Effects of Maternal Chewing on Prenatal Stress-Induced Cognitive Impairments in the Offspring via Multiple Molecular Pathways
Abstract
1. Introduction
2. Results
2.1. Maternal Chewing during Prenatal Stress Ameliorated Spatial Learning Impairment in the Offspring Induced by Prenatal Stress
2.2. Maternal Chewing during Prenatal Stress Ameliorated Hippocampal Neurogenesis Defects in Offspring Induced by Prenatal Stress
2.3. Maternal Chewing during Prenatal Stress Increased BDNF Expression in the Hippocampus in the Offspring
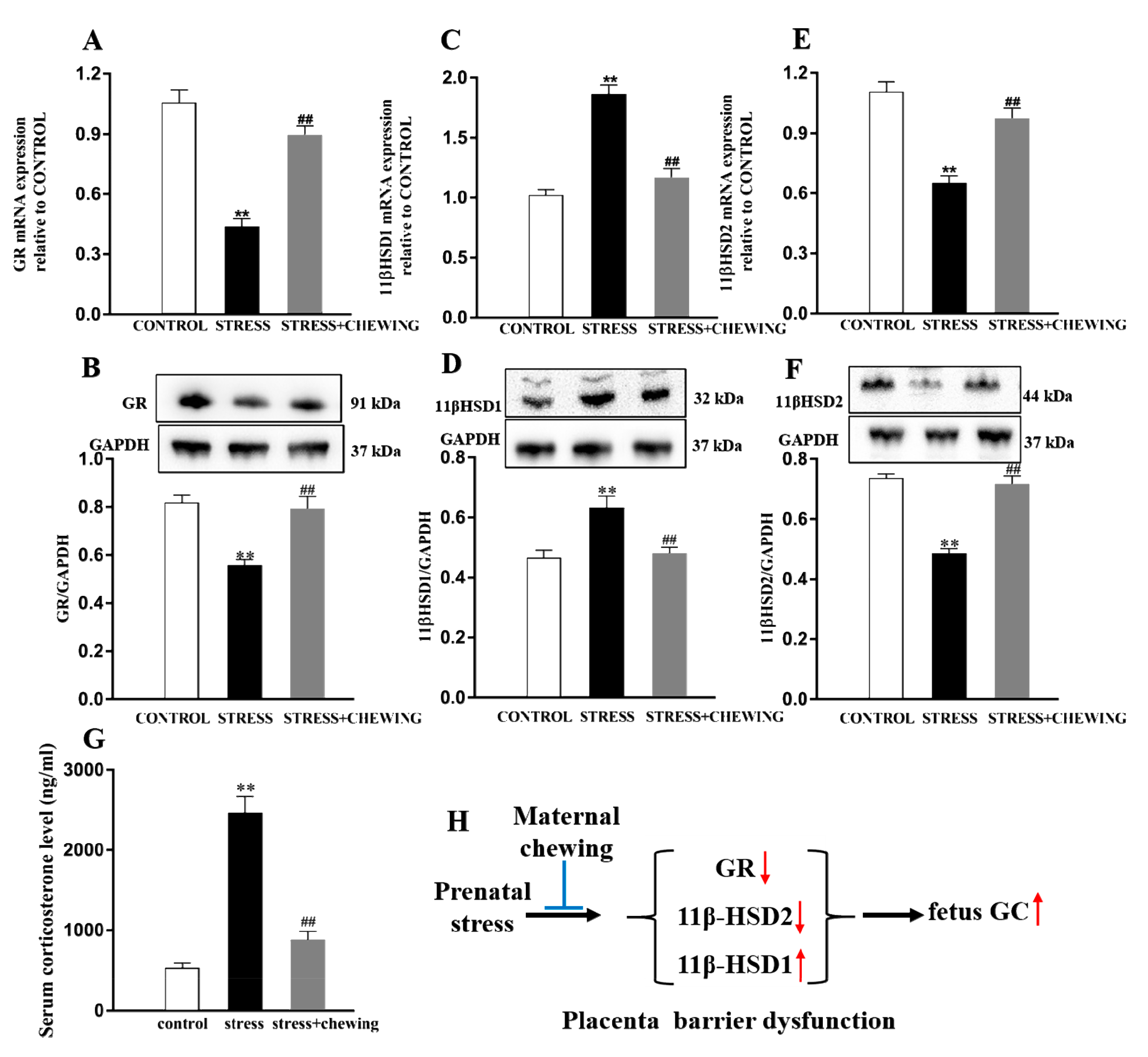
2.4. Maternal Chewing during Prenatal Stress Deceased the GC Level in the Offspring by Protecting the Placenta Barrier Function
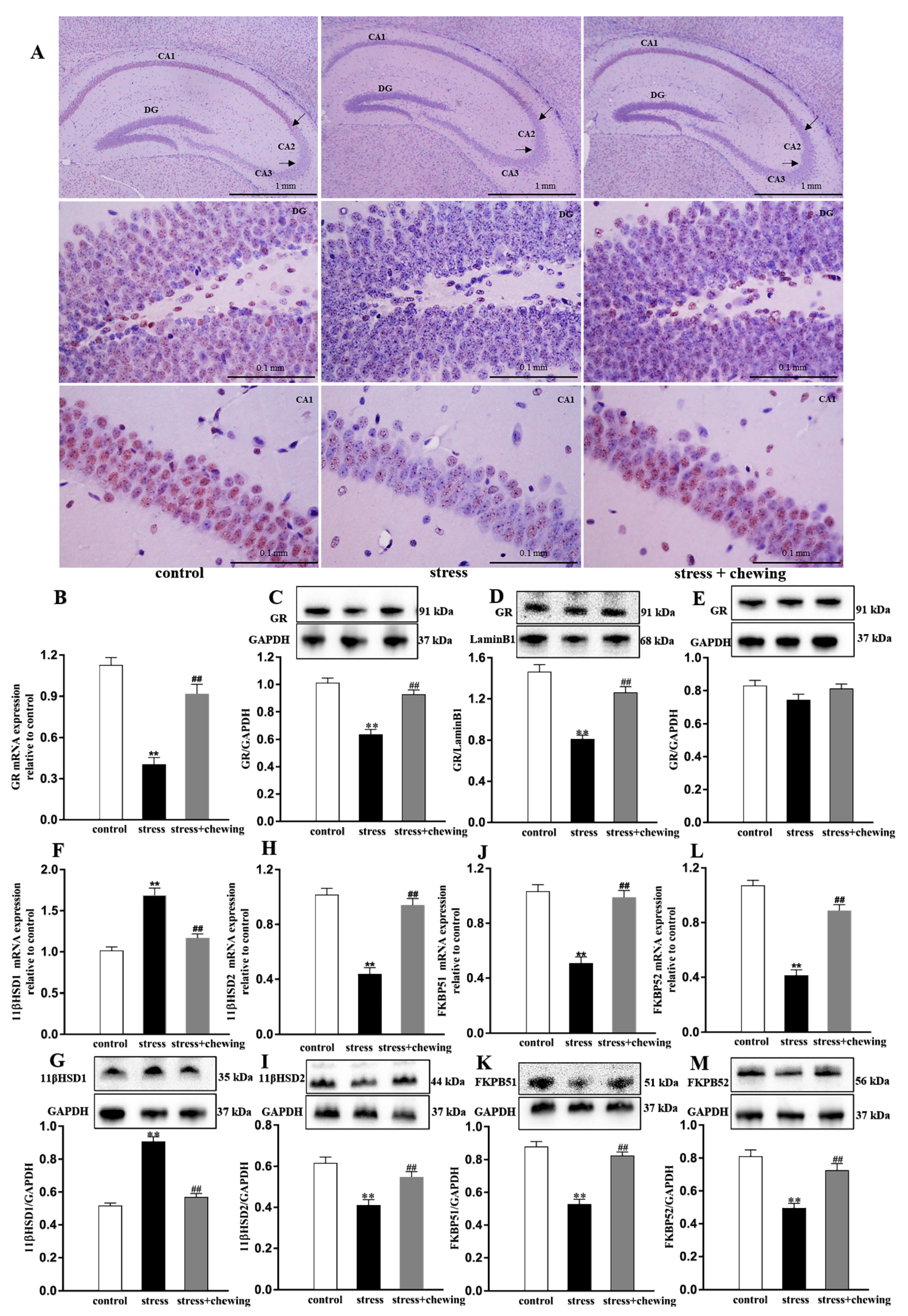
2.5. Maternal Chewing during Prenatal Stress Ameliorated the Nuclear GR Transport Impairment Induced by Prenatal Stress
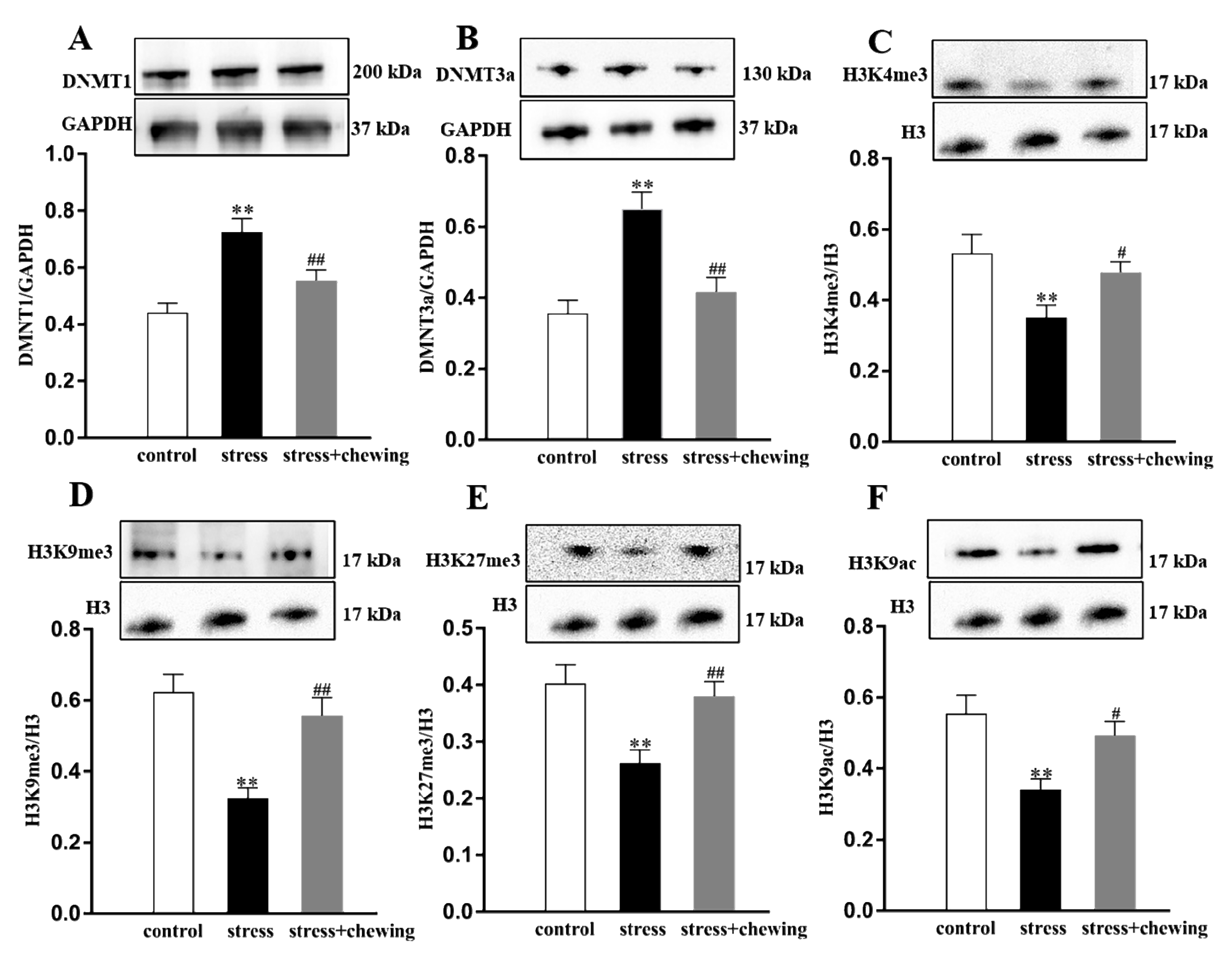
2.6. Maternal Chewing during Prenatal Stress Regulated Enzymes of DNA Methylation as Well as Histone H3 Methylation and Acetylation in the Offspring
3. Discussion
4. Materials and Methods
4.1. Animal Models
4.2. Morris Water Maze Test
4.3. Corticosterone Assay
4.4. Real-Time PCR
4.5. Western Blot Analyses
4.6. Immunohistochemical Staining
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BDNF | Brain-derived neurotrophic factor |
| DG | Dentate gyrus |
| GC | Glucocorticoid |
| GR | Glucocorticoid receptor |
| FKBP51 | FK506-binding protein 51 |
| FKBP52 | FK506-binding protein 52 |
| HPA | Hypothalamic–pituitary–adrenal |
| 11β-HSD1 | 11β-hydroxysteroid dehydrogenase type 1 |
| 11β-HSD2 | 11β-hydroxysteroid dehydrogenase type 2 |
References
- Blair, M.M.; Glynn, L.M.; Sandman, C.A.; Davis, E.P. Prenatal maternal anxiety and early childhood temperament. Stress 2011, 14, 644–651. [Google Scholar] [CrossRef]
- Pierce, L.J.; Thompson, B.L.; Gharib, A.; Schlueter, L.; Reilly, E.; Valdes, V.; Roberts, S.; Conroy, K.; Levitt, P.; Nelson, C.A. Association of Perceived Maternal Stress During the Perinatal Period With Electroencephalography Patterns in 2-Month-Old Infants. JAMA Pediatr. 2019, 173, 561–570. [Google Scholar] [CrossRef]
- Schuurmans, C.; Kurrasch, D.M. Neurodevelopmental consequences of maternal distress: What do we really know? Clin. Genet. 2012, 83, 108–117. [Google Scholar] [CrossRef]
- Modir, F.; Salmani, M.E.; Goudarzi, I.; Lashkarboluki, T.; Abrari, K. Prenatal stress decreases spatial learning and memory retrieval of the adult male offspring of rats. Physiol. Behav. 2014, 129, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shang, Y.; Xiao, X.; Yu, M.; Zhang, T. Prenatal stress-induced impairments of cognitive flexibility and bidirectional synaptic plasticity are possibly associated with autophagy in adolescent male-offspring. Exp. Neurol. 2017, 298, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Q.; Wang, Y.; Hu, J.; Jiang, H.; Cheng, W.; Ma, Y.; Liu, M.; Sun, A.; Zhang, X.; et al. Duloxetine prevents the effects of prenatal stress on depressive-like and anxiety-like behavior and hippocampal expression of pro-inflammatory cytokines in adult male offspring rats. Int. J. Dev. Neurosci. 2016, 55, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Belnoue, L.; Grosjean, N.; Ladevèze, E.; Abrous, D.N.; Koehl, M. Prenatal Stress Inhibits Hippocampal Neurogenesis but Spares Olfactory Bulb Neurogenesis. PLoS ONE 2013, 8, e72972. [Google Scholar] [CrossRef]
- Bock, J.; Murmu, M.S.; Biala, Y.; Weinstock, M.; Braun, K. Prenatal stress and neonatal handling induce sex-specific changes in dendritic complexity and dendritic spine density in hippocampal subregions of prepubertal rats. Neuroscience 2011, 193, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, B.; Yan, C.; Hu, H.; Cai, S.; Liu, J.; Wu, M.; Ouyang, F.; Shen, X. Effects of duration and timing of prenatal stress on hippocampal myelination and synaptophysin expression. Brain Res. 2013, 1527, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.H.S.; Hadinedoushan, H. Prenatal stress induces learning deficits and is associated with a decrease in granules and CA3 cell dendritic tree size in rat hippocampus. Anat. Sci. Int. 2007, 82, 211–217. [Google Scholar] [CrossRef]
- Azuma, K.; Zhou, Q.; Niwa, M.; Kubo, K.-Y. Association between Mastication, the Hippocampus, and the HPA Axis: A Comprehensive Review. Int. J. Mol. Sci. 2017, 18, 1687. [Google Scholar] [CrossRef] [PubMed]
- Suarez, E.B.; Caldwell, A.L.M.; Allen, N.J. Role of astrocyte-synapse interactions in CNS disorders. J. Physiol. 2016, 595, 1903–1916. [Google Scholar] [CrossRef]
- Packer, A. Neocortical neurogenesis and the etiology of autism spectrum disorder. Neurosci. Biobehav. Rev. 2016, 64, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Nishigawa, K.; Suzuki, Y.; Matsuka, Y. Masticatory performance alters stress relief effect of gum chewing. J. Prosthodont. Res. 2015, 59, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.P. Chewing gum and stress reduction. J. Clin. Transl. Res. 2016, 2, 52–54. [Google Scholar] [CrossRef] [PubMed]
- Kubo, K.-Y.; Iinuma, M.; Chen, H. Mastication as a Stress-Coping Behavior. BioMed Res. Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Ono, Y.; Yamamoto, T.; Kubo, K.-Y.; Onozuka, M. Occlusion and brain function: Mastication as a prevention of cognitive dysfunction. J. Oral Rehabil. 2010, 37, 624–640. [Google Scholar] [CrossRef] [PubMed]
- Kubo, K.-Y.; Kotachi, M.; Suzuki, A.; Iinuma, M.; Azuma, K. Chewing during prenatal stress prevents prenatal stress-induced suppression of neurogenesis, anxiety-like behavior and learning deficits in mouse offspring. Int. J. Med Sci. 2018, 15, 849–858. [Google Scholar] [CrossRef]
- Kubo, K.-Y.; Suzuki, A.; Iinuma, M.; Sato, Y.; Nagashio, R.; Ohta, E.; Azuma, K. Vulnerability to stress in mouse offspring is ameliorated when pregnant dams are provided a chewing stick during prenatal stress. Arch. Oral Boil. 2019, 97, 150–155. [Google Scholar] [CrossRef]
- Onishi, M.; Iinuma, M.; Tamura, Y.; Kubo, K.-Y. Learning deficits and suppression of the cell proliferation in the hippocampal dentate gyrus of offspring are attenuated by maternal chewing during prenatal stress. Neurosci. Lett. 2014, 560, 77–80. [Google Scholar] [CrossRef]
- Suzuki, A.; Iinuma, M.; Hayashi, S.; Sato, Y.; Azuma, K.; Kubo, K.-Y. Maternal chewing during prenatal stress ameliorates stress-induced hypomyelination, synaptic alterations, and learning impairment in mouse offspring. Brain Res. 2016, 1651, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.P.; McKlveen, J.M.; Ghosal, S.; Kopp, B.; Wulsin, A.; Makinson, R.; Scheimann, J.; Myers, B. Regulation of the Hypothalamic-Pituitary-Adrenocortical Stress Response. Compr. Physiol. 2016, 6, 603. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.; Gomez, R.; Williams, G.; Lembke, A.; Lazzeroni, L.C.; Murphy, G.M.; Schatzberg, A.F. HPA axis in major depression: Cortisol, clinical symptomatology and genetic variation predict cognition. Mol. Psychiatry 2016, 22, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Nishi, M.; Kawata, M. Brain Corticosteroid Receptor Dynamics and Trafficking: Implications from Live Cell Imaging. Neuroscience 2006, 12, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.B.; Riggs, D.L.; Hessling, M.; Schumacher, F.; Buchner, J.; Smith, D.F. FK506-Binding Protein 52 Phosphorylation: A Potential Mechanism for Regulating Steroid Hormone Receptor Activity. Mol. Endocrinol. 2007, 21, 2956–2967. [Google Scholar] [CrossRef]
- Fries, G.R.; Gassen, N.C.; Rein, T. The FKBP51 Glucocorticoid Receptor Co-Chaperone: Regulation, Function, and Implications in Health and Disease. Int. J. Mol. Sci. 2017, 18, 2614. [Google Scholar] [CrossRef] [PubMed]
- De Quervain, D.; Schwabe, L.; Roozendaal, B. Stress, glucocorticoids and memory: Implications for treating fear-related disorders. Nat. Rev. Neurosci. 2016, 18, 7–19. [Google Scholar] [CrossRef]
- Judd, L.L.; Schettler, P.J.; Brown, E.S.; Wolkowitz, O.M.; Sternberg, E.M.; Bender, B.G.; Bulloch, K.; Cidlowski, J.A.; De Kloet, E.R.; Fardet, L.; et al. Adverse Consequences of Glucocorticoid Medication: Psychological, Cognitive, and Behavioral Effects. Am. J. Psychiatry 2014, 171, 1045–1051. [Google Scholar] [CrossRef]
- Vyas, S.; Rodrigues, A.J.; Silva, J.M.; Tronche, F.; Almeida, O.F.X.; Sousa, N.; Sotiropoulos, I. Chronic Stress and Glucocorticoids: From Neuronal Plasticity to Neurodegeneration. Neural Plast. 2016, 2016, 1–15. [Google Scholar] [CrossRef]
- Gross, M.; Romi, H.; Gilimovich, Y.; Drori, E.; Pinhasov, A. Placental glucocorticoid receptor and 11beta-hydroxysteroid dehydrogenase-2 recruitment indicates impact of prenatal adversity upon postnatal development in mice. Stress 2018, 21, 474–483. [Google Scholar] [CrossRef]
- Ni, L.; Pan, Y.; Tang, C.; Xiong, W.; Wu, X.; Zou, C. Antenatal exposure to betamethasone induces placental 11beta-hydroxysteroid dehydrogenase type 2 expression and the adult metabolic disorders in mice. PLoS ONE 2018, 13, e0203802. [Google Scholar] [CrossRef] [PubMed]
- Raikkonen, K.; Pesonen, A.-K.; O’Reilly, J.R.; Tuovinen, S.; Lahti-Pulkkinen, M.; Kajantie, E.; Villa, P.; Laivuori, H.; Hämäläinen, E.; Seckl, J.R.; et al. Maternal depressive symptoms during pregnancy, placental expression of genes regulating glucocorticoid and serotonin function and infant regulatory behaviors. Psychol. Med. 2015, 45, 3217–3226. [Google Scholar] [CrossRef]
- Sakkiah, S.; Meganathan, C.; Sohn, Y.S.; Namadevan, S.; Lee, K.W. Identification of important chemical features of 11beta-hydroxysteroid dehydrogenase type1 inhibitors: Application of ligand based virtual screening and density functional theory. Int. J. Mol. Sci. 2012, 13, 5138–5162. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Kipari, T.M.J.; Zhang, Z.; Man, T.Y.; Forster, T.; Homer, N.Z.M.; Seckl, J.R.; Holmes, M.C.; Chapman, K.E. 11beta-hydroxysteroid dehydrogenase-1 deficiency alters brain energy metabolism in acute systemic inflammation. Brain Behav. Immun. 2018, 69, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Jeanneteau, F.; Chao, M.V. Are BDNF and glucocorticoid activities calibrated? Neuroscience 2013, 239, 173–195. [Google Scholar] [CrossRef]
- Daskalakis, N.P.; De Kloet, E.R.; Yehuda, R.; Malaspina, D.; Kranz, T.M. Early Life Stress Effects on Glucocorticoid—BDNF Interplay in the Hippocampus. Front. Mol. Neurosci. 2015, 8, 68. [Google Scholar] [CrossRef]
- Benoit, J.D.; Rakic, P.; Frick, K.M. Prenatal stress induces spatial memory deficits and epigenetic changes in the hippocampus indicative of heterochromatin formation and reduced gene expression. Behav. Brain Res. 2015, 281, 1–8. [Google Scholar] [CrossRef]
- Kwapis, J.L.; Alaghband, Y.; Kramár, E.A.; López, A.J.; Vogel-Ciernia, A.; White, A.O.; Shu, G.; Rhee, D.; Michael, C.M.; Montellier, E.; et al. Epigenetic regulation of the circadian gene Per1 contributes to age-related changes in hippocampal memory. Nat. Commun. 2018, 9, 3323. [Google Scholar] [CrossRef]
- Singh, P.; Konar, A.; Kumar, A.; Srivas, S.; Thakur, M.K. Hippocampal chromatin-modifying enzymes are pivotal for scopolamine-induced synaptic plasticity gene expression changes and memory impairment. J. Neurochem. 2015, 134, 642–651. [Google Scholar] [CrossRef]
- Chen, Y.; Öztürk, N.C.; Zhou, F.C. DNA Methylation Program in Developing Hippocampus and Its Alteration by Alcohol. PLoS ONE 2013, 8, e60503. [Google Scholar] [CrossRef]
- Gu, X.; Xu, Y.; Xue, W.-Z.; Wu, Y.; Ye, Z.; Xiao, G.; Wang, H.-L. Interplay of miR-137 and EZH2 contributes to the genome-wide redistribution of H3K27me3 underlying the Pb-induced memory impairment. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kim, S.Y.; Artis, S.; Molfese, D.L.; Schumacher, A.; Sweatt, J.D.; Paylor, R.E.; Lubin, F.D. Histone methylation regulates memory formation. J. Neurosci. 2010, 30, 3589–3599. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, P.; Garcia, V.; Dornelles, A.D.S.; Da Silva, V.; Maurmann, N.; Portal, B.; Ferreira, R.; Piazza, F.; Roesler, R.; Schröder, N. Memory impairment induced by brain iron overload is accompanied by reduced H3K9 acetylation and ameliorated by sodium butyrate. Neuroscience 2012, 200, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Anacker, C.; Hen, R. Adult hippocampal neurogenesis and cognitive flexibility—linking memory and mood. Nat. Rev. Neurosci. 2017, 18, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.-L.; Ma, X.-T.; Wang, J.-J.; Liu, H.; Chen, Y.; Yang, Y. Physical exercise induces hippocampal neurogenesis and prevents cognitive decline. Behav. Brain Res. 2017, 317, 332–339. [Google Scholar] [CrossRef]
- Cho, K.-O.; Lybrand, Z.R.; Ito, N.; Brulet, R.; Tafacory, F.; Zhang, L.; Good, L.; Ure, K.; Kernie, S.G.; Birnbaum, S.G.; et al. Aberrant hippocampal neurogenesis contributes to epilepsy and associated cognitive decline. Nat. Commun. 2015, 6, 6606. [Google Scholar] [CrossRef]
- Poulose, S.M.; Miller, M.G.; Scott, T.; Shukitt-Hale, B. Nutritional Factors Affecting Adult Neurogenesis and Cognitive Function. Adv. Nutr. 2017, 8, 804–811. [Google Scholar] [CrossRef]
- Barbie-Shoshani, Y.; Shoham, S.; Bejar, C.; Weinstock, M. Sex-Specific Effects of Prenatal Stress on Memory and Markers of Neuronal Activity in Juvenile Rats. Dev. Neurosci. 2016, 38, 206–219. [Google Scholar] [CrossRef]
- Grundwald, N.J.; Brunton, P.J. Prenatal stress programs neuroendocrine stress responses and affective behaviors in second generation rats in a sex-dependent manner. Psychoneuroendocrinology 2015, 62, 204–216. [Google Scholar] [CrossRef]
- Schwabe, L.; Bohbot, V.D.; Wolf, O.T. Prenatal stress changes learning strategies in adulthood. Hippocampus 2012, 22, 2136–2143. [Google Scholar] [CrossRef]
- Kowiański, P.; Lietzau, G.; Czuba, E.; Waśkow, M.; Steliga, A.; Moryś, J. BDNF: A Key Factor with Multipotent Impact on Brain Signaling and Synaptic Plasticity. Cell. Mol. Neurobiol. 2017, 38, 579–593. [Google Scholar] [CrossRef] [PubMed]
- McGowan, P.O.; Matthews, S.G. Prenatal Stress, Glucocorticoids, and Developmental Programming of the Stress Response. Endocrinology 2017, 159, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Plant, D.; Pawlby, S.; Sharp, D.; Zunszain, P.A.; Pariante, C.M. Prenatal maternal depression is associated with offspring inflammation at 25 years: A prospective longitudinal cohort study. Transl. Psychiatry 2016, 6, e936. [Google Scholar] [CrossRef] [PubMed]
- Boero, G.; Pisu, M.G.; Biggio, F.; Muredda, L.; Carta, G.; Banni, S.; Paci, E.; Follesa, P.; Concas, A.; Porcu, P.; et al. Impaired glucocorticoid-mediated HPA axis negative feedback induced by juvenile social isolation in male rats. Neuropharmacology 2018, 133, 242–253. [Google Scholar] [CrossRef]
- Gjerstad, J.K.; Lightman, S.; Spiga, F. Role of glucocorticoid negative feedback in the regulation of HPA axis pulsatility. Stress 2018, 21, 403–416. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, W.-S.; Zuo, R.; Sun, K. Mechanisms for establishment of the placental glucocorticoid barrier, a guard for life. Cell. Mol. Life Sci. 2018, 76, 13–26. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, W.; Li, W.; Liu, C.; Chen, Y.; Yang, Q.; Wang, Y.; Sun, K. Inhibition of 11beta-HSD2 expression by triclosan via induction of apoptosis in human placental syncytiotrophoblasts. J. Clin. Endocrinol. Metab. 2015, 100, E542–E549. [Google Scholar] [CrossRef][Green Version]
- Clifton, V.L.; Cuffe, J.S.; Moritz, K.M.; Cole, T.; Fuller, P.J.; Lu, N.; Kumar, S.; Chong, S.; Saif, Z. Review: The role of multiple placental glucocorticoid receptor isoforms in adapting to the maternal environment and regulating fetal growth. Placenta 2017, 54, 24–29. [Google Scholar] [CrossRef]
- Chapman, K.; Holmes, M.; Seckl, J. 11beta-hydroxysteroid dehydrogenases: Intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 2013, 93, 1139–1206. [Google Scholar] [CrossRef]
- Sivils, J.C.; Storer, C.L.; Galigniana, M.D.; Cox, M.B. Regulation of steroid hormone receptor function by the 52-kDa FK506-binding protein (FKBP52). Curr. Opin. Pharmacol. 2011, 11, 314–319. [Google Scholar] [CrossRef]
- Nishi, M.; Ogawa, H.; Ito, T.; Matsuda, K.I.; Kawata, M. Dynamic changes in subcellular localization of mineralocorticoid receptor in living cells: In comparison with glucocorticoid receptor using dual-color labeling with green fluorescent protein spectral variants. Mol. Endocrinol. 2001, 15, 1077–1092. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rensel, M.A.; Ding, J.A.; Pradhan, D.S.; Schlinger, B.A. 11beta-HSD Types 1 and 2 in the Songbird Brain. Front. Endocrinol. (Lausanne) 2018, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Chater-Diehl, E.; Laufer, B.I.; Singh, S.M. Changes to histone modifications following prenatal alcohol exposure: An emerging picture. Alcohol 2017, 60, 41–52. [Google Scholar] [CrossRef]
- Richmond, R.C.; Simpkin, A.; Woodward, G.; Gaunt, T.R.; Lyttleton, O.; McArdle, W.L.; Ring, S.M.; Smith, A.D.; Timpson, N.J.; Tilling, K.; et al. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: Findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum. Mol. Genet. 2014, 24, 2201–2217. [Google Scholar] [CrossRef] [PubMed]
- Chomyk, A.M.; Volsko, C.; Tripathi, A.K.; Deckard, S.A.; Trapp, B.D.; Fox, R.J.; Dutta, R. DNA methylation in demyelinated multiple sclerosis hippocampus. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Brusslan, J.A.; Bonora, G.; Rus-Canterbury, A.M.; Tariq, F.; Jaroszewicz, A.; Pellegrini, M. A Genome-Wide Chronological Study of Gene Expression and Two Histone Modifications, H3K4me3 and H3K9ac, during Developmental Leaf Senescence. Plant Physiol. 2015, 168, 1246–1261. [Google Scholar] [CrossRef] [PubMed]
- Igolkina, A.A.; Zinkevich, A.; Karandasheva, K.O.; Popov, A.A.; Selifanova, M.V.; Buzdin, A.; Tkachev, V.; Penzar, D.D.; Nikitin, D.; Buzdin, A.A. H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 Histone Tags Suggest Distinct Regulatory Evolution of Open and Condensed Chromatin Landmarks. Cells 2019, 8, 1034. [Google Scholar] [CrossRef]
- Hunter, R.G.; McCarthy, K.J.; Milne, T.A.; Pfaff, D.; McEwen, B.S. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc. Natl. Acad. Sci. 2009, 106, 20912–20917. [Google Scholar] [CrossRef]
- Abbott, P.W.; Gumusoglu, S.B.; Bittle, J.; Beversdorf, D.Q.; Stevens, E.H. Prenatal stress and genetic risk: How prenatal stress interacts with genetics to alter risk for psychiatric illness. Psychoneuroendocrinology 2018, 90, 9–21. [Google Scholar] [CrossRef]
- Mueller, B.R.; Bale, T.L. Sex-specific programming of offspring emotionality after stress early in pregnancy. J. Neurosci. 2008, 28, 9055–9065. [Google Scholar] [CrossRef]
- Zohsel, K.; Buchmann, A.F.; Blomeyer, D.; Hohm, E.; Schmidt, M.H.; Esser, G.; Brandeis, D.; Banaschewski, T.; Laucht, M. Mothers’ prenatal stress and their children’s antisocial outcomes—a moderating role for the Dopamine D4 Receptor (DRD4) gene. J. Child Psychol. Psychiatry 2013, 55, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Choi, B.Y.; Kho, A.R.; Jeong, J.H.; Hong, D.K.; Kang, D.H.; Kang, B.S.; Song, H.K.; Choi, H.-C.; Suh, S.W. Inhibition of NADPH Oxidase Activation by Apocynin Rescues Seizure-Induced Reduction of Adult Hippocampal Neurogenesis. Int. J. Mol. Sci. 2018, 19, 3087. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Q.; Suzuki, A.; Iinuma, M.; Wang, K.-Y.; Kubo, K.-y.; Azuma, K. Effects of Maternal Chewing on Prenatal Stress-Induced Cognitive Impairments in the Offspring via Multiple Molecular Pathways. Int. J. Mol. Sci. 2020, 21, 5627. https://doi.org/10.3390/ijms21165627
Zhou Q, Suzuki A, Iinuma M, Wang K-Y, Kubo K-y, Azuma K. Effects of Maternal Chewing on Prenatal Stress-Induced Cognitive Impairments in the Offspring via Multiple Molecular Pathways. International Journal of Molecular Sciences. 2020; 21(16):5627. https://doi.org/10.3390/ijms21165627
Chicago/Turabian StyleZhou, Qian, Ayumi Suzuki, Mitsuo Iinuma, Ke-Yong Wang, Kin-ya Kubo, and Kagaku Azuma. 2020. "Effects of Maternal Chewing on Prenatal Stress-Induced Cognitive Impairments in the Offspring via Multiple Molecular Pathways" International Journal of Molecular Sciences 21, no. 16: 5627. https://doi.org/10.3390/ijms21165627
APA StyleZhou, Q., Suzuki, A., Iinuma, M., Wang, K.-Y., Kubo, K.-y., & Azuma, K. (2020). Effects of Maternal Chewing on Prenatal Stress-Induced Cognitive Impairments in the Offspring via Multiple Molecular Pathways. International Journal of Molecular Sciences, 21(16), 5627. https://doi.org/10.3390/ijms21165627




