. An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 2005, 102, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Ojwang, J.O.; Buckheit, R.W.; Pommier, Y.; Mazumder, A.; De Vreese, K.; Este, J.A.; Reymen, D.; Pallansch, L.A.; Lackman-Smith, C.; Wallace, T.L.; et al. T30177, an oligonucleotide stabilized by an intramolecular guanosine octet, is a potent inhibitor of laboratory strains and clinical isolates of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 1995, 39, 2426–2435. [Google Scholar] [CrossRef]
- Rando, R.F.; Ojwang, J.; Elbaggari, A.; Reyes, G.R.; Tinder, R.; McGrath, M.S.; Hogan, M.E. Suppression of human immunodeficiency virus type 1 activity in vitro by oligonucleotides which form intramolecular tetrads. J. Biol. Chem. 1995, 270, 1754–1760. [Google Scholar] [CrossRef]
- Mazumder, A.; Neamati, N.; Ojwang, J.O.; Sunder, S.; Rando, R.F.; Pommier, Y. Inhibition of the human immunodeficiency virus type 1 integrase by guanosine quartet structures. Biochemistry 1996, 35, 13762–13771. [Google Scholar] [CrossRef]
- Bishop, J.S.; Guy-Caffey, J.K.; Ojwang, J.O.; Smith, S.R.; Hogan, M.E.; Cossum, P.A.; Rando, R.F.; Chaudhary, N. Intramolecular G-quartet motifs confer nuclease resistance to a potent anti-HIV oligonucleotide. J. Biol. Chem. 1996, 271, 5698–5703. [Google Scholar] [CrossRef]
- Cherepanov, P.; Esté, J.A.; Rando, R.F.; Ojwang, J.O.; Reekmans, G.; Steinfeld, R.; David, G.; De Clercq, E.; Debyser, Z. Mode of interaction of G-quartets with the integrase of human immunodeficiency virus type 1. Mol. Pharmacol. 1997, 52, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Jing, N.; Hogan, M.E. Structure-activity of tetrad-forming oligonucleotides as a potent anti- HIV therapeutic drug. J. Biol. Chem. 1998, 273, 34992–34999. [Google Scholar] [CrossRef]
- Jing, N.; Marchand, C.; Liu, J.; Mitra, R.; Hogan, M.E.; Pommier, Y. Mechanism of inhibition of HIV-1 integrase by G-tetrad-forming oligonucleotides in vitro. J. Biol. Chem. 2000, 275, 21460–21467. [Google Scholar] [CrossRef]
- Magbanua, E.; Zivkovic, T.; Hansen, B.; Beschorner, N.; Meyer, C.; Lorenzen, I.; Grötzinger, J.; Hauber, J.; Torda, A.E.; Mayer, G.; et al. d(GGGT) 4 and r(GGGU) 4 are both HIV-1 inhibitors and interleukin-6 receptor aptamers. RNA Biol. 2013, 10, 216–227. [Google Scholar] [CrossRef]
- Kelley, S.; Boroda, S.; Musier-Forsyth, K.; Kankia, B.I. HIV-integrase aptamer folds into a parallel quadruplex: A thermodynamic study. Biophys. Chem. 2011, 155, 82–88. [Google Scholar] [CrossRef]
- Mukundan, V.T.; Do, N.Q.; Phan, A.T. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011, 39, 8984–8991. [Google Scholar] [CrossRef] [PubMed]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef]
- Do, N.Q.; Phan, A.T. Monomer-dimer equilibrium for the 5′-5′ stacking of propeller-type parallel-stranded G-quadruplexes: NMR structural study. Chemistry (Easton) 2012, 18, 14752–14759. [Google Scholar] [CrossRef] [PubMed]
- Marchand, C.; Maddali, K.; Metifiot, M.; Pommier, Y. HIV-1 IN Inhibitors: 2010 Update and Perspectives. Curr. Top. Med. Chem. 2009, 9, 1016–1037. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, A.; Petraccone, L.; Scuotto, M.; Vellecco, V.; Bucci, M.; Mayol, L.; Varra, M.; Esposito, V.; Galeone, A. 5-Hydroxymethyl-2′-deoxyuridine residues in the thrombin binding aptamer: Investigating anticoagulant activity by making a tiny chemical modification. ChemBioChem 2014, 15, 2427–2434. [Google Scholar] [CrossRef]
- Virgilio, A.; Petraccone, L.; Vellecco, V.; Bucci, M.; Varra, M.; Irace, C.; Santamaria, R.; Pepe, A.; Mayol, L.; Esposito, V.; et al. Site-specific replacement of the thymine methyl group by fluorine in thrombin binding aptamer significantly improves structural stability and anticoagulant activity. Nucleic Acids Res. 2015, 43, 10602–10611. [Google Scholar] [CrossRef][Green Version]
- Esposito, V.; Pirone, L.; Mayol, L.; Pedone, E.; Virgilio, A.; Galeone, A. Exploring the binding of d(GGGT)4 to the HIV-1 integrase: An approach to investigate G-quadruplex aptamer/target protein interactions. Biochimie 2016, 127, 19–22. [Google Scholar] [CrossRef]
- Virgilio, A.; Amato, T.; Petraccone, L.; Esposito, F.; Grandi, N.; Tramontano, E.; Romero, R.; Haider, S.; Gomez-Monterrey, I.; Novellino, E.; et al. Improvement of the activity of the anti-HIV-1 integrase aptamer T30175 by introducing a modified thymidine into the loops. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Monsen, R.C.; Trent, J.O. G-quadruplex virtual drug screening: A review. Biochimie 2018, 152, 134–148. [Google Scholar] [CrossRef]
- Martino, L.; Virno, A.; Pagano, B.; Virgilio, A.; Di Micco, S.; Galeone, A.; Giancola, C.; Bifulco, G.; Mayol, L.; Randazzo, A. Structural and thermodynamic studies of the interaction of distamycin A with the parallel quadruplex structure [d(TGGGGT)]4. J. Am. Chem. Soc. 2007, 129, 16048–16056. [Google Scholar] [CrossRef] [PubMed]
- Cosconati, S.; Marinelli, L.; Trotta, R.; Virno, A.; De Tito, S.; Romagnoli, R.; Pagano, B.; Limongelli, V.; Giancola, C.; Baraldi, P.G.; et al. Structural and conformational requisites in DNA quadruplex groove binding: Another piece to the puzzle. J. Am. Chem. Soc. 2010, 132, 6425–6433. [Google Scholar] [CrossRef] [PubMed]
- Carcelli, M.; Rogolino, D.; Gatti, A.; Pala, N.; Corona, A.; Caredda, A.; Tramontano, E.; Pannecouque, C.; Naesens, L.; Esposito, F. Chelation motifs affecting metal-dependent viral enzymes: N’-acylhydrazone ligands as dual target inhibitors of HIV-1 integrase and reverse transcriptase ribonuclease h domain. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Esposito, F.; Tramontano, E. Past and future. Current drugs targeting HIV-1 integrase and reverse transcriptase-associated ribonuclease H activity: Single and dual active site inhibitors. Antivir. Chem. Chemother. 2014, 23, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Carcelli, M.; Rogolino, D.; Sechi, M.; Rispoli, G.; Fisicaro, E.; Compari, C.; Grandi, N.; Corona, A.; Tramontano, E.; Pannecouque, C.; et al. Antiretroviral activity of metal-chelating HIV-1 integrase inhibitors. Eur. J. Med. Chem. 2014, 83, 594–600. [Google Scholar] [CrossRef]
- Corona, A.; di Leva, F.S.; Rigogliuso, G.; Pescatori, L.; Madia, V.N.; Subra, F.; Delelis, O.; Esposito, F.; Cadeddu, M.; Costi, R.; et al. New insights into the interaction between pyrrolyl diketoacids and HIV-1 integrase active site and comparison with RNase H. Antivir. Res. 2016, 134, 236–243. [Google Scholar] [CrossRef]
- Sala, M.; Spensiero, A.; Esposito, F.; Scala, M.C.; Vernieri, E.; Bertamino, A.; Manfra, M.; Carotenuto, A.; Grieco, P.; Novellino, E.; et al. Development and identification of a novel anti-HIV-1 peptide derived by modification of the N-terminal domain of HIV-1 integrase. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Esposito, F.; Tintori, C.; Martini, R.; Christ, F.; Debyser, Z.; Ferrarese, R.; Cabiddu, G.; Corona, A.; Ceresola, E.R.; Calcaterra, A.; et al. Kuwanon-L as a New Allosteric HIV-1 Integrase Inhibitor: Molecular Modeling and Biological Evaluation. ChemBioChem 2015, 16, 2507–2512. [Google Scholar] [CrossRef]
- Tintori, C.; Esposito, F.; Morreale, F.; Martini, R.; Tramontano, E.; Botta, M. Investigation on the sucrose binding pocket of HIV-1 Integrase by molecular dynamics and synergy experiments. Bioorganic Med. Chem. Lett. 2015, 25, 3013–3016. [Google Scholar] [CrossRef]
- Martini, R.; Esposito, F.; Corona, A.; Ferrarese, R.; Ceresola, E.R.; Visconti, L.; Tintori, C.; Barbieri, A.; Calcaterra, A.; Iovine, V.; et al. Natural Product Kuwanon-L Inhibits HIV-1 Replication through Multiple Target Binding. ChemBioChem 2017, 18, 374–377. [Google Scholar] [CrossRef]
- Sgobba, M.; Olubiyi, O.; Ke, S.; Haider, S. Molecular dynamics of HIV1-integrase in coomplex with 93del-A Structural Perspective on the Mechanism of Inhibition. J. Biomol. Struct. Dyn. 2012, 29, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, A.; Esposito, V.; Randazzo, A.; Mayol, L.; Galeone, A. Effects of 8-methyl-2′-deoxyadenosine incorporation into quadruplex forming oligodeoxyribonucleotides. Bioorganic Med. Chem. 2005, 13, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Thao Tran, P.L.; Virgilio, A.; Esposito, V.; Citarella, G.; Mergny, J.L.; Galeone, A. Effects of 8-methylguanine on structure, stability and kinetics of formation of tetramolecular quadruplexes. Biochimie 2011, 93, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, A.; Esposito, V.; Citarella, G.; Pepe, A.; Mayol, L.; Galeone, A. The insertion of two 8-methyl-2′-deoxyguanosine residues in tetramolecular quadruplex structures: Trying to orientate the strands. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef]
- Avino, A.; Fabrega, C.; Tintore, M.; Eritja, R. Thrombin binding aptamer, more than a simple aptamer: Chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef]
- He, G.X.; Krawczyk, S.H.; Swaminathan, S.; Shea, R.G.; Dougherty, J.P.; Terhorst, T.; Law, V.S.; Griffin, L.C.; Coutré, S.; Bischofberger, N. N2- and C8-substituted oligodeoxynucleotides with enhanced thrombin inhibitory activity in vitro and in vivo. J. Med. Chem. 1998, 41, 2234–2242. [Google Scholar] [CrossRef]
- Dalvit, C. Efficient multiple-solvent suppression for the study of the interactions of organic solvents with biomolecules. J. Biomol. NMR 1998, 11, 437–444. [Google Scholar] [CrossRef]
- Maple, J.R.; Hwang, M.J.; Stockfisch, T.P.; Dinur, U.; Waldman, M.; Ewig, C.S.; Hagler, A.T. Derivation of class II force fields. I. Methodology and quantum force field for the alkyl functional group and alkane molecules. J. Comput. Chem. 1994, 15, 162–182. [Google Scholar] [CrossRef]
- Rudnicki, W.R.; Lesyng, B. Applicability of commonly used atom-atom type potential energy functions in structural analysis of nucleic acids. The role of electrostatic interactions. Comput. Chem. 1995, 19, 253–258. [Google Scholar] [CrossRef]
- Weiner, S.J.; Kollman, P.A.; Singh, U.C.; Case, D.A.; Ghio, C.; Alagona, G.; Profeta, S.; Weiner, P. A New Force Field for Molecular Mechanical Simulation of Nucleic Acids and Proteins. J. Am. Chem. Soc. 1984, 106, 765–784. [Google Scholar] [CrossRef]
- Esposito, F.; Ambrosio, F.A.; Maleddu, R.; Costa, G.; Rocca, R.; Maccioni, E.; Catalano, R.; Romeo, I.; Eleftheriou, P.; Karia, D.C.; et al. Chromenone derivatives as a versatile scaffold with dual mode of inhibition of HIV-1 reverse transcriptase-associated Ribonuclease H function and integrase activity. Eur. J. Med. Chem. 2019, 182. [Google Scholar] [CrossRef] [PubMed]


| Oligonucleotide | Sequence | Tm (°C) ± 1 | a IC50 IN LEDGF-Independent Activity (µM) |
|---|---|---|---|
| INT-M2 | 5′-GMGTGGGTGGGTGGGT-3′ | 75 | 0.275 ± 0.025 |
| INT-M6 | 5′-GGGTGMGTGGGTGGGT-3′ | 67 | 0.096 ± 0.001 |
| INT-M10 | 5′-GGGTGGGTGMGTGGGT-3′ | 71 | 0.228 ± 0.031 |
| INT-M14 | 5′-GGGTGGGTGGGTGGMGT-3′ | 75 | 0.280 ± 0.060 |
| INT-nat (T30923) | 5′-GGGTGGGTGGGTGGGT-3′ | 88 | 0.088 ± 0.003 |
| Raltegravir | N.A. | N.A. | 0.058 ± 0.002 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esposito, V.; Esposito, F.; Pepe, A.; Gomez Monterrey, I.; Tramontano, E.; Mayol, L.; Virgilio, A.; Galeone, A. Probing the Importance of the G-Quadruplex Grooves for the Activity of the Anti-HIV-Integrase Aptamer T30923. Int. J. Mol. Sci. 2020, 21, 5637. https://doi.org/10.3390/ijms21165637
Esposito V, Esposito F, Pepe A, Gomez Monterrey I, Tramontano E, Mayol L, Virgilio A, Galeone A. Probing the Importance of the G-Quadruplex Grooves for the Activity of the Anti-HIV-Integrase Aptamer T30923. International Journal of Molecular Sciences. 2020; 21(16):5637. https://doi.org/10.3390/ijms21165637
Chicago/Turabian StyleEsposito, Veronica, Francesca Esposito, Antonietta Pepe, Isabel Gomez Monterrey, Enzo Tramontano, Luciano Mayol, Antonella Virgilio, and Aldo Galeone. 2020. "Probing the Importance of the G-Quadruplex Grooves for the Activity of the Anti-HIV-Integrase Aptamer T30923" International Journal of Molecular Sciences 21, no. 16: 5637. https://doi.org/10.3390/ijms21165637
APA StyleEsposito, V., Esposito, F., Pepe, A., Gomez Monterrey, I., Tramontano, E., Mayol, L., Virgilio, A., & Galeone, A. (2020). Probing the Importance of the G-Quadruplex Grooves for the Activity of the Anti-HIV-Integrase Aptamer T30923. International Journal of Molecular Sciences, 21(16), 5637. https://doi.org/10.3390/ijms21165637











