Phylogenomic proof of Recurrent Demipolyploidization and Evolutionary Stalling of the “Triploid Bridge” in Arundo (Poaceae)
Abstract
1. Introduction
2. Results
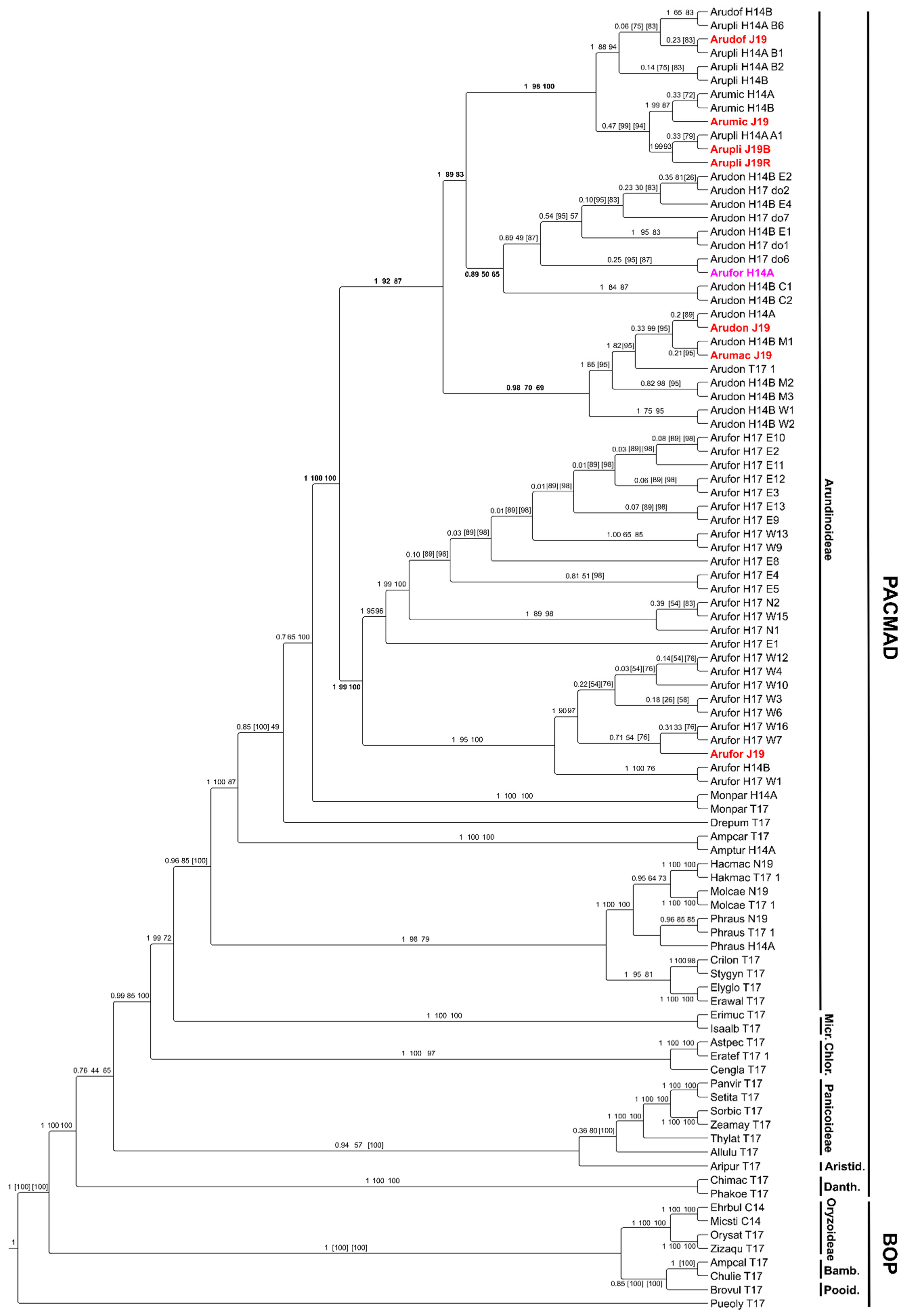
2.1. Ten New Transcriptomes for Arundo and Arundinoideae Taxa
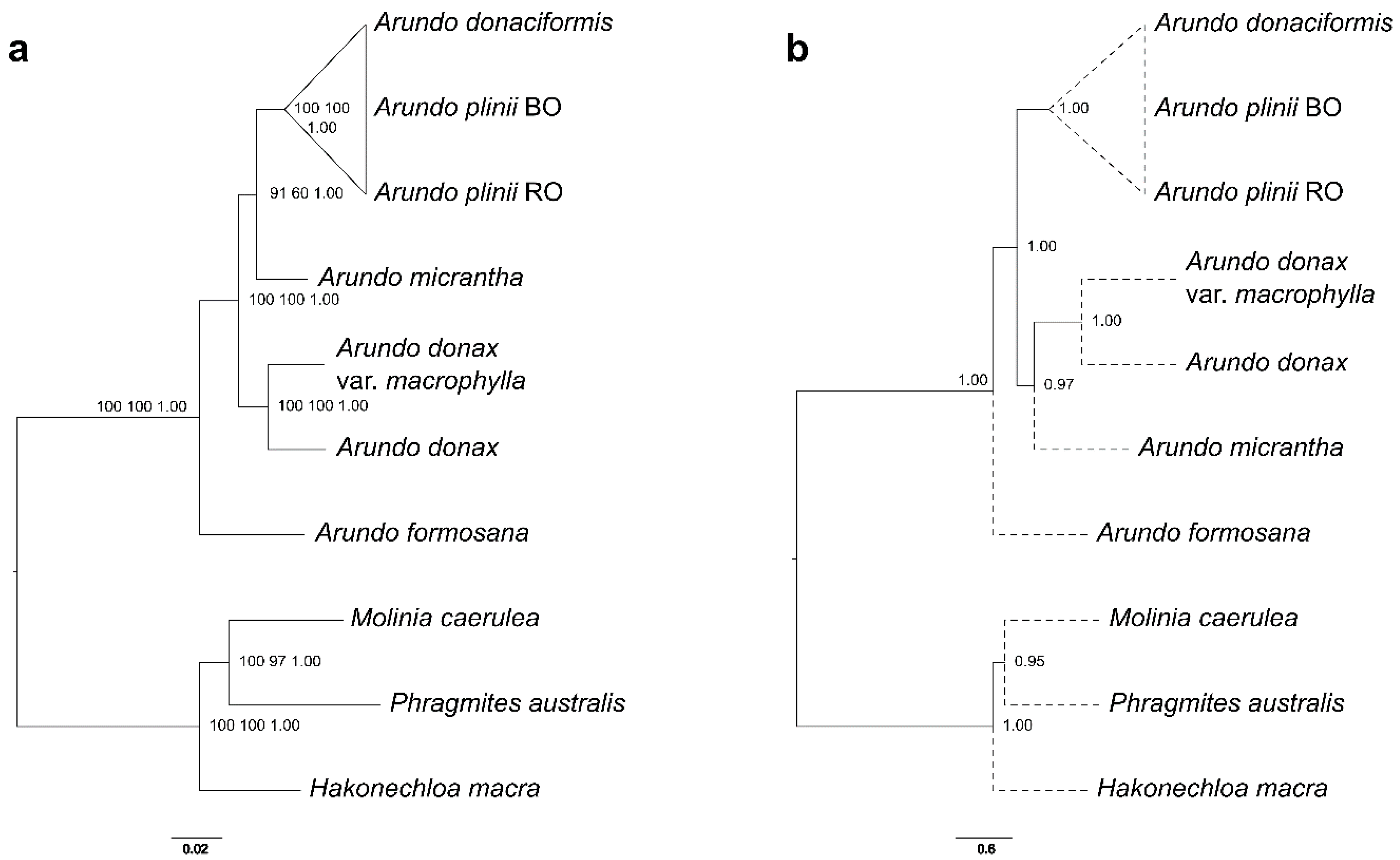
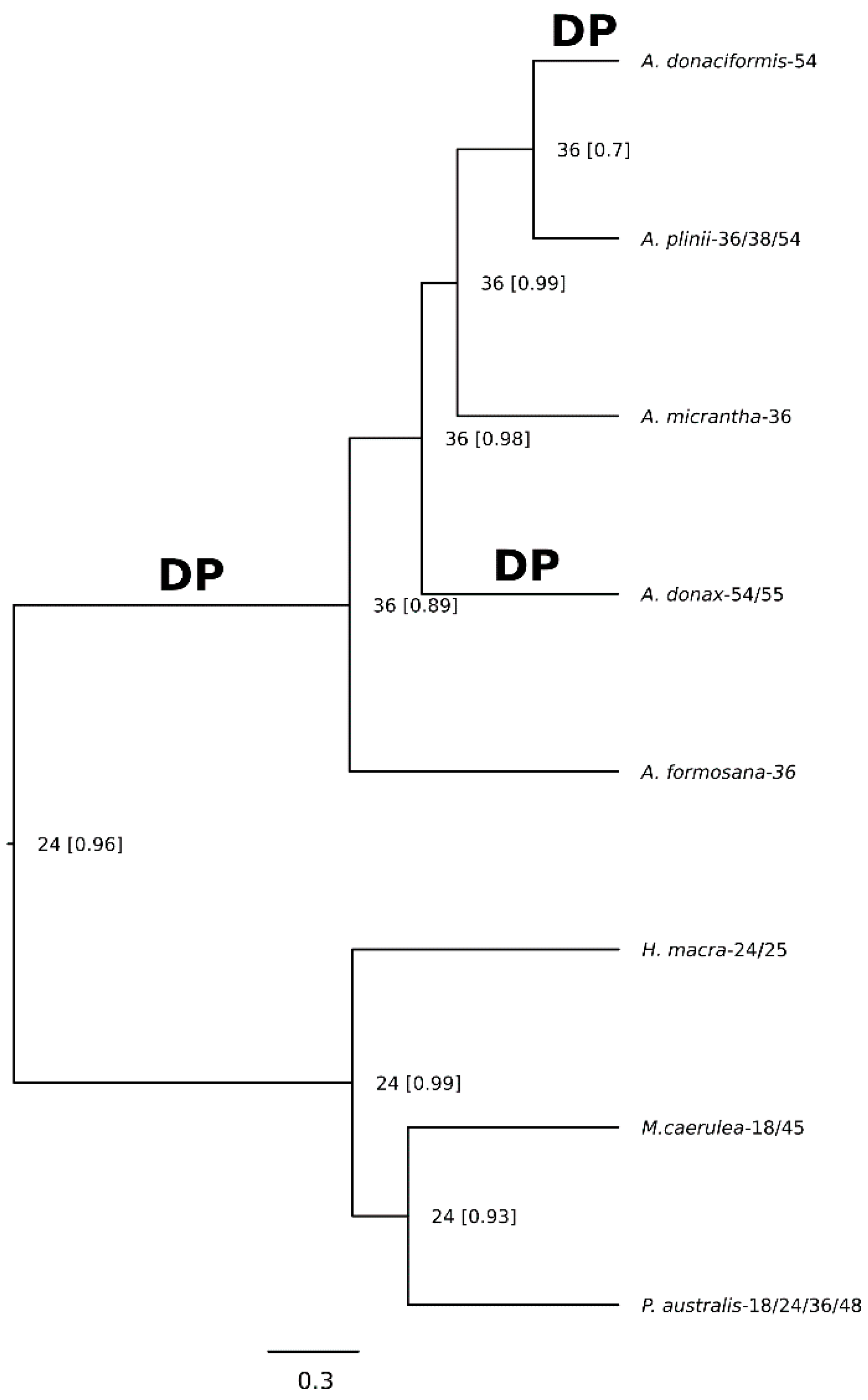
2.2. A Nuclear Phylogenomic Reconstruction of Arundo Species Reveals A. Formosana as Basal and A. Micrantha as Likely Hybrid
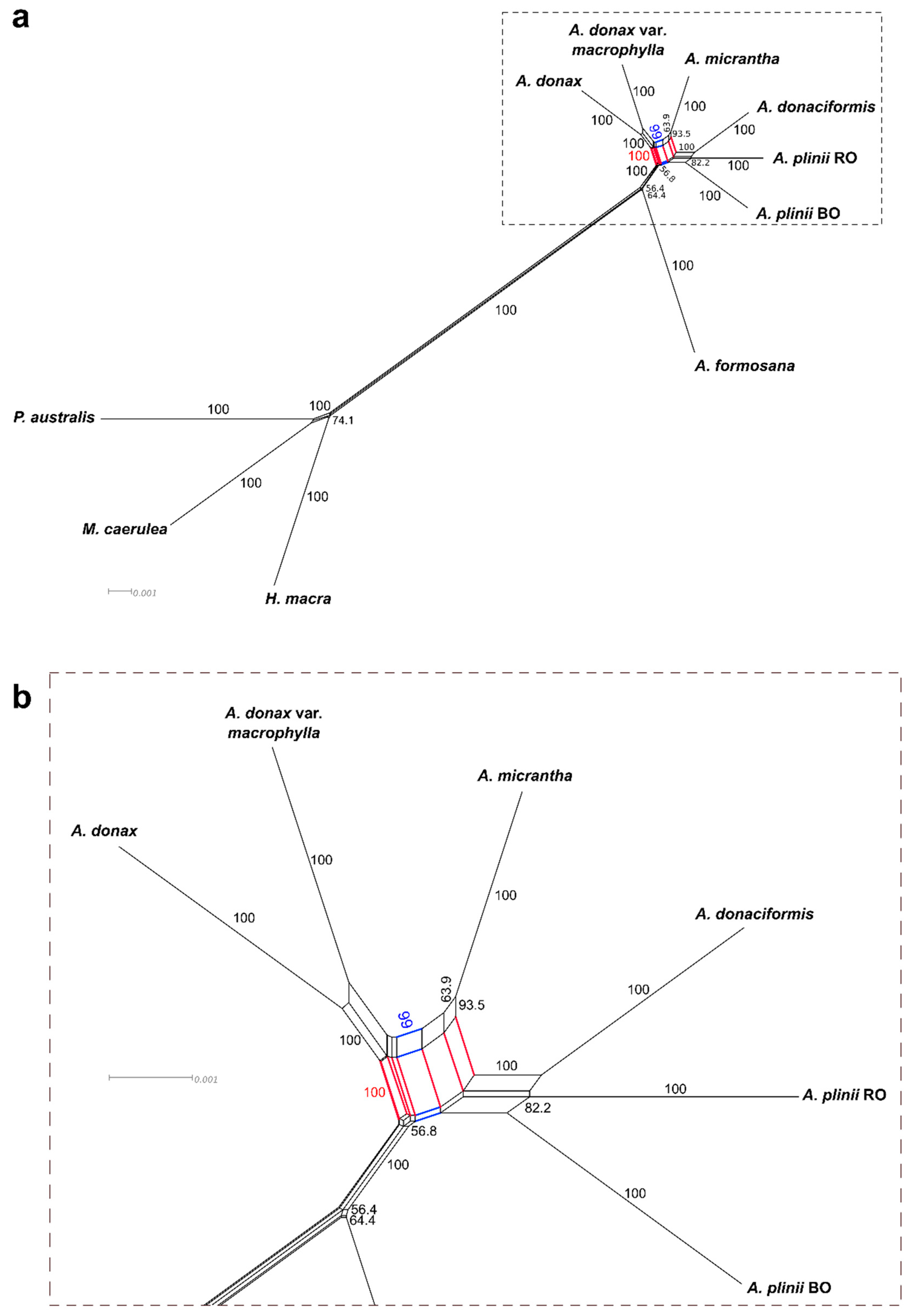
2.3. Extended Plastidial Dataset Confirms Basal A. Formosana and Supports Parphyletic A. Donax
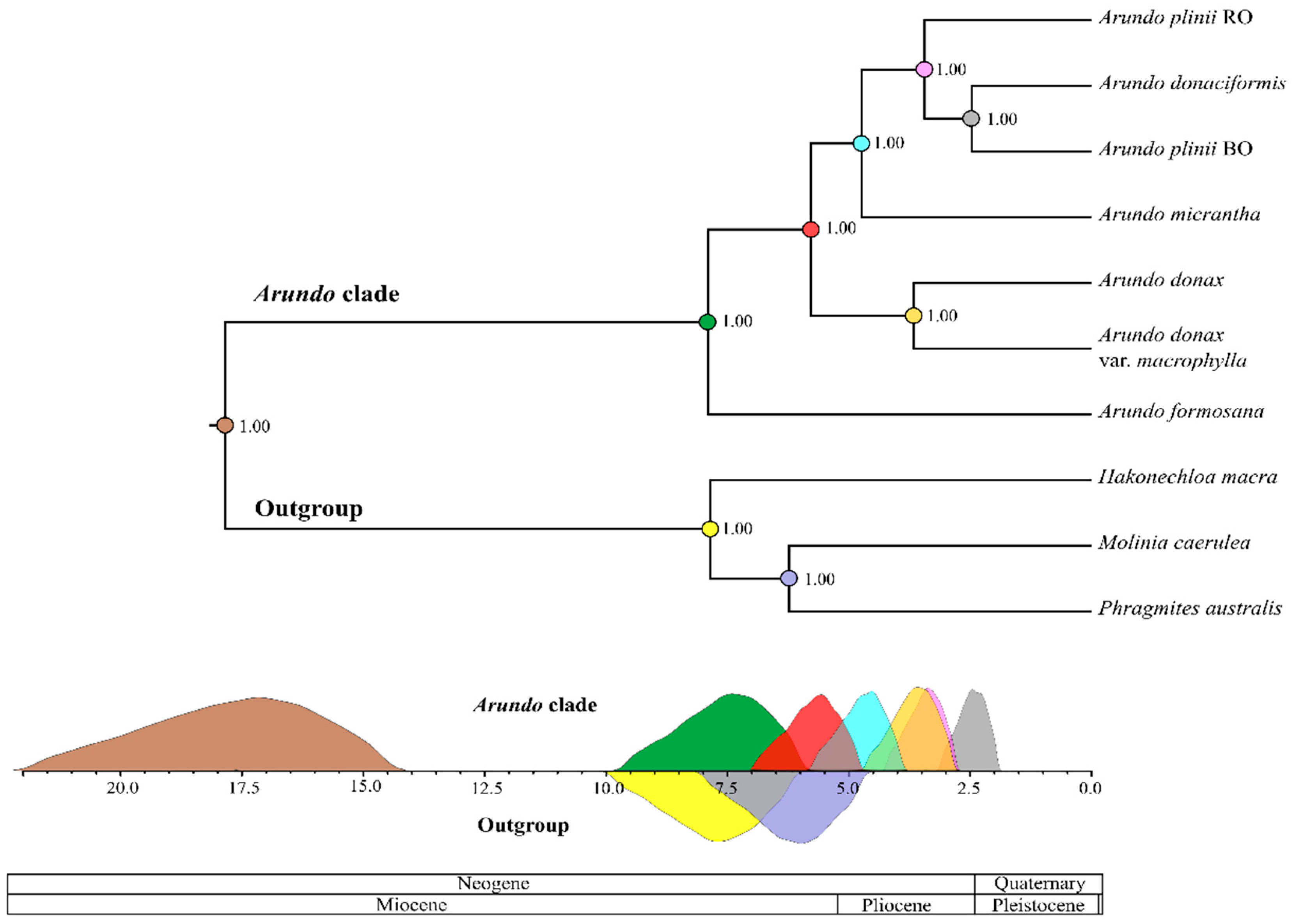
2.4. Molecular Dating of Arundo Origin
2.5. Demiduplication Is the Most Prominent Mechanism of Chromosome Number Evolution in Arundo
2.6. Patterns of Molecular Evolution Identify Candidate Genes under Positive Selection in Arundo and Arundinoideae
3. Discussion
3.1. A New Scenario of Arundo Phylogeny
3.2. Hybridization Events Clarified
3.3. Recurrent Demipolyploidization in the Arundo Genus Does Not Lead to Tetraplodization
3.4. Implications for Genetic Improvement in Arundo
4. Materials and Methods
4.1. Transcriptome de Novo Assembly
4.2. Gene Functional Annotation
4.3. Orthologous Groups Identification and Supermatrix Construction
4.4. Phylogenomic Reconstruction of Arundo Species with Nuclear Genes
4.5. Phylogenetic Reconstruction of Arundinoideae Using the Plastidial Dataset
4.6. Molecular Dating
4.7. Inference of Chromosome-Number Change
4.8. Molecular Evolution Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MYA | Million years ago |
| GO | Gene ontology |
| ML | Maxim likelihood |
| MP | Maxim parsimony |
| BI | Bayesian inference |
| gCF | Gene concordance factor |
| sCF | Site concordance factor |
| gEF_p | Probability of equal frequencies rejection for genes |
| sEF_p | Probability of equal frequencies rejection for genes |
| gIC | Gene Internode Certainty |
| sIC | Site Internode Certainty |
| PP | Posterior probability |
| UFBOOT | Ultrafast bootstrap |
| npBS | Nonparametric bootstrap |
| HPD | High Posterior Densities |
References
- Hardion, L.; Verlaque, R.; Rbaumel, A.; Juin, M.; Vila, B. Revised systematics of Mediterranean Arundo (Poaceae) based on AFLP fingerprints and morphology. Taxon 2012, 61, 1217–1226. [Google Scholar] [CrossRef]
- Hardion, L.; Verlaque, R.; Vorontsova, M.S.; Combroux, I.; Chen, C.W.; Takamizo, T.; Vila, B. Does infraspecific taxonomy match species evolutionary history? A phylogeographic study of Arundo formosana (Poaceae). Bot. J. Linn. Soc. 2017, 183, 236–249. [Google Scholar] [CrossRef]
- Lin, W.T.; Lin, C.Y.; Chou, W.C. Assessment of vegetation recovery and soil erosion at landslides caused by a catastrophic earthquake: A case study in Central Taiwan. Ecol. Eng. 2006, 28, 79–89. [Google Scholar] [CrossRef]
- Hardion, L.; Baumel, A.; Verlaque, R.; Vila, B. Distinct evolutionary histories of lowland biota on Italian and Balkan peninsulas revealed by the phylogeography of Arundo plinii (Poaceae). J. Biogeogr. 2014, 41, 2150–2161. [Google Scholar] [CrossRef]
- Danin, A. Arundo (Gramineae ) in the Mediterranean reconsidered. Willdenowia 2004, 34, 361–369. [Google Scholar] [CrossRef]
- Danin, A.; Raus, T.; Scholz, H. Contribution to the Flora of Greece: A New Species of Arundo (Poaceae). Willdenowia 2002, 32, 191–194. [Google Scholar] [CrossRef]
- Mascia, F.; Fenu, G.; Angius, R.; Bacchetta, G. Arundo micrantha, a new reed species for Italy, threatened in the freshwater habitat by the congeneric invasive A. donax. Plant Biosyst. 2013, 147, 717–729. [Google Scholar] [CrossRef]
- Lewandowski, I.; Scurlock, J.M.O.; Lindvall, E.; Christou, M. The development and current status of perennial rhizomatous grasses as energy crops in the US and Europe. Biomass Bioenergy 2003, 25, 335–361. [Google Scholar] [CrossRef]
- Balogh, E.; Herr, J.M.; Czakó, M.; Márton, L. Defective development of male and female gametophytes in Arundo donax L. (POACEAE). Biomass Bioenergy 2012, 45, 265–269. [Google Scholar] [CrossRef]
- Hardion, L.; Verlaque, R.; Rosato, M.; Rosselló, J.A.; Vila, B. Impact of polyploidy on fertility variation of Mediterranean Arundo L. (Poaceae). Comptes Rendus-Biol. 2015, 338, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Hardion, L.; Verlaque, R.; Saltonstall, K.; Leriche, A.; Vila, B. Origin of the invasive Arundo donax (Poaceae): A trans-Asian expedition in herbaria. Ann. Bot. 2014, 114, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Pilu, R.; Bucci, A.; Cerino Badone, F.; Landoni, M. Giant reed (Arundo donax L.): A weed plant or a promising energy crop? AFRICAN J. Biotechnol. 2012, 11, 9163–9174. [Google Scholar]
- Mariani, C.; Cabrini, R.; Danin, A.; Piffanelli, P.; Fricano, A.; Gomarasca, S.; Dicandilo, M.; Grassi, F.; Soave, C. Origin, diffusion and reproduction of the giant reed (Arundo donax L.): A promising weedy energy crop. Ann. Appl. Biol. 2010, 157, 191–202. [Google Scholar] [CrossRef]
- Bell, C.E. Giant reed (Arundo donax L.) response to glyphosate and imazapyr. J. Aquat. Plant Manag. 2011, 49, 111–113. [Google Scholar]
- Angelini, L.G.; Ceccarini, L.; Nassi o Di Nasso, N.; Bonari, E. Comparison of Arundo donax L. and Miscanthus x giganteus in a long-term field experiment in Central Italy: Analysis of productive characteristics and energy balance. Biomass Bioenergy 2009, 33, 635–643. [Google Scholar] [CrossRef]
- Renny-Byfield, S.; Wendel, J.F. Doubling down on genomes: Polyploidy and crop plants. Am. J. Bot. 2014, 101, 1711–1725. [Google Scholar] [CrossRef]
- Bretagnolle, F.; Thompson, J.D. Gametes with the somatic chromosome number: Mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 1995, 129, 1–22. [Google Scholar] [CrossRef]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.; Sankoff, D.; DePamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009, 96, 336–348. [Google Scholar] [CrossRef]
- Cui, L.; Wall, P.K.; Leebens-Mack, J.H.; Lindsay, B.G.; Soltis, D.E.; Doyle, J.J.; Soltis, P.S.; Carlson, J.E.; Arumuganathan, K.; Barakat, A.; et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006, 16, 738–749. [Google Scholar] [CrossRef]
- Ramsey, J.; Schemske, D.W. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 2002, 33, 589–639. [Google Scholar] [CrossRef]
- Wei, N.; Cronn, R.; Liston, A.; Ashman, T.L. Functional trait divergence and trait plasticity confer polyploid advantage in heterogeneous environments. New Phytol. 2019, 221, 2286–2297. [Google Scholar] [CrossRef]
- Triplett, J.K.; Clark, L.G.; Fisher, A.E.; Wen, J. Independent allopolyploidization events preceded speciation in the temperate and tropical woody bamboos. New Phytol. 2014, 204, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Bucci, A.; Cassani, E.; Landoni, M.; Cantaluppi, E.; Pilu, R. Analysis of chromosome number and speculations on the origin of Arundo donax L. (Giant Reed). Cytol. Genet. 2013, 47, 237–241. [Google Scholar] [CrossRef]
- Buschiazzo, E.; Ritland, C.; Bohlmann, J.; Ritland, K. Slow but not low: Genomic comparisons reveal slower evolutionary rate and higher dN/dS in conifers compared to angiosperms. BMC Evol. Biol. 2012, 12, 8. [Google Scholar] [CrossRef]
- Huson, D.H.; Klöpper, T.; Lockhart, P.J.; Steel, M.A. Reconstruction of reticulate networks from gene trees. Lect. Notes Bioinform. Subser. Lect. Notes Comput. Sci. 2005, 3500, 233–249. [Google Scholar]
- Pillon, Y.; Johansen, J.B.; Sakishima, T.; Roalson, E.H.; Price, D.K.; Stacy, E.A. Gene discordance in phylogenomics of recent plant radiations, an example from Hawaiian Cyrtandra (Gesneriaceae). Mol. Phylogenet. Evol. 2013, 69, 293–298. [Google Scholar] [CrossRef]
- Yi, T.S.; Jin, G.H.; Wen, J. Chloroplast capture and intra- and inter-continental biogeographic diversification in the Asian—New World disjunct plant genus Osmorhiza (Apiaceae). Mol. Phylogenet. Evol. 2015, 85, 10–21. [Google Scholar] [CrossRef]
- Zimmer, E.A.; Wen, J. Using nuclear gene data for plant phylogenetics: Progress and prospects. Mol. Phylogenet. Evol. 2012, 65, 774–785. [Google Scholar] [CrossRef]
- Lambertini, C. Why are tall-statured energy grasses of polyploid species complexes potentially invasive? A review of their genetic variation patterns and evolutionary plasticity. Biol. Invasions 2019, 21, 3019–3041. [Google Scholar] [CrossRef]
- Rice, A.; Glick, L.; Abadi, S.; Einhorn, M.; Kopelman, N.M.; Salman-Minkov, A.; Mayzel, J.; Chay, O.; Mayrose, I. The Chromosome Counts Database (CCDB)—A community resource of plant chromosome numbers. New Phytol. 2015, 206, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Marchant, C.J.; Macfarlane, R.M. Chromosome polymorphism in triploid populations of Fritillaria lanceolata Pursh (Liliaceae) in California. Bot. J. Linn. Soc. 1980, 81, 135–154. [Google Scholar] [CrossRef]
- Kovalsky, I.E.; Roggero Luque, J.M.; Elías, G.; Fernández, S.A.; Solís Neffa, V.G. The role of triploids in the origin and evolution of polyploids of Turnera sidoides complex (Passifloraceae, Turneroideae). J. Plant Res. 2018, 131, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Hair, J.B. Contributions to a chromosome atlas of the New Zealand flora. 12. Poa. New Zeal. J. Bot. 1968, 6, 267–276. [Google Scholar] [CrossRef]
- Sauer, W. Die Gattung Avenula (Dumort.) Dumort., Poaceae, auf der Balkanhalbinsel. Acta Bot. Croat. 1984, 43, 315–328. [Google Scholar]
- Gould, F.W.; Soderstrom, T.R. Chromosome numbers of some Ceylon grasses. Can. J. Bot. 1974, 52, 1075–1090. [Google Scholar] [CrossRef]
- Chae, W.B.; Hong, S.J.; Gifford, J.M.; Lane Rayburn, A.; Widholm, J.M.; Juvik, J.A. Synthetic polyploid production of miscanthus sacchariflorus, miscanthus sinensis, and miscanthus x giganteus. GCB Bioenergy 2013, 5, 338–350. [Google Scholar] [CrossRef]
- Valli, F.; Trebbi, D.; Zegada-Lizarazu, W.; Monti, A.; Tuberosa, R.; Salvi, S. In vitro physical mutagenesis of giant reed (Arundo donax L.). GCB Bioenergy 2017, 9, 1380–1389. [Google Scholar] [CrossRef]
- Sweetman, C.; Waterman, C.D.; Rainbird, B.M.; Smith, P.M.C.; Jenkins, C.D.; Day, D.A.; Soole, K.L. AtNDB2 Is the Main External NADH Dehydrogenase in Mitochondria and Is Important for Tolerance to Environmental Stress. Plant Physiol. 2019, 181, 774–788. [Google Scholar] [CrossRef]
- Maruyama, D.; Endo, T.; Nishikawa, S.I. BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 1684–1689. [Google Scholar] [CrossRef]
- Marks, G.E. The Origin and Significance of Intraspecific Polyploidy: Experimental Evidence from Solanum chacoense. Evolution (N. Y.) 1966, 20, 552–557. [Google Scholar]
- Lü, S.; Zhao, H.; Des Marais, D.L.; Parsons, E.P.; Wen, X.; Xu, X.; Bangarusamy, D.K.; Wang, G.; Rowland, O.; Juenger, T.; et al. Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 2012, 159, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Doblas, V.G.; Amorim-Silva, V.; Posé, D.; Rosado, A.; Esteban, A.; Arró, M.; Azevedo, H.; Bombarely, A.; Borsani, O.; Valpuesta, V.; et al. The SUD1 gene encodes a putative E3 ubiquitin ligase and is a positive regulator of 3-hydroxy-3-methylglutaryl coenzyme a reductase activity in Arabidopsis. Plant Cell 2013, 25, 728–743. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Poli, M.; Sablok, G.; Wang, B.; Liang, Y.; La Porta, N.; Velikova, V.; Loreto, F.; Li, M.; Varotto, C. Dissection of early transcriptional responses to water stress in Arundo donax L. by unigene-based RNA-seq. Biotechnol. Biofuels 2016, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Breese, M.R.; Liu, Y. NGSUtils: A software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics 2013, 29, 494–496. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Chen, T.W.; Gan, R.C.R.; Wu, T.H.; Huang, P.J.; Lee, C.Y.; Chen, Y.Y.M.; Chen, C.C.; Tang, P. FastAnnotator--an efficient transcript annotation web tool. BMC Genomics 2012, 13, S9. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Smith, S.A.; Dunn, C.W. Phyutility: A phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 2008, 24, 715–716. [Google Scholar] [CrossRef]
- Yang, Y.; Smith, S.A. Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: Improving accuracy and matrix occupancy for phylogenomics. Mol. Biol. Evol. 2014, 31, 3081–3092. [Google Scholar] [CrossRef]
- Whelan, S.; Allen, J.E.; Blackburne, B.P.; Talavera, D. ModelOMatic: Fast and automated model selection between ry, nucleotide, amino acid, and codon substitution models. Syst. Biol. 2015, 64, 42–55. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. Partitionfinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; Von Haeseler, A.; Minh, B.Q. Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 2018, 19, 15–30. [Google Scholar] [CrossRef]
- Teisher, J.K.; McKain, M.R.; Schaal, B.A.; Kellogg, E.A. Polyphyly of Arundinoideae (Poaceae) and evolution of the twisted geniculate lemma awn. Ann. Bot. 2017, 120, 725–738. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, 1–6. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, 699–710. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.R.; Drummond, A.J. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 2017, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Lewis, P.O.; Fan, Y.; Kuo, L.; Chen, M.H. Improving marginal likelihood estimation for bayesian phylogenetic model selection. Syst. Biol. 2011, 60, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Baele, G.; Lemey, P.; Bedford, T.; Rambaut, A.; Suchard, M.A.; Alekseyenko, A.V. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 2012, 29, 2157–2167. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, S.; Tamura, M.N. Evolutionary timescale of monocots determined by the fossilized birth-death model using a large number of fossil records. Evolution (N. Y.) 2016, 70, 1136–1144. [Google Scholar] [CrossRef]
- Saarela, J.M.; Burke, S.V.; Wysocki, W.P.; Barrett, M.D.; Clark, L.G.; Craine, J.M.; Peterson, P.M.; Soreng, R.J.; Vorontsova, M.S.; Duvall, M.R. A 250 plastome phylogeny of the grass family (Poaceae): Topological support under different data partitions. PeerJ 2018, 2018, 1–71. [Google Scholar] [CrossRef]
- Burke, S.V.; Lin, C.S.; Wysocki, W.P.; Clark, L.G.; Duvall, M.R. Phylogenomics and plastome evolution of tropical forest grasses (Leptaspis, streptochaeta: Poaceae). Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Prasad, V.; Strömberg, C.A.E.; Leaché, A.D.; Samant, B.; Patnaik, R.; Tang, L.; Mohabey, D.M.; Ge, S.; Sahni, A. Late Cretaceous origin of the rice tribe provides evidence for early diversification in Poaceae. Nat. Commun. 2011, 2, 1–9. [Google Scholar] [CrossRef]
- Junier, T.; Zdobnov, E.M. The Newick utilities: High-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 2010, 26, 1669–1670. [Google Scholar] [CrossRef]
- Gibson, D.J. Grasses and Grassland Ecology. In Oxford Biology; OUP Oxford: Oxford, UK, 2009; ISBN 9780198529187. [Google Scholar]
- Kosakovsky Pond, S.L.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef]
- Ranwez, V.; Harispe, S.; Delsuc, F.; Douzery, E.J.P. MACSE: Multiple alignment of coding SEquences accounting for frameshifts and stop codons. PLoS ONE 2011, 6, e22594. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D.W. GARD: A genetic algorithm for recombination detection. Bioinformatics 2006, 22, 3096–3098. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Weaver, S.; Smith, M.D.; Wertheim, J.O.; Murrell, S.; Aylward, A.; Eren, K.; Pollner, T.; Martin, D.P.; Smith, D.M.; et al. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015, 32, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D.; Wertheim, J.O.; Weaver, S.; Murrell, B.; Scheffler, K.; Kosakovsky Pond, S.L. Less is more: An adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 2015, 32, 1342–1353. [Google Scholar] [CrossRef]
- Wertheim, J.O.; Murrell, B.; Smith, M.D.; Pond, S.L.K.; Scheffler, K. RELAX: Detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 2015, 32, 820–832. [Google Scholar] [CrossRef]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. FUBAR: A fast, unconstrained bayesian AppRoximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
| MODEL | Parameters | Ln(Likelihood) | AIC | ΔAIC |
|---|---|---|---|---|
| CONST_RATE_DEMI_EST * | 4 | −14.92 | 37.83 | 0.00 |
| CONST_RATE_DEMI | 3 | −17.83 | 41.67 | 3.83 |
| BASE_NUM | 4 | −20.30 | 48.61 | 10.77 |
| BASE_NUM_DUPL | 5 | −20.18 | 50.36 | 12.53 |
| CONST_RATE | 3 | −29.81 | 65.61 | 27.78 |
| CONST_RATE_NO_DUPL | 2 | −31.15 | 66.30 | 28.47 |
| LINEAR_RATE_DEMI | 5 | −29.61 | 69.22 | 31.39 |
| LINEAR_RATE | 5 | −29.99 | 69.98 | 32.14 |
| LINEAR_RATE_NO_DUPL | 4 | −31.02 | 70.03 | 32.20 |
| LINEAR_RATE_DEMI_EST | 6 | −29.49 | 70.98 | 33.14 |
| OG ID | Sites | LR | p-Value | Fdr | TAIR_ID | Name | Description |
|---|---|---|---|---|---|---|---|
| OG0018377 | 1531 | 39.72 | 2.38 × 10−9 | 2.88 × 10−7 | AT5G47690 | PDS5A | PO76/PDS5 cohesin cofactor |
| OG0018357 | 964 | 29.35 | 4.23 × 10−7 | 3.05 × 10−5 | AT4G34100 | CER9 | RING/U-box-superfamily-protein |
| OG0018102 | 258 | 22.17 | 1.53 × 10−5 | 0.000735 | AT5G48300 | ADG1 | ADP glucose pyrophosphorylase 1 |
| OG0018342 | 973 | 17.99 | 0.000124 | 0.004471 | AT4G11420 | EIF3A | Eukaryotic translation initiation factor 3 subunit A |
| OG0018364 | 899 | 13.89 | 0.000965 | 0.027788 | AT4G16150 | CAMTA5 | Calmodulin-binding; transcription-regulators |
| OG0018157 | 445 | 12.44 | 0.001986 | 0.047669 | AT4G35140 | NA | Transducin/WD40-repeat-like-superfamily-protein |
| OG0018070 | 126 | 12.10 | 0.002359 | 0.048518 | NA | NA | NA |
| OG ID | Test Nodes | K | LR | FDR p-Val a | TAIR ID | Name | Description |
|---|---|---|---|---|---|---|---|
| OG0017620 | hm | 4.35 | 6.19 | 0.030 | AT5G19210 | NA | P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| OG0018017 | ami | 3.69 | 10.02 | 0.014 | AT1G64710 | NA | GroES-like zinc-binding alcohol dehydrogenase family protein |
| OG0018069 | adf | 39.10 | 8.39 | 0.014 | AT5G42020 | BIP2 | Luminal binding protein involved in polar nuclei fusion during proliferation of endosperm nuclei. |
| OG0018170 | pa | 3.79 | 9.98 | 0.009 | AT2G01680 | NA | Ankyrin repeat family protein |
| OG0018204 | adf | 4.73 | 6.09 | 0.028 | AT5G08530 | CI51 | 51 kDa subunit of mitochondrial complex I |
| OG0018205 | mc | 9.25 | 15.13 | 0 | AT3G15140 | ERI-1 | Ribonuclease H-like superfamily functioning as siRNA exonuclease. It affects post-transcriptional gene silencing and growth rate /biomass. |
| OG0018357 * | ama, mc | 3.67 | 7.93 | 0.014 | AT4G34100 | CER9 | Involved in cuticular wax biosynthesis. Arabidopsis mutants have leaf waxes nearly pure C24 and C26 acid, weakly glaucous stem surface, and reduced fertility in early flowers. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jike, W.; Li, M.; Zadra, N.; Barbaro, E.; Sablok, G.; Bertorelle, G.; Rota-Stabelli, O.; Varotto, C. Phylogenomic proof of Recurrent Demipolyploidization and Evolutionary Stalling of the “Triploid Bridge” in Arundo (Poaceae). Int. J. Mol. Sci. 2020, 21, 5247. https://doi.org/10.3390/ijms21155247
Jike W, Li M, Zadra N, Barbaro E, Sablok G, Bertorelle G, Rota-Stabelli O, Varotto C. Phylogenomic proof of Recurrent Demipolyploidization and Evolutionary Stalling of the “Triploid Bridge” in Arundo (Poaceae). International Journal of Molecular Sciences. 2020; 21(15):5247. https://doi.org/10.3390/ijms21155247
Chicago/Turabian StyleJike, Wuhe, Mingai Li, Nicola Zadra, Enrico Barbaro, Gaurav Sablok, Giorgio Bertorelle, Omar Rota-Stabelli, and Claudio Varotto. 2020. "Phylogenomic proof of Recurrent Demipolyploidization and Evolutionary Stalling of the “Triploid Bridge” in Arundo (Poaceae)" International Journal of Molecular Sciences 21, no. 15: 5247. https://doi.org/10.3390/ijms21155247
APA StyleJike, W., Li, M., Zadra, N., Barbaro, E., Sablok, G., Bertorelle, G., Rota-Stabelli, O., & Varotto, C. (2020). Phylogenomic proof of Recurrent Demipolyploidization and Evolutionary Stalling of the “Triploid Bridge” in Arundo (Poaceae). International Journal of Molecular Sciences, 21(15), 5247. https://doi.org/10.3390/ijms21155247







