Hsp22 with an N-Terminal Domain Truncation Mediates a Reduction in Tau Protein Levels
Abstract
1. Introduction
2. Results
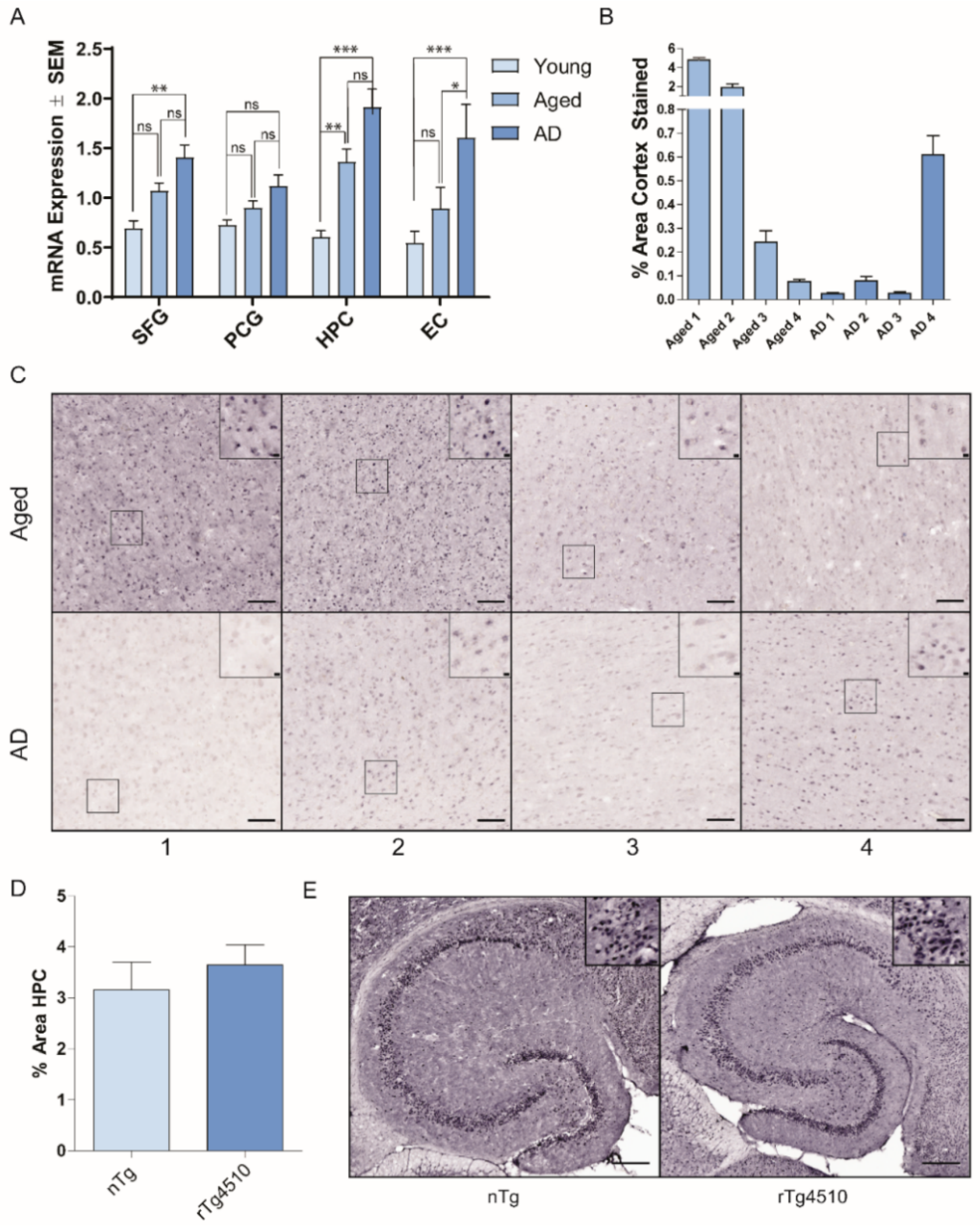
2.1. Hsp22 Expression with Age and in AD
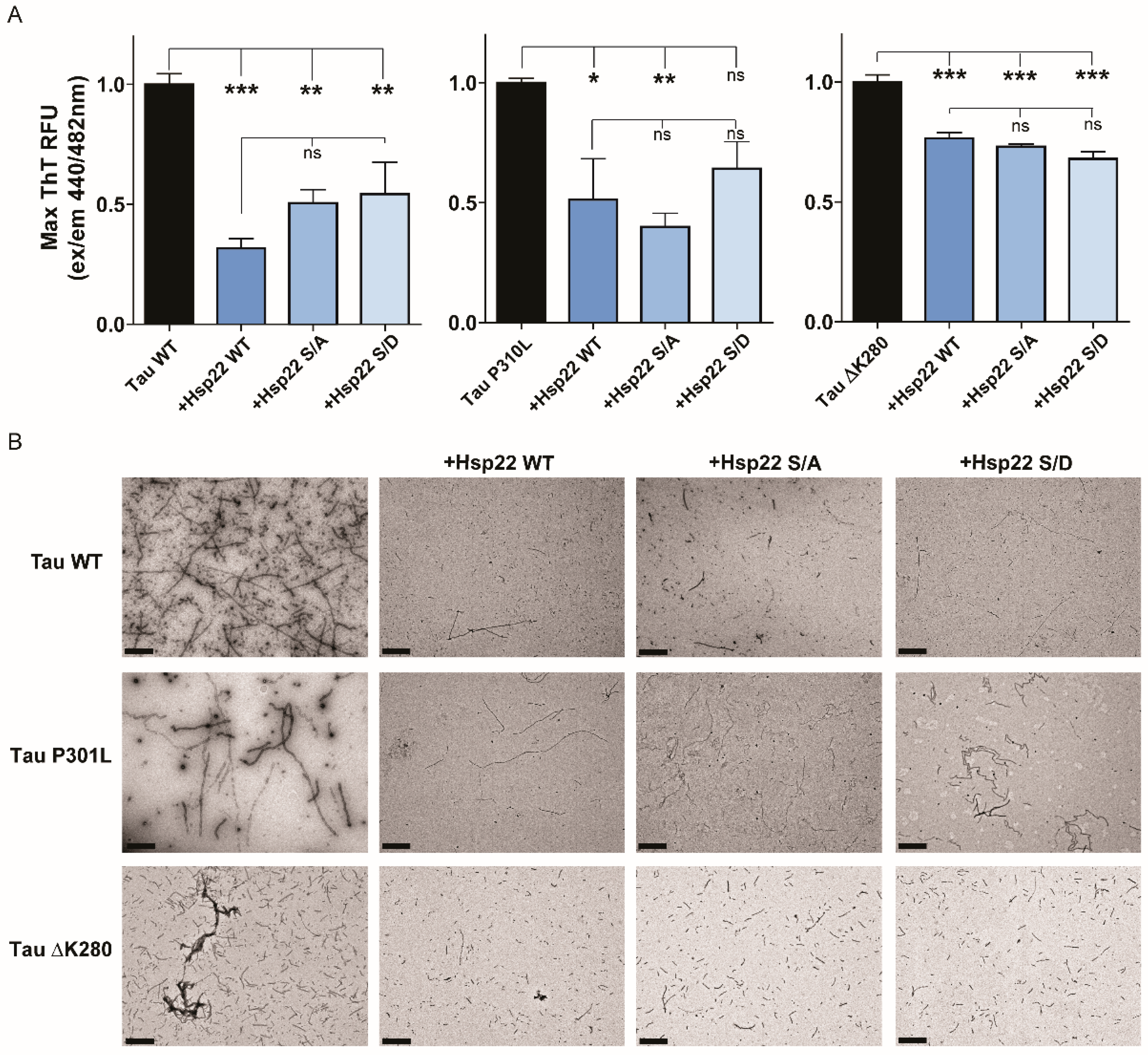
2.2. Hsp22 Prevents the Formation of Tau Aggregates In Vitro
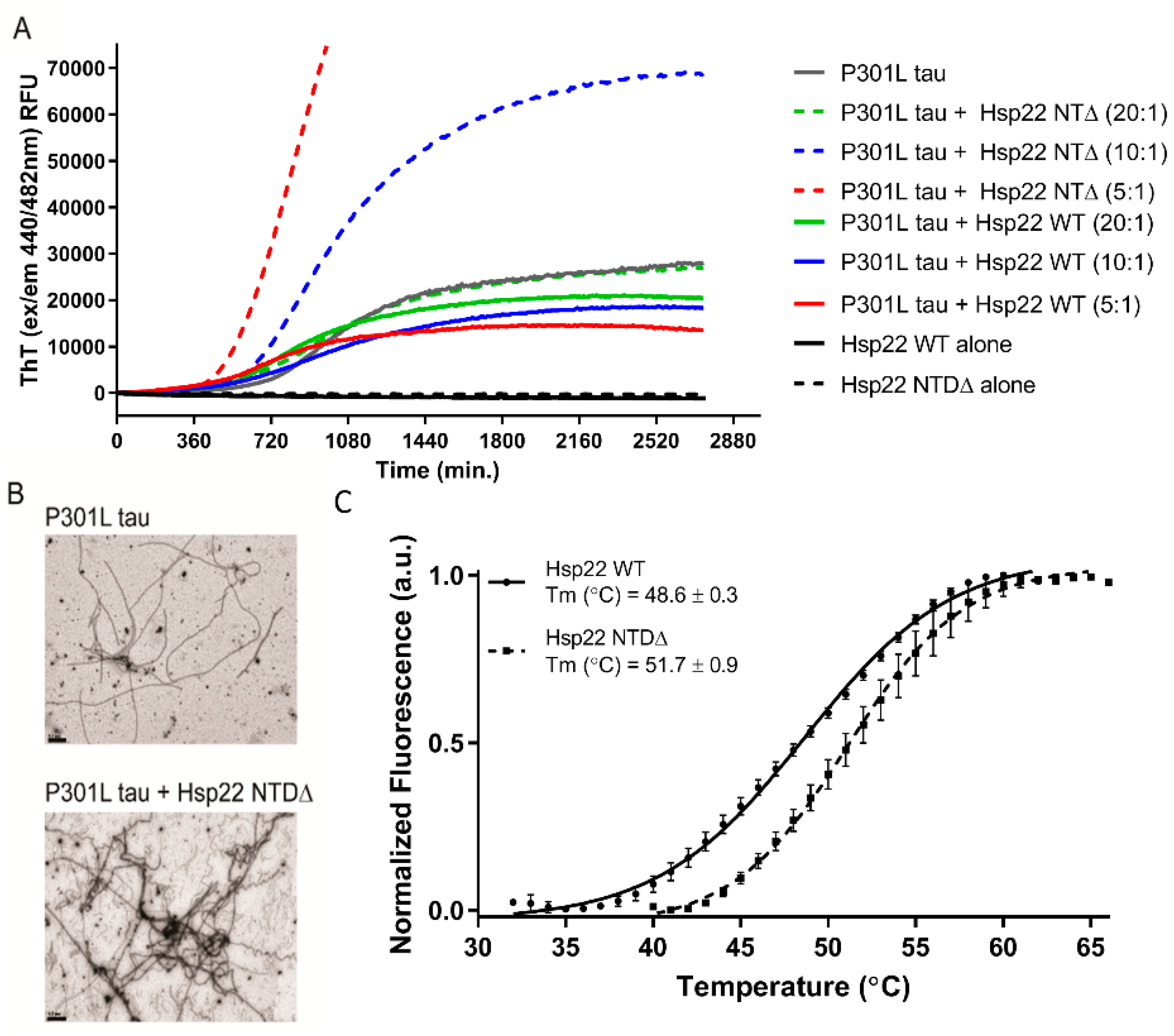
2.3. Hsp22 NTDΔ Exacerbates Tau P301L Aggregate Formation In Vitro
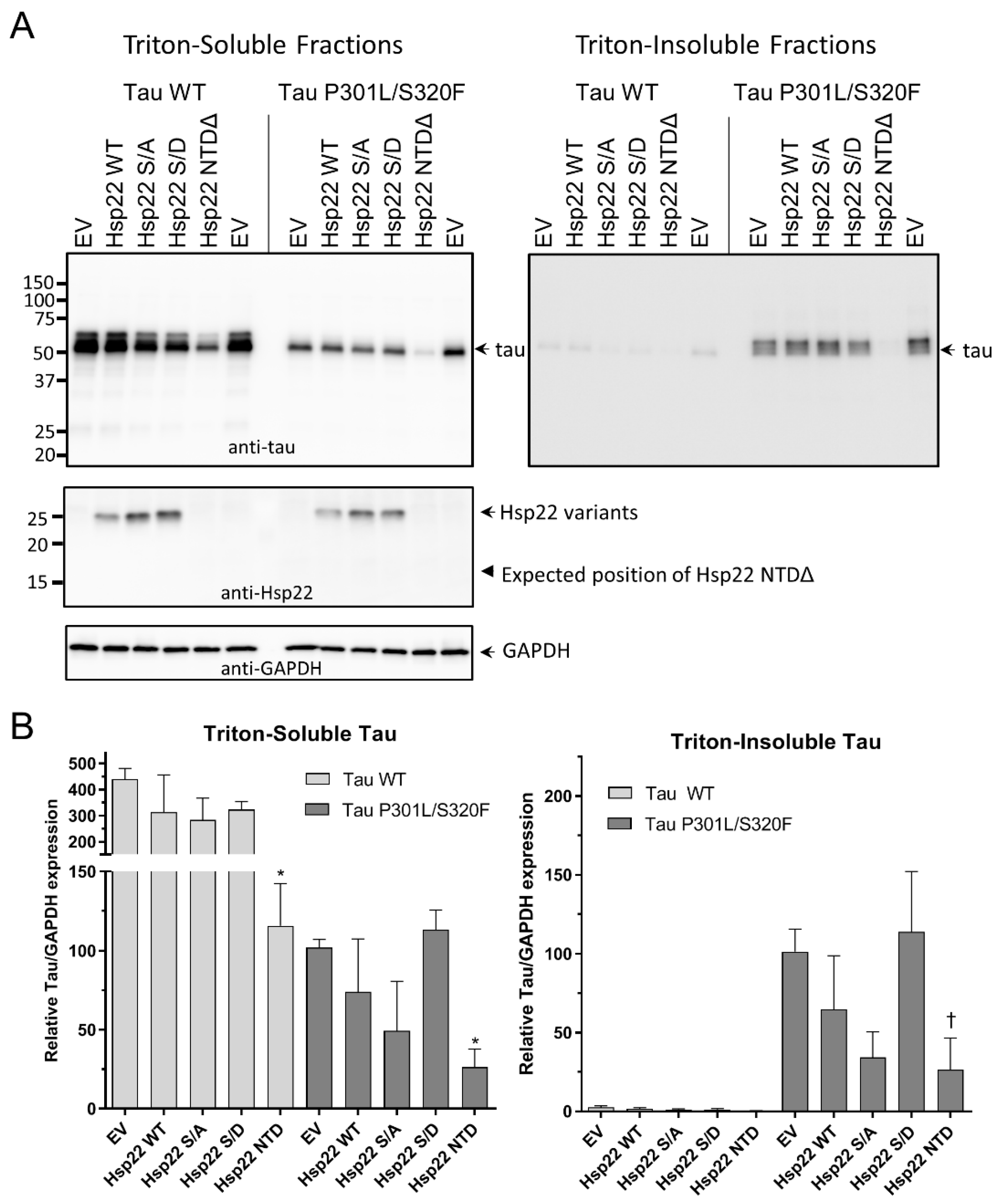
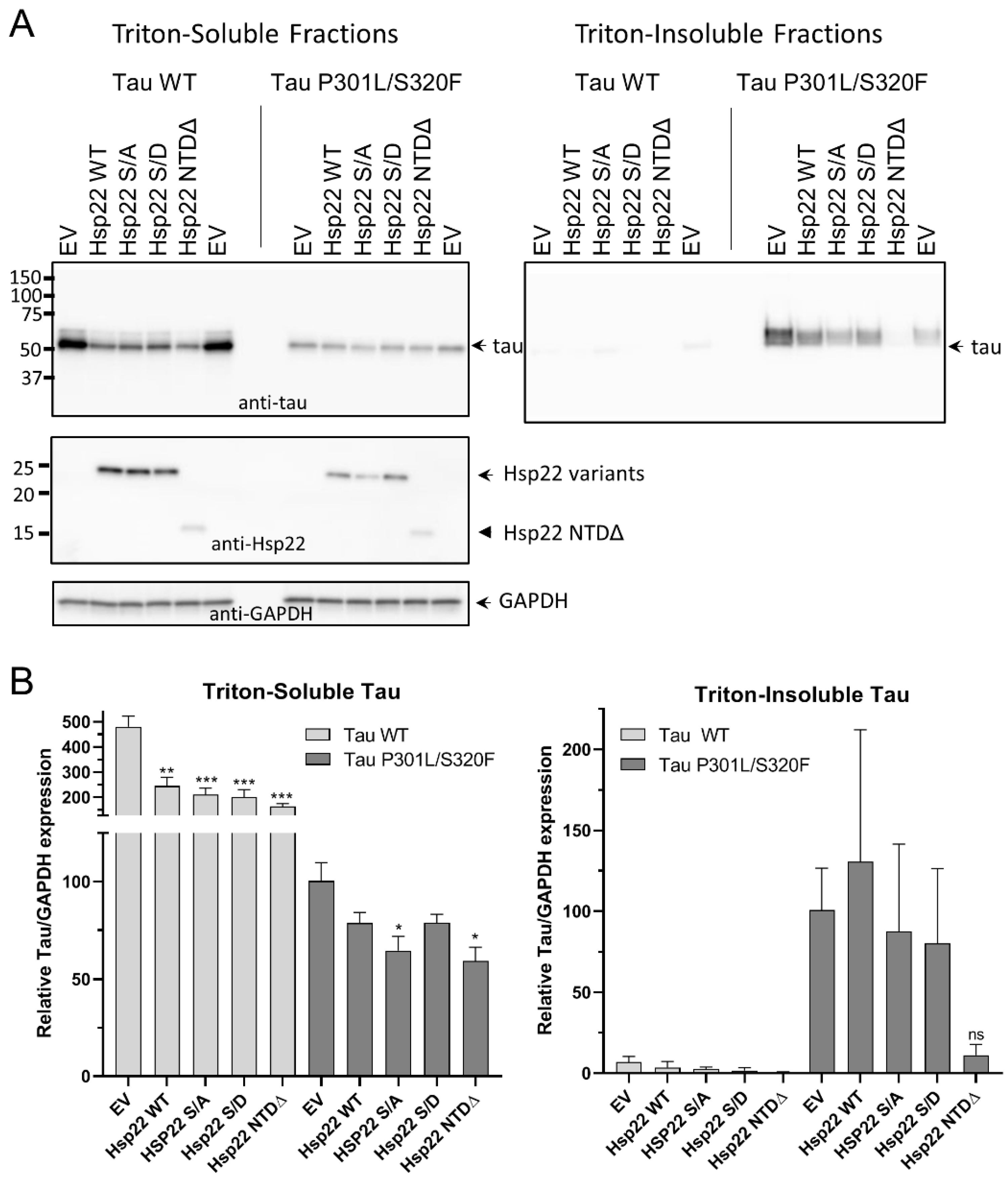
2.4. Hsp22 NTDΔ Enhances the Clearance of Tau Protein in HEK293T Cells
2.5. Hsp22 WT and Phosphorylation Site Mutants Enhance the Clearance of Tau Protein in HEK293T Cells at High sHsp Ratios
3. Discussion
4. Materials and Methods
4.1. Hsp22 mRNA Expression in Human Brain Tissue
4.2. Transgenic Animal Brain Tissue Preparation
4.3. Anti-Hsp22 Immunohistochemistry Analysis
4.4. Analysis of Intrinsic Disorder Predisposition
4.5. Molecular Cloning
4.6. Protein Expression and Purification
4.7. Thioflavin T Fluorescence Assay
4.8. Transmission Electron Microscopy
4.9. Differential Scanning Fluorimetry
4.10. HEK 293T Co-Transfection and Fractionation
4.11. Western Blots
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Powers, E.T.; Morimoto, R.I.; Dillin, A.; Kelly, J.W.; Balch, W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009, 78, 959–991. [Google Scholar] [CrossRef] [PubMed]
- Klaips, C.L.; Jayaraj, G.G.; Hartl, F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2018, 217, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Fox, S.G.; Cavallini, A.; Kerridge, C.; O’Neill, M.J.; Wolak, J.; Bose, S.; Morimoto, R.I. Tau protein aggregates inhibit the protein-folding and vesicular trafficking arms of the cellular proteostasis network. J. Biol. Chem. 2019, 294, 7917–7930. [Google Scholar] [CrossRef] [PubMed]
- Skibinski, G.; Finkbeiner, S. Longitudinal measures of proteostasis in live neurons: Features that determine fate in models of neurodegenerative disease. FEBS Lett. 2013, 587, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.L.; Li, W.; Cheetham, M.E. Molecular chaperones and neuronal proteostasis. Semin. Cell Dev. Biol. 2015, 40, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Yerbury, J.J.; Ooi, L.; Dillin, A.; Saunders, D.N.; Hatters, D.M.; Beart, P.M.; Cashman, N.R.; Wilson, M.R.; Ecroyd, H. Walking the tightrope: Proteostasis and neurodegenerative disease. J. Neurochem. 2016, 137, 489–505. [Google Scholar] [CrossRef]
- Kurtishi, A.; Rosen, B.; Patil, K.S.; Alves, G.W.; Møller, S.G. Cellular Proteostasis in Neurodegeneration. Mol. Neurobiol. 2019, 56, 3676–3689. [Google Scholar] [CrossRef]
- Sweeney, P.; Park, H.; Baumann, M.; Dunlop, J.; Frydman, J.; Kopito, R.; McCampbell, A.; Leblanc, G.; Venkateswaran, A.; Nurmi, A.; et al. Protein misfolding in neurodegenerative diseases: Implications and strategies. Transl. Neurodegener. 2017, 6, 6. [Google Scholar] [CrossRef]
- Jagust, W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat. Rev. Neurosci. 2018, 19, 687–700. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef] [PubMed]
- Balch, W.E.; Morimoto, R.I.; Dillin, A.; Kelly, J.W. Adapting Proteostasis for Disease Intervention. Science 2008, 319, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Eisele, Y.S.; Monteiro, C.; Fearns, C.; Encalada, S.E.; Wiseman, R.L.; Powers, E.T.; Kelly, J.W. Targeting Protein Aggregation for the Treatment of Degenerative Diseases. Nat. Rev. Drug Discov. 2015, 14, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Shelton, L.B.; Baker, J.D.; Zheng, D.; Sullivan, L.E.; Solanki, P.K.; Webster, J.M.; Sun, Z.; Sabbagh, J.J.; Nordhues, B.A.; Koren, J.; et al. Hsp90 activator Aha1 drives production of pathological tau aggregates. Proc. Natl. Acad. Sci. USA 2017, 114, 9707–9712. [Google Scholar] [CrossRef]
- Baker, J.D.; Shelton, L.B.; Zheng, D.; Favretto, F.; Nordhues, B.A.; Darling, A.; Sullivan, L.E.; Sun, Z.; Solanki, P.K.; Martin, M.D.; et al. Human cyclophilin 40 unravels neurotoxic amyloids. PLoS Biol. 2017, 15, e2001336. [Google Scholar] [CrossRef]
- Blair, L.J.; Sabbagh, J.J.; Dickey, C.A. Targeting Hsp90 and its co-chaperones to treat Alzheimer’s disease. Expert Opin. Ther. Targets 2014, 18, 1219–1232. [Google Scholar] [CrossRef]
- Muchowski, P.J.; Wacker, J.L. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 2005, 6, 11–22. [Google Scholar] [CrossRef]
- Lindberg, I.; Shorter, J.; Wiseman, R.L.; Chiti, F.; Dickey, C.A.; McLean, P.J. Chaperones in Neurodegeneration. J. Neurosci. 2015, 35, 13853–13859. [Google Scholar] [CrossRef]
- Bakthisaran, R.; Tangirala, R.; Rao, C.M. Small heat shock proteins: Role in cellular functions and pathology. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2015, 1854, 291–319. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A First Line of Stress Defense: Small Heat Shock Proteins and their function in protein homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef]
- Sudnitsyna, M.V.; Mymrikov, E.V.; Seit-Nebi, A.S.; Gusev, N.B. The role of intrinsically disordered regions in the structure and functioning of small heat shock proteins. Curr. Protein Pept. Sci. 2012, 13, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Haslbeck, M.; Franzmann, T.; Weinfurtner, D.; Buchner, J. Some like it hot: The structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 2005, 12, 842–846. [Google Scholar] [CrossRef] [PubMed]
- Freilich, R.; Betegon, M.; Tse, E.; Mok, S.-A.; Julien, O.; Agard, D.A.; Southworth, D.R.; Takeuchi, K.; Gestwicki, J.E. Competing protein-protein interactions regulate binding of Hsp27 to its client protein tau. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Haslbeck, M.; Weinkauf, S.; Buchner, J. Small heat shock proteins: Simplicity meets complexity. J. Biol. Chem. 2019, 294, 2121–2132. [Google Scholar] [CrossRef] [PubMed]
- Mymrikov, E.V.; Daake, M.; Richter, B.; Haslbeck, M.; Buchner, J. The Chaperone Activity and Substrate Spectrum of Human Small Heat Shock Proteins. J. Biol. Chem. 2017, 292, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Abisambra, J.F.; Blair, L.J.; Hill, S.E.; Jones, J.R.; Kraft, C.; Rogers, J.; Koren, J.; Jinwal, U.K.; Lawson, L.; Johnson, A.G.; et al. Phosphorylation Dynamics Regulate Hsp27-Mediated Rescue of Neuronal Plasticity Deficits in Tau Transgenic Mice. J. Neurosci. 2010, 30, 15374–15382. [Google Scholar] [CrossRef]
- Baughman, H.E.R.; Clouser, A.F.; Klevit, R.E.; Nath, A. HspB1 and Hsc70 chaperones engage distinct tau species and have different inhibitory effects on amyloid formation. J. Biol. Chem. 2018, 293, 2687–2700. [Google Scholar] [CrossRef]
- Mok, S.-A.; Condello, C.; Freilich, R.; Gillies, A.; Arhar, T.; Oroz, J.; Kadavath, H.; Julien, O.; Assimon, V.A.; Rauch, J.N.; et al. Mapping interactions with the chaperone network reveals factors that protect against tau aggregation. Nat. Struct. Mol. Biol. 2018, 25, 384–393. [Google Scholar] [CrossRef]
- Kametani, F.; Hasegawa, M. Reconsideration of Amyloid Hypothesis and Tau Hypothesis in Alzheimer’s Disease. Front. Neurosci. 2018, 12. [Google Scholar] [CrossRef]
- Wolfe, M.S. The Role of Tau in Neurodegenerative Diseases and Its Potential as a Therapeutic Target. Available online: https://www.hindawi.com/journals/scientifica/2012/796024/ (accessed on 23 December 2019).
- Behl, C. BAG3 and friends: Co-chaperones in selective autophagy during aging and disease. Autophagy 2011, 7, 795–798. [Google Scholar] [CrossRef]
- Li, F.; Xiao, H.; Hu, Z.; Zhou, F.; Yang, B. Exploring the multifaceted roles of heat shock protein B8 (HSPB8) in diseases. Eur. J. Cell Biol. 2018, 97, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Seidel, K.; Vinet, J.; den Dunnen, W.F.A.; Brunt, E.R.; Meister, M.; Boncoraglio, A.; Zijlstra, M.P.; Boddeke, H.W.G.M.; Rüb, U.; Kampinga, H.H.; et al. The HSPB8-BAG3 chaperone complex is upregulated in astrocytes in the human brain affected by protein aggregation diseases. Neuropathol. Appl. Neurobiol. 2012, 38, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Cristofani, R.; Crippa, V.; Vezzoli, G.; Rusmini, P.; Galbiati, M.; Cicardi, M.E.; Meroni, M.; Ferrari, V.; Tedesco, B.; Piccolella, M.; et al. The small heat shock protein B8 (HSPB8) efficiently removes aggregating species of dipeptides produced in C9ORF72-related neurodegenerative diseases. Cell Stress Chaperones 2018, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bouhy, D.; Juneja, M.; Katona, I.; Holmgren, A.; Asselbergh, B.; De Winter, V.; Hochepied, T.; Goossens, S.; Haigh, J.J.; Libert, C.; et al. A knock-in/knock-out mouse model of HSPB8-associated distal hereditary motor neuropathy and myopathy reveals toxic gain-of-function of mutant Hspb8. Acta Neuropathol. 2018, 135, 131–148. [Google Scholar] [CrossRef]
- Shemetov, A.A.; Seit-Nebi, A.S.; Gusev, N.B. Phosphorylation by cyclic AMP-dependent protein kinase inhibits chaperone-like activity of human HSP22 in vitro | SpringerLink. Mol. Cell Biochem. 2011, 355, 47–55. [Google Scholar] [CrossRef]
- Lei, Z.; Brizzee, C.; Johnson, G.V.W. BAG3 facilitates the clearance of endogenous tau in primary neurons. Neurobiol. Aging 2015, 36, 241–248. [Google Scholar] [CrossRef]
- Fuchs, M.; Poirier, D.J.; Seguin, S.J.; Lambert, H.; Carra, S.; Charette, S.J.; Landry, J. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem. J. 2009, 425, 245–255. [Google Scholar] [CrossRef]
- Hayes, D.; Napoli, V.; Mazurkie, A.; Stafford, W.F.; Graceffa, P. Phosphorylation dependence of hsp27 multimeric size and molecular chaperone function. J. Biol. Chem. 2009, 284, 18801–18807. [Google Scholar] [CrossRef]
- Jovcevski, B.; Kelly, M.A.; Rote, A.P.; Berg, T.; Gastall, H.Y.; Benesch, J.L.P.; Aquilina, J.A.; Ecroyd, H. Phosphomimics destabilize Hsp27 oligomeric assemblies and enhance chaperone activity. Chem. Biol. 2015, 22, 186–195. [Google Scholar] [CrossRef]
- Strang, K.H.; Croft, C.L.; Sorrentino, Z.A.; Chakrabarty, P.; Golde, T.E.; Giasson, B.I. Distinct differences in prion-like seeding and aggregation between tau protein variants provide mechanistic insights into tauopathies. J. Biol. Chem. 2017. [Google Scholar] [CrossRef]
- Webster, J.M.; Darling, A.L.; Uversky, V.N.; Blair, L.J. Small Heat Shock Proteins, Big Impact on Protein Aggregation in Neurodegenerative Disease. Front. Pharmacol. 2019, 10, 1047. [Google Scholar] [CrossRef] [PubMed]
- Rusmini, P.; Cristofani, R.; Galbiati, M.; Cicardi, M.E.; Meroni, M.; Ferrari, V.; Vezzoli, G.; Tedesco, B.; Messi, E.; Piccolella, M.; et al. The Role of the Heat Shock Protein B8 (HSPB8) in Motoneuron Diseases. Front. Mol. Neurosci. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Possenti, A.; Freer, R.; Nakano, Y.; Villegas, N.C.H.; Tang, M.; Cauhy, P.V.M.; Lassus, B.A.; Chen, S.; Fowler, S.L.; et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat. Neurosci. 2019, 22, 47. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, Y.; Shodai, A.; Morimura, T.; Hikiami, R.; Minamiyama, S.; Ayaki, T.; Tooyama, I.; Furukawa, Y.; Takahashi, R.; Urushitani, M. Elimination of TDP-43 inclusions linked to amyotrophic lateral sclerosis by a misfolding-specific intrabody with dual proteolytic signals. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Chatterjee, D.; Bhatt, M.; Butler, D.; Genst, E.D.; Dobson, C.M.; Messer, A.; Kordower, J.H. Proteasome-targeted nanobodies alleviate pathology and functional decline in an α-synuclein-based Parkinson’s disease model. NPJ Parkinson’s Dis. 2018, 4, 1–10. [Google Scholar] [CrossRef]
- Santacruz, K.; Lewis, J.; Spires, T.; Paulson, J.; Kotilinek, L.; Ingelsson, M.; Guimaraes, A.; DeTure, M.; Ramsden, M.; McGowan, E.; et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science 2005, 309, 476–481. [Google Scholar] [CrossRef]
- Cribbs, D.H.; Berchtold, N.C.; Perreau, V.; Coleman, P.D.; Rogers, J.; Tenner, A.J.; Cotman, C.W. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: A microarray study. J. Neuroinflamm. 2012, 9, 179. [Google Scholar] [CrossRef]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztányi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D2P2: Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Webster, J.M.; Darling, A.L.; Sanders, T.A.; Blazier, D.M.; Vidal-Aguiar, Y.; Beaulieu-Abdelahad, D.; Plemmons, D.G.; Hill, S.E.; Uversky, V.N.; Bickford, P.C.; et al. Hsp22 with an N-Terminal Domain Truncation Mediates a Reduction in Tau Protein Levels. Int. J. Mol. Sci. 2020, 21, 5442. https://doi.org/10.3390/ijms21155442
Webster JM, Darling AL, Sanders TA, Blazier DM, Vidal-Aguiar Y, Beaulieu-Abdelahad D, Plemmons DG, Hill SE, Uversky VN, Bickford PC, et al. Hsp22 with an N-Terminal Domain Truncation Mediates a Reduction in Tau Protein Levels. International Journal of Molecular Sciences. 2020; 21(15):5442. https://doi.org/10.3390/ijms21155442
Chicago/Turabian StyleWebster, Jack M., April L. Darling, Taylor A. Sanders, Danielle M. Blazier, Yamile Vidal-Aguiar, David Beaulieu-Abdelahad, Drew G. Plemmons, Shannon E. Hill, Vladimir N. Uversky, Paula C. Bickford, and et al. 2020. "Hsp22 with an N-Terminal Domain Truncation Mediates a Reduction in Tau Protein Levels" International Journal of Molecular Sciences 21, no. 15: 5442. https://doi.org/10.3390/ijms21155442
APA StyleWebster, J. M., Darling, A. L., Sanders, T. A., Blazier, D. M., Vidal-Aguiar, Y., Beaulieu-Abdelahad, D., Plemmons, D. G., Hill, S. E., Uversky, V. N., Bickford, P. C., Dickey, C. A., & Blair, L. J. (2020). Hsp22 with an N-Terminal Domain Truncation Mediates a Reduction in Tau Protein Levels. International Journal of Molecular Sciences, 21(15), 5442. https://doi.org/10.3390/ijms21155442





