Molecular and Hormonal Regulation of Leaf Morphogenesis in Arabidopsis
Abstract
1. Introduction
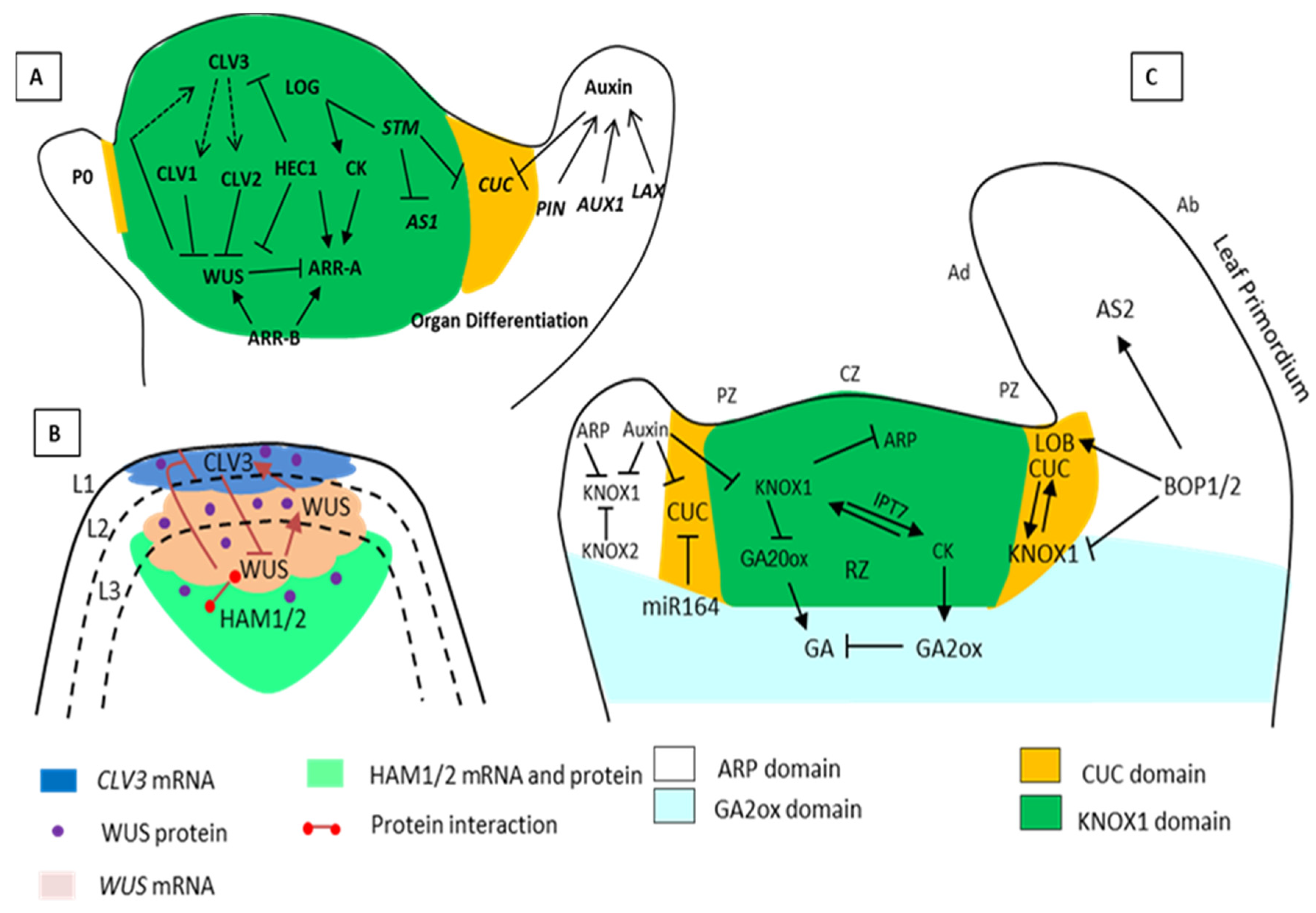
1.1. Maintenance of Shoot Apical Meristems (SAM) and the Leaf Initiation Gene Network
1.2. Gene Functioning in Leaf Initiation
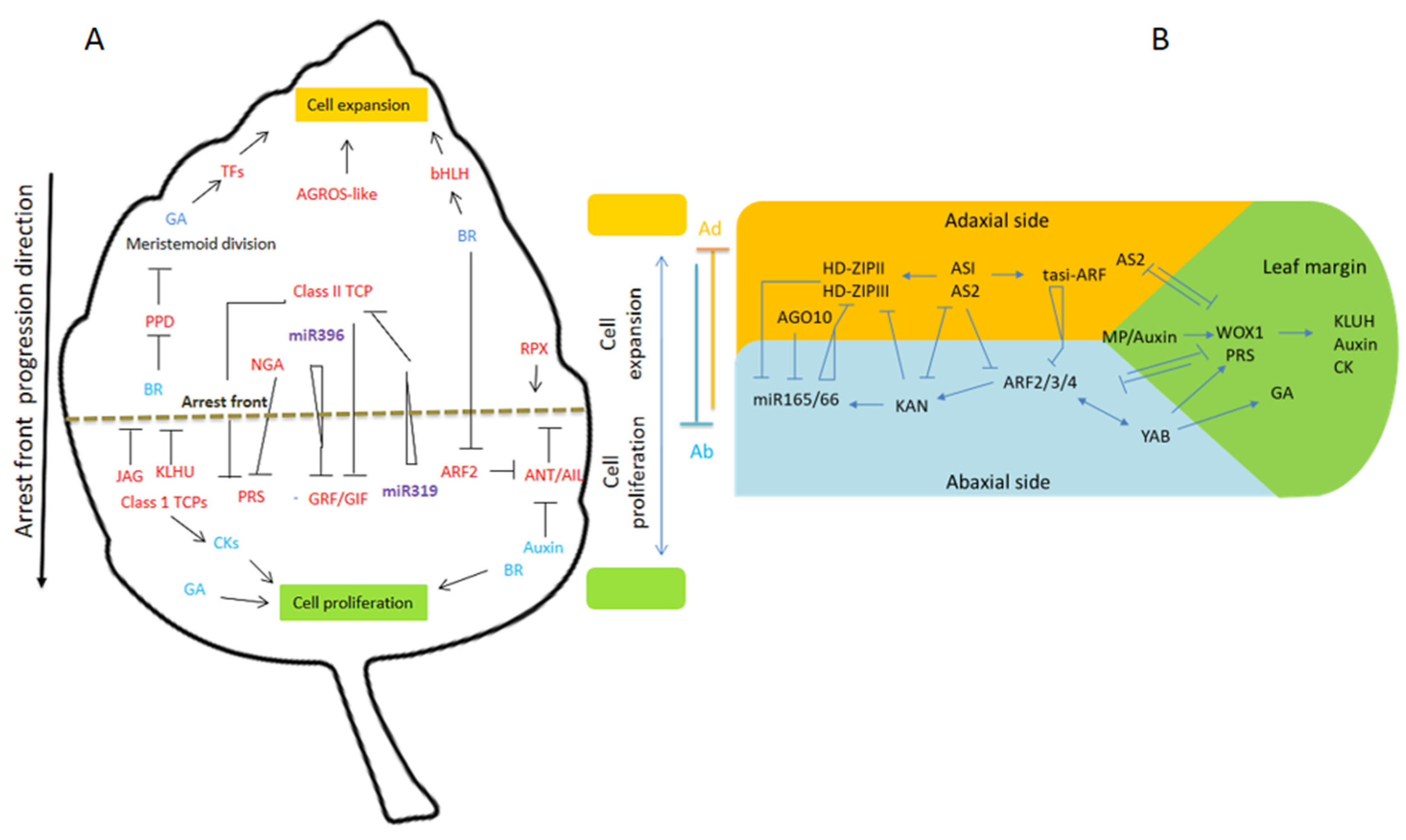
2. Leaf Outgrowth and Expansion
3. Adaxial/Abaxial Patterning
4. Leaf Margins Serrations
5. Developmental Functions of Micrornas
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Cho, K.H.; Jun, S.E.; Jeong, S.J.; Lee, Y.K.; Kim, G.T. Developmental processes of leaf morphogenesis in arabidopsis. J. Plant Biol. 2007, 50, 282–290. [Google Scholar] [CrossRef]
- Du, F.; Guan, C.; Jiao, Y. Molecular mechanisms of leaf morphogenesis. Molec. Plant 2018, 11, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Li, L.C.; Kang, D.M.; Chen, Z.L.; Qu, L.J. Hormonal regulation of leaf morphogenesis in Arabidopsis. J. Integ. Plant Biol. 2007, 49, 75–80. [Google Scholar] [CrossRef]
- Efroni, I.; Eshed, Y.; Lifschitz, E. Morphogenesis of simple and compound leaves: A critical review. Plant Cell 2010, 22, 1019–1032. [Google Scholar] [CrossRef]
- Jiao, K.; Li, X.; Su, S.; Guo, W.; Guo, Y.; Hu, Z.; Shen, Z.; Luo, D. Genetic control of compound leaf development in the mungbean (Vigna radiata L.). Hortic. Res. 2019, 6, 23. [Google Scholar] [CrossRef]
- Barton, M.K. Cell type specifi cation and selfrenewal in the vegetative shoot apical meristem. Curr. Opin. Plant Biol. 1998, 1, 37–42. [Google Scholar] [CrossRef]
- Itoh, J.I.; Kitano, H.; Matsuoka, M.; Nagato, Y. SHOOT ORGANIZATION genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 2000, 12, 2161–2174. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Deb, Y.; Marti, D.; Frenz, M.; Kuhlemeier, C.; Reinhardt, D. Phyllotaxis involves auxin drainage through leaf primordia. Development 2015, 142, 1992–2001. [Google Scholar] [CrossRef]
- Bhatia, N.; Heisler, M.G. Self-organizing periodicity in development: Organ positioning in plants. Development 2018, 145, dev149336. [Google Scholar] [CrossRef]
- Yonekura, T.; Iwamoto, A.; Fujita, H.; Sugiyama, M. Mathematical model studies of the comprehensive generation of major and minor phyllotactic patterns in plants with a predominant focus on orixate phyllotaxis. PLoS Comput. Biol. 2019, 15, e1007044. [Google Scholar] [CrossRef]
- Traas, J. Phyllotaxis. Development 2013, 140, 249–253. [Google Scholar] [CrossRef]
- Barton, M.K. Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 2010, 341, 95–113. [Google Scholar] [CrossRef]
- Kucypera, K.; Lipowczan, M.; Piekarska-Stachowiak, A.; Nakielski, J. A method to generate the surface cell layer of the 3D virtual shoot apex from apical initials. Plant Methods 2017, 13, 110. [Google Scholar] [CrossRef]
- Tax, F.E.; Durbak, A. Meristems in the movies: Live imaging as a tool for decoding intercellular signaling in shoot apical meristems. Plant Cell 2006, 18, 1331–1337. [Google Scholar] [CrossRef]
- Adibi, M.; Yoshida, S.; Weijers, D.; Fleck, C. Centering the organizing center in the Arabidopsis thaliana shoot apical meristem by a combination of cytokinin signaling and self-organization. PLoS ONE 2016, 11, e0147830. [Google Scholar] [CrossRef]
- Bustamante, M.; Matus, J.T.; Riechmann, J.L. Genome-wide analyses for dissecting gene regulatory networks in the shoot apical meristem. J. Exp. Bot. 2016, 67, 1639–1648. [Google Scholar] [CrossRef]
- Fouracre, J.P.; Poethig, R.S. Role for the shoot apical meristem in the specification of juvenile leaf identity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 10168–10177. [Google Scholar] [CrossRef]
- Rinne, P.L.; Kaikuranta, P.M.; Van Der Schoot, C. The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 2001, 26, 249–264. [Google Scholar] [CrossRef]
- Sager, R.; Lee, J.Y. Plasmodesmata in integrated cell signalling: Insights from development and environmental signals and stresses. J. Exp. Bot. 2014, 65, 6337–6358. [Google Scholar] [CrossRef]
- Tian, C.; Wang, Y.; Yu, H.; He, J.; Wang, J.; Shi, B.; Jiao, Y. A gene expression map of shoot domains reveals regulatory mechanisms. Nat. Commum. 2019, 10, 141. [Google Scholar] [CrossRef]
- Reddy, G.V.; Meyerowitz, E.M. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 2005, 310, 663–667. [Google Scholar] [CrossRef]
- Yadav, R.K.; Girke, T.; Pasala, S.; Xie, M.; Reddy, G.V. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 2009, 106, 4941–4946. [Google Scholar] [CrossRef]
- Shi, B.; Vernoux, T. Patterning at the shoot apical meristem and phyllotaxis. Curr. Top. Dev. Biol. 2019, 131, 81–107. [Google Scholar]
- Ha, C.M.; Jun, J.H.; Fletcher, J.C. Shoot apical meristem form and function. Curr. Top. Dev. Biol. 2010, 91, 103–140. [Google Scholar]
- Vernoux, T.; Besnard, F.; Traas, J. Auxin at the shoot apical meristem. Cold Spring Harb. 2010, 2, a001487. [Google Scholar] [CrossRef]
- Vernoux, T.; Brunoud, G.; Farcot, E.; Morin, V.; Van den Daele, H.; Legrand, J.; Guédon, Y. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011, 7, 508. [Google Scholar] [CrossRef]
- Souer, E.; Van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The No Apical Meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef]
- Hibara, K.I.; Karim, M.R.; Takada, S.; Taoka, K.I.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Scofield, S.; Dewitte, W.; Nieuwland, J.; Murray, J.A. The A rabidopsis homeobox gene SHOOT MERISTEMLESS has cellular and meristem-organisational roles with differential requirements for cytokinin and CYCD3 activity. Plant J. 2013, 75, 53–66. [Google Scholar] [CrossRef]
- Clark, S.E.; Jacobsen, S.E.; Levin, J.Z.; Meyerowitz, E.M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 1996, 122, 1565–1575. [Google Scholar]
- Timmermans, M.C.; Hudson, A.; Becraft, P.W.; Nelson, T. ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 1999, 284, 151–153. [Google Scholar] [CrossRef]
- Byrne, M.E.; Barley, R.; Curtis, M.; Arroyo, J.M.; Dunham, M.; Hudson, A.; Martienssen, R.A. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 2000, 408, 967–971. [Google Scholar] [CrossRef]
- Simmons, A.R.; Bergmann, D.C. Transcriptional control of cell fate in the stomatal lineage. Curr. Opin. Plant Biol. 2016, 29, 1–8. [Google Scholar] [CrossRef]
- Sun, X.; Wang, C.; Xiang, N.; Li, X.; Yang, S.; Du, J.; Yang, Y.; Yang, Y. Activation of secondary cell wall biosynthesis by miR319-targeted TCP4 transcription factor. Plant Biotechnol. J. 2017, 15, 1284–1294. [Google Scholar] [CrossRef]
- Lu, Y.; Feng, Z.; Liu, X.; Bian, L.; Xie, H.; Zhang, C.; Liang, J. MiR393 and miR390 synergistically regulate lateral root growth in rice under different conditions. BMC Plant Biol. 2018, 18, 261. [Google Scholar] [CrossRef]
- Iwakawa, H.; Iwasaki, M.; Kojima, S.; Ueno, Y.; Soma, T.; Tanaka, H.; Machida, C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007, 51, 173–184. [Google Scholar] [CrossRef]
- Martin-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Yuan, S.; Zhao, J.; Li, Z.; Hu, Q.; Yuan, N.; Zhou, M.; Luo, H. MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Hortic. Res. 2019, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yan, A.; Han, H.; Li, T.; Geng, Y.; Liu, X.; Meyerowitz, E.M. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science (N.Y.) 2018, 361, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Fan, Z.; Shan, W.; Yin, X.R.; Kuang, J.F.; Lu, W.J.; Chen, J. Association of BrERF72 with methyl jasmonate-induced leaf senescence of Chinese flowering cabbage through activating JA biosynthesis-related genes. Hortic. Res. 2018, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, B.; Wang, H.; Li, J.; Huang, H.; Xu, L. YUCCA genes are expressed in response to leaf adaxial–abaxial juxtaposition and are required for leaf margin development. Plant Physiol. 2011, 157, 1805–1819. [Google Scholar] [CrossRef]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef]
- White, D.W. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 13238–13243. [Google Scholar] [CrossRef]
- Ikeda, M.; Ohme-Takagi, M. TCPs, WUSs, and WINDs: Families of transcription factors that regulate shoot meristem formation, stem cell maintenance, and somatic cell differentiation. Front Plant Sci. 2014, 5, 427. [Google Scholar] [CrossRef]
- Tsiantis, M.; Schneeberger, R.; Golz, J.F.; Freeling, M.; Langdale, J.A. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 1999, 284, 154–156. [Google Scholar] [CrossRef]
- Kerstetter, R.A.; Laudencia-Chingcuanco, D.; Smith, L.G.; Hake, S. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 1997, 124, 3045–3054. [Google Scholar]
- Murray, J.A.; Jones, A.; Godin, C.; Traas, J. Systems analysis of shoot apical meristem growth and development: Integrating hormonal and mechanical signaling. Plant Cell 2012, 24, 3907–3919. [Google Scholar] [CrossRef] [PubMed]
- Kuchen, E.E.; Fox, S.; De Reuille, P.B.; Kennaway, R.; Bensmihen, S.; Avondo, J.; Coen, E. Generation of leaf shape through early patterns of growth and tissue polarity. Science 2012, 335, 1092–1096. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yong, X.; Yu, J.; Cheng, T.; Wang, J.; Yang, W.; Zhang, Q. Identification of candidate adaxial–abaxial related genes regulating petal expansion during flower opening in Rosa chinensis ‘Old Blush’. Front. Plant Sci. 2019, 10, 1098. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Barkoulas, M.; Tsiantis, M. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 2006, 133, 3955–3961. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Meng, Y.; Liu, H.; Wang, H.; Liu, Y.; Zhu, B.; Wang, Z.; Hou, Y.; Zhang, P.; Wen, J.; Yang, H.; et al. HEADLESS, a WUSCHEL homolog, uncovers novel aspects of shoot meristem regulation and leaf blade development in Medicago truncatula. J. Exp. Bot. 2019, 70, 149–163. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.; Berger, J.; Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar]
- Busch, W.; Miotk, A.; Ariel, F.D.; Zhao, Z.; Forner, J.; Daum, G.; Suzaki, T.; Schuster, C.; Schultheiss, S.J.; Leibfried, A.; et al. Transcriptional control of a plant stem cell niche. Dev. Cell. 2010, 18, 841–853. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef]
- Leibfried, A.; To, J.P.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005, 438, 1172–1175. [Google Scholar] [CrossRef]
- Xie, M.; Chen, H.; Huang, L.; O’Neil, R.C.; Shokhirev, M.N.; Ecker, J.R. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat Commun. 2018, 9, 1604. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Honma, T.; Aoyama, T.; Sato, S.; Kato, T.; Tabata, S.; Oka, A. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 2001, 294, 1519–1521. [Google Scholar] [CrossRef] [PubMed]
- Besnard, F.; Refahi, Y.; Morin, V.; Marteaux, B.; Brunoud, G.; Chambrier, P.; Rozier, F.; Mirabet, V.; Legrand, J.; Lainé, S.; et al. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 2014, 505, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Besnard, F.; Rozier, F.; Vernoux, T. The AHP6 cytokinin signaling inhibitor mediates an auxin-cytokinin crosstalk that regulates the timing of organ initiation at the shoot apical meristem. Plant Signal. Behav. 2014, 9, e28788. [Google Scholar] [CrossRef] [PubMed]
- Brandstatter, I.; Kieber, J.J. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 1998, 10, 1009–1020. [Google Scholar] [CrossRef]
- To, J.P.; Haberer, G.; Ferreira, F.J.; Deruere, J.; Mason, M.G.; Schaller, G.E.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 2004, 16, 658–671. [Google Scholar] [CrossRef]
- Ma, Y.; Miotk, A.; Šutiković, Z.; Ermakova, O.; Wenzl, C.; Medzihradszky, A.; Gaillochet, C.; Forner, J.; Utan, G.; Brackmann, K.; et al. WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat. Commun. 2019, 10, 5093. [Google Scholar] [CrossRef]
- Gremski, K.; Ditta, G.; Yanofsky, M.F. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 2007, 134, 3593–3601. [Google Scholar] [CrossRef]
- Schuster, C.; Gaillochet, C.; Medzihradszky, A.; Busch, W.; Daum, G.; Krebs, M.; Kehle, A.; Lohmann, J.U. A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 2014, 28, 438–449. [Google Scholar] [CrossRef]
- MacArthur, B.D.; Ma’ayan, A.; Lemischka, I.R. Systems biology of stem cell fate and cellular reprogramming. Nat. Rev. Mol. Cell Biol. 2009, 10, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhao, X.; Hsieh, J.; Wichterle, H.; Impey, S.; Banerjee, S.; Neveu, P.; Kosik, K.S. MicroRNA regulation of neural stem cells and neurogenesis. J. Neurosci. 2010, 30, 14931–14936. [Google Scholar] [CrossRef] [PubMed]
- Heidstra, R.; Sabatini, S. Plant and animal stem cells: Similar yet different. Nat. Rev. Mol. Cell Biol. 2014, 15, 301–312. [Google Scholar] [CrossRef]
- Kinoshita, A.; Betsuyaku, S.; Osakabe, Y.; Mizuno, S.; Nagawa, S.; Stahl, Y.; Simon, R.; Yamaguchi-Shinozaki, K.; Fukuda, H.; Sawa, S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 2010, 137, 3911–3920. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Xu, Y.; Ng, K.H.; Ito, T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009, 23, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef]
- Daum, G.; Medzihradszky, A.; Suzaki, T.; Lohmann, J.U. A mechanistic framework for non-cell autonomous stem cell induction in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14619–14624. [Google Scholar] [CrossRef]
- Gruel, J.; Deichmann, J.; Landrein, B.; Hitchcock, T.; Jönsson, H. The interaction of transcription factors controls the spatial layout of plant aerial stem cell niches. Npj. Syst. Biol. Appl. 2018, 4, 36. [Google Scholar] [CrossRef]
- Lee, C.; Clark, S.E. A WUSCHEL-Independent Stem Cell Specification Pathway Is Repressed by PHB, PHV and CNA in Arabidopsis. PLoS ONE 2015, 10, e0126006. [Google Scholar] [CrossRef]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 1993, 119, 397–418. [Google Scholar]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 1995, 121, 2057–2067. [Google Scholar]
- Hu, C.; Zhu, Y.; Cui, Y.; Cheng, K.; Liang, W.; Wei, Z.; Zhu, M.; Yin, H.; Zeng, L.; Xiao, Y.; et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 2018, 4, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Perales, M.; Gruel, J.; Ohno, C.; Heisler, M.; Girke, T.; Reddy, G.V. Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 2013, 9, 654. [Google Scholar] [CrossRef] [PubMed]
- Magnani, E.; Jiménez-Gómez, J.M.; Soubigou-Taconnat, L.; Lepiniec, L.; Fiume, E. Profiling the onset of somatic embryogenesis in Arabidopsis. BMC Genom. 2017, 18, 998. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J.; Luo, J. The PIN-FORMED auxin efflux carriers in plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Bhosale, R. Developmental roles of AUX1/LAX auxin influx carriers in plants. Front. Plant Sci. 2019, 10, 1306. [Google Scholar] [CrossRef]
- Van Berkel, K.; de Boer, R.J.; Scheres, B.; ten Tusscher, K. Polar auxin transport: Models and mechanisms. Development 2013, 140, 2253–2268. [Google Scholar] [CrossRef]
- Bennett, T.; Hines, G.; van Rongen, M.; Waldie, T.; Sawchuk, M.G.; Scarpella, E.; Leyser, O. Connective auxin transport in the shoot facilitates communication between shoot apices. PLoS Biol. 2016, 14, e1002446. [Google Scholar] [CrossRef]
- Guenot, B.; Bayer, E.; Kierzkowski, D.; Smith, R.S.; Mandel, T.; Žádníková, P.; Kuhlemeier, C. Pin1-independent leaf initiation in Arabidopsis. Plant Physiol. 2012, 159, 1501–1510. [Google Scholar] [CrossRef]
- De Reuille, P.B.; Bohn-Courseau, I.; Ljung, K.; Morin, H.; Carraro, N.; Godin, C.; Traas, J. Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 1627–1632. [Google Scholar] [CrossRef]
- Lincoln, C.; Long, J.; Yamaguchi, J.; Serikawa, K.; Hake, S. A Knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 1994, 6, 1859–1876. [Google Scholar] [PubMed]
- Kierzkowski, D.; Runions, A.; Vuolo, F.; Strauss, S.; Lymbouridou, R.; Routier-Kierzkowska, A.L.; Zhang, Z. A Growth-Based Framework for Leaf Shape Development and Diversity. Cell 2019, 177, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Gälweiler, L.; Guan, C.; Müller, A.; Wisman, E.; Mendgen, K.; Yephremov, A.; Palme, K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 1998, 282, 2226–2230. [Google Scholar] [CrossRef] [PubMed]
- Banasiak, A.; Biedron, M.; Dolzblasz, A.; Berezowsk, M.A. Ontogenetic changes in auxin biosynthesis and distribution determine the organogenic activity of the shoot apical meristem in pin1 mutants. Int. J. Mol. Sci. 2019, 20, 180. [Google Scholar] [CrossRef]
- Vidaurre, D.P.; Ploense, S.; Krogan, N.T.; Berleth, T. AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 2007, 134, 2561–2567. [Google Scholar] [CrossRef]
- Zhao, Z.; Andersen, S.U.; Ljung, K.; Dolezal, K.; Miotk, A.; Schultheiss, S.J.; Lohmann, J.U. Hormonal control of the shoot stem-cell niche. Nature 2010, 465, 1089–1092. [Google Scholar] [CrossRef]
- Rademacher, E.H.; Möller, B.; Lokerse, A.S.; Llavata-Peris, C.I.; van den Berg, W.; Weijers, D. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011, 68, 597–606. [Google Scholar] [CrossRef]
- Denay, G.; Chahtane, H.; Tichtinsky, G.; Parcy, F. A flower is born: An update on Arabidopsis floral meristem formation. Curr. Opin. Plant Biol. 2017, 35, 15–22. [Google Scholar] [CrossRef]
- Krogan, N.T.; Marcos, D.; Weiner, A.I.; Berleth, T. The auxin response factor MONOPTEROS controls meristem function and organogenesis in both the shoot and root through the direct regulation of PIN genes. New Phytol. 2016, 212, 42–50. [Google Scholar] [CrossRef]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A Molecular Framework for Auxin-Controlled Homeostasis of Shoot Stem Cells in Arabidopsis. Mol. Plant 2018, 11, 819–891. [Google Scholar] [CrossRef]
- Bayer, E.M.; Smith, R.S.; Mandel, T.; Nakayama, N.; Sauer, M.; Prusinkiewicz, P.; Kuhlemeier, C. Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 2009, 23, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Capua, Y.; Eshed, Y. Coordination of auxin-triggered leaf initiation by tomato LEAFLESS. Proc. Natl. Acad. Sci. USA 2017, 114, 3246–3251. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Jiao, Y. The Diverse Roles of Auxin in Regulating Leaf Development. Plants 2019, 8, 243. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Mandel, T.; Kuhlemeier, C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 2000, 12, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Pesce, E.R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowska, D. Surface growth at the reproductive shoot apex of Arabidopsis thaliana pin-formed 1 and wild type. J. Exp. Bot. 2004, 55, 1021–1032. [Google Scholar] [CrossRef]
- Gendron, J.M.; Liu, J.S.; Fan, M.; Bai, M.Y.; Wenkel, S.; Springer, P.S.; Barton, M.K.; Wang, Z.Y. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 21152–21157. [Google Scholar] [CrossRef]
- Kawamura, E.; Horiguchi, G.; Tsukaya, H. Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 2010, 62, 429–441. [Google Scholar] [CrossRef]
- Bilsborough, G.D.; Runions, A.; Barkoulas, M.; Jenkins, H.W.; Hasson, A.; Galinha, C.; Laufs, P.; Hay, A.; Prusinkiewicz, P.; Tsiantis, M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 2011, 108, 3424–3429. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.; Hedden, P.; Tsiantis, M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005, 15, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Machida, C.; Nakagawa, A.; Kojima, S.; Takahashi, H.; Machida, Y. The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis. Wiley interdisciplinary reviews. Dev. Bio. 2015, 4, 655–671. [Google Scholar] [CrossRef]
- Grigg, S.P.; Canales, C.; Hay, A.; Tsiantis, M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 2005, 437, 1022–1026. [Google Scholar] [CrossRef]
- Li, Z.; Li, B.; Liu, J.; Guo, Z.; Liu, Y.; Li, Y.; Shen, W.-H.; Huang, Y.; Huang, H.; Zhang, Y.; et al. Transcription factors AS1 and AS2 interact with LHP1 to repress KNOX genes in Arabidopsis. J. Integr. Plant Biol. 2016, 58, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Mei, G.; Hu, Y.; Deng, J.; Zhang, T. LMI1-like and KNOX1 genes coordinately regulate plant leaf development in dicotyledons. Plant Mol. Biol. 2019, 99, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Satterlee, J.W.; Scanlon, M.J. Coordination of Leaf Development across Developmental Axes. Plants 2019, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.M.; Jun, J.H.; Nam, H.G.; Fletcher, J.C. BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 2007, 19, 1809–1825. [Google Scholar] [CrossRef]
- Jun, J.H.; Ha, C.M.; Fletcher, J.C. BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 2010, 22, 62–76. [Google Scholar] [CrossRef]
- Furumizu, C.; Alvarez, J.P.; Sakakibara, K.; Bowman, J.L. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet. 2015, 11, e1004980. [Google Scholar] [CrossRef]
- Qi, J.; Wu, B.; Feng, S.; Lu, S.; Guan, C.; Zhang, X.; Qiu, D.; Hu, Y.; Zhou, Y.; Li, C.; et al. Mechanical regulation of organ asymmetry in leaves. Nat. Plants 2017, 3, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Kazama, T.; Ichihashi, Y.; Murata, S.; Tsukaya, H. The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Andriankaja, M.; Dhondt, S.; De Bodt, S.; Vanhaeren, H.; Coppens, F.; De Milde, L.; Muhlenbock, P.; Skirycz, A.; Gonzalez, N.; Beemster, G.T.; et al. Exit from proliferation during leaf development in Arabidopsis thaliana: A not-so-gradual process. Dev. Cell 2012, 22, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- Sablowski, R.; Carnier Dornelas, M. Interplay between cell growth and cell cycle in plants. J. Exp. Bot. 2014, 65, 2703–2714. [Google Scholar] [CrossRef]
- Czesnick, H.; Lenhard, M. Size control in plants–lessons from leaves and flowers. Cold Spring Harb. Perspect. Biol. 2015, 7, a019190. [Google Scholar] [CrossRef]
- Mizukami, Y.; Fischer, R.L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 942–947. [Google Scholar] [CrossRef]
- Anastasiou, E.; Kenz, S.; Gerstung, M.; MacLean, D.; Timmer, J.; Fleck, C.; Lenhard, M. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 2007, 13, 843–856. [Google Scholar] [CrossRef]
- Lee, B.H.; Ko, J.H.; Lee, S.; Lee, Y.; Pak, J.H.; Kim, J.H. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 2009, 151, 655–668. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 2010, 137, 103–112. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008, 22, 1331–1336. [Google Scholar] [CrossRef] [PubMed]
- Challa, K.R.; Rath, M.; Nath, U. The CIN-TCP transcription factors promote commitment to differentiation in Arabidopsis leaf pavement cells via both auxin-dependent and independent pathways. PLoS Genet. 2019, 15, e1007988. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, G.; Kim, G.T.; Tsukaya, H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005, 43, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kende, H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 13374–13379. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef]
- Das Gupta, M.; Nath, U. Divergence in patterns of leaf growth polarity is associated with the expression divergence of miR396. Plant Cell 2015, 27, 2785–2799. [Google Scholar] [CrossRef]
- Vercruyssen, L.; Verkest, A.; Gonzalez, N.; Heyndrickx, K.S.; Eeckhout, D.; Han, S.K.; Jégu, T.; Archacki, R.; Van Leene, J.; Andriankaja, M.; et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodelling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 2014, 26, 210–229. [Google Scholar] [CrossRef]
- Efroni, I.; Blum, E.; Goldshmidt, A.; Eshed, Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 2008, 20, 2293–2306. [Google Scholar] [CrossRef]
- Yin, Z.; Li, Y.; Zhu, W.; Fu, X.; Han, X.; Wang, J.; Lin, H.; Ye, W. Identification, Characterization, and Expression Patterns of TCP Genes and microRNA319 in Cotton. Int. J. Mol. Sci. 2018, 19, 3655. [Google Scholar] [CrossRef]
- Nath, U.; Crawford, B.C.; Carpenter, R.; Coen, E. Genetic control of surface curvature. Science 2003, 299, 1404–1407. [Google Scholar] [CrossRef]
- Ori, N.; Cohen, A.R.; Etzioni, A.; Brand, A.; Yanai, O.; Shleizer, S.; Menda, N.; Amsellem, Z.; Efroni, I.; Pekker, I.; et al. Regulation of LANCEOLATE by miR319 is required for compound leaf development in tomato. Nat. Genet. 2007, 39, 787–791. [Google Scholar] [CrossRef]
- Kim, J.H. Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants. BMB Rep. 2019, 52, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.P.; Goldshmidt, A.; Efroni, I.; Bowman, J.L.; Eshed, Y. The NGATHA distal organ development genes are essential for style specification in Arabidopsis. Plant Cell 2009, 21, 1373–1393. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Poh, H.M.; Chua, N.-H. The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 2006, 47, 1–9. [Google Scholar] [CrossRef]
- John, F.; Roffler, S.; Wicker, T.; Ringli, C. Plant TOR signaling components. Plant Signal Behav. 2011, 6, 1700–1705. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xie, Q.; Chua, N.H. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 2003, 15, 1951–1961. [Google Scholar] [CrossRef]
- Okushima, Y.; Mitina, I.; Quach, H.L.; Theologis, A. AUXIN RESPONSE FACTOR 2 (ARF2): A pleiotropic developmental regulator. Plant J. 2005, 43, 29–46. [Google Scholar] [CrossRef]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef]
- Trigueros, M.; Navarrete-Gomez, M.; Sato, S.; Christensen, S.K.; Pelaz, S.; Weigel, D.; Yanofsky, M.F.; Ferrandiz, C. The NGATHA genes direct style development in the Arabidopsis gynoecium. Plant Cell 2009, 21, 1394–1409. [Google Scholar] [CrossRef]
- Martínez-Fernández, I.; Sanchís, S.; Marini, N.; Balanzá, V.; Ballester, P.; Navarrete-Gómez, M.; Oliveira, A.C.; Colombo, L.; Ferrándiz, C. The effect of NGATHA altered activity on auxin signaling pathways within the Arabidopsis gynoecium. Front Plant Sci. 2014, 5, 21. [Google Scholar]
- Bresso, E.G.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommer, C. Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 2018, 176, 1694–1708. [Google Scholar] [CrossRef] [PubMed]
- Nakata, M.; Matsumoto, N.; Tsugeki, R.; Rikirsch, E.; Laux, T.; Okada, K. Roles of the middle domain-specific WUSCHELRELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 2012, 24, 519–535. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Raman, A.S.; Ream, J.E.; Fujiwara, H.; Cerny, R.E.; Brown, S.M. Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 1998, 118, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Gusti, A.; Cheminant, S.; Alioua, M.; Dhondt, S.; Coppens, F.; Beemster, G.T.; Genschik, P. Gibberellin signalling controls cell proliferation rate in Arabidopsis. Curr. Biol. 2009, 19, 1188–1193. [Google Scholar] [CrossRef]
- Choe, S.; Fujioka, S.; Noguchi, T.; Takatsuto, S.; Yoshida, S.; Feldmann, K.A. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001, 26, 573–582. [Google Scholar] [CrossRef]
- Zhiponova, M.K.; Vanhoutte, I.; Boudolf, V.; Betti, C.; Dhondt, S.; Coppens, F.; Mylle, E.; Maes, S.; Gonzalez-Garcia, M.P.; Cano- Delgado, A.I.; et al. Brassinosteroid production and signalling differentially control cell division and expansion in the leaf. New Phytol. 2013, 197, 490–502. [Google Scholar] [CrossRef]
- Gonzalez, N.; Pauwels, L.; Baekelandt, A.; De Milde, L.; Van Leene, J.; Besbrugge, N.; Heyndrickx, K.S.; Cuellar Perez, A.; Durand, A.N.; De Clercq, R.; et al. A repressor protein complex regulates leaf growth in Arabidopsis. Plant Cell 2015, 27, 2273–2287. [Google Scholar] [CrossRef]
- Ikeda, M.; Fujiwara, S.; Mitsuda, N.; Ohme-Takagi, M. A triantagonistic basic helix–loop–helix system regulates cell elongation in Arabidopsis. Plant Cell 2012, 24, 4483–4497. [Google Scholar] [CrossRef]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef]
- Moon, J.; Hake, S. How a leaf gets its shape? Curr. Opin. Plant Biol. 2011, 14, 24–30. [Google Scholar] [CrossRef]
- Ochando, I.; González-Reig, S.; Ripoll, J.J.; Vera, A.; Martínez-Laborda, A. Alteration of the shoot radial pattern in Arabidopsis thaliana by a gain-of-function allele of the class III HD-Zip gene INCURVATA4. Int. J. Dev. Biol. 2008, 52, 953–961. [Google Scholar] [CrossRef]
- Waites, R.; Selvadurai, H.R.; Oliver, I.R.; Hudson, A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 1998, 93, 779–789. [Google Scholar] [CrossRef]
- Pekker, I.; John, P.A.; Yuval, E. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef] [PubMed]
- Sarojam, R.; Sappl, P.G.; Goldshmidt, A.; Efroni, I.; Floyd, S.K.; Eshed, Y.; Bowman, J.L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22, 2113–2130. [Google Scholar] [CrossRef]
- Kumaran, M.K.; Bowman, J.L.; Sundaresan, V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 2002, 14, 2761–2770. [Google Scholar] [CrossRef] [PubMed]
- Husbands, A.Y.; Benkovics, A.H.; Nogueira, F.T.; Lodha, M.; Timmermans, M.C. The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell 2015, 27, 3321–3335. [Google Scholar] [CrossRef]
- Williams, L.; Grigg, S.P.; Xie, M.; Christensen, S.; Fletcher, J.C. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166 g and its AtHD-ZIP target genes. Development 2005, 132, 3657–3668. [Google Scholar] [CrossRef]
- Singh, A.; Singh, S.; Panigrahi, K.C.; Reski, R.; Sarkar, A.K. Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 945–953. [Google Scholar] [CrossRef]
- Yu, T.; Guan, C.; Wang, J.; Sajjad, M.; Ma, L.; Jiao, Y. Dynamic patterns of gene expression during leaf initiation. J. Genet. Genom. 2017, 44, 599–601. [Google Scholar] [CrossRef]
- Eshed, Y.; Baum, S.F.; Perea, J.V.; Bowman, J.L. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001, 11, 1251–1260. [Google Scholar] [CrossRef]
- Kerstetter, R.A.; Bollman, K.; Taylor, R.A.; Bomblies, K.; Poethig, R.S. KANADI regulates organ polarity in Arabidopsis. Nature 2001, 411, 706–709. [Google Scholar] [CrossRef] [PubMed]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Roy, S.; Singh, S.; Das, S.S.; Gautam, V.; Yadav, S.; Sarkar, A.K. Phytohormonal crosstalk modulates the expression of miR166/165s, target Class III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana. Sci. Rep. 2017, 7, 3408. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, N.; Kim, H.B.; Park, N.I.; Kim, H.S.; Kim, Y.K.; Park, Y.I.; Choi, S.B. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010, 64, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Ohbayashi, I.; Takahashi, H.; Kojima, S.; Ishibashi, N.; Keta, S.; Sugiyama, M. A genetic link between epigenetic repressor AS1-AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis. Biol. Open 2016, 5, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.Q.; Keta, S.; Asai, T.; Kojima, S.; Nakagawa, A.; Micol, J.L.; Machida, C. A genetic link between epigenetic repressor AS1-AS2 and DNA replication factors in establishment of adaxial-abaxial leaf polarity of Arabidopsis. Plant Biotechnol. (Tokyo) 2018, 35, 39–49. [Google Scholar] [CrossRef]
- de Felippes, F.F.; Marchais, A.; Sarazin, A.; Oberlin, S.; Voinnet, O. A single miR390 targeting event is sufficient for triggering TAS3-tasiRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2017, 45, 5539–5554. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.H.; Liou, L.W.; Zhang, X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef]
- Xia, R.; Xu, J.; Meyers, B.C. The emergence, evolution, and diversification of the miR390–TAS3–ARF pathway in land plants. Plant Cell 2017, 29, 1232–1247. [Google Scholar] [CrossRef]
- Sawa, S.S.; Watanabe, K.; Goto, K.; Kanaya, E.; Morita, E.H.; Okada, K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domain. Gene Dev. 1999, 13, 1079–1088. [Google Scholar] [CrossRef]
- Guan, C.; Wu, B.; Yu, T.; Wang, Q.; Krogan, N.T.; Liu, X.; Jiao, Y. Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis. Curr. Biol. CB 2017, 27, 2940–2950. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Eshed, Y.; Baum, S.F.; Otsuga, D.; Drews, G.N.; Bowman, J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 1999, 126, 4117–4128. [Google Scholar] [PubMed]
- Li, H.; Xu, L.; Wang, H.; Yuan, Z.; Cao, X.; Yang, Z.; Zhang, D.; Xu, Y.; Huang, H. The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA165/166 in Arabidopsis leaf development. Plant Cell 2005, 17, 2157–2171. [Google Scholar] [CrossRef]
- Garcia, D.; Collier, S.A.; Byrne, M.E.; Martienssen, R.A. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 2006, 16, 933–938. [Google Scholar] [CrossRef]
- Eshed, Y.; Izhaki, A.; Baum, S.F.; Floyd, S.K.; Bowman, J.L. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 2004, 131, 2997–3006. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, A.; Yockteng, R.; Schnable, J.; Alvarez-Buylla, E.R.; Freeling, M.; Specht, C.D. Co-option of the polarity gene network shapes filament morphology in angiosperms. Sci. Rep. 2015, 4, 6194. [Google Scholar] [CrossRef] [PubMed]
- Takahiro, Y.; Akira, N.; Hirokazu, T. Leaf adaxial–abaxial polarity specification and lamina outgrowth: Evolution and development. Plant Cell Physiol. 2012, 53, 1180–1194. [Google Scholar] [CrossRef]
- Floyd, S.K.; Bowman, J.L. The ancestral developmental tool kit of land plants. Int. J. Plant Sci. 2007, 168, 1–35. [Google Scholar] [CrossRef]
- Floyd, S.K.; Bowman, J.L. Gene expression patterns in seed plant shoot meristems and leaves: Homoplasy or homology? J. Plant Res. 2010, 123, 43–55. [Google Scholar] [CrossRef]
- Nardmann, J.; Ji, J.; Werr, W.; Scanlon, M.J. The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 2004, 131, 2827–2839. [Google Scholar] [CrossRef]
- Aichinger, E.; Kornet, N.; Friedrich, T.; Laux, T. Plant stem cell niches. Annu. Rev. Plant Biol. 2012, 63, 615–636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tucker, E.; Hermann, M.; Laux, T. A Molecular Framework for the Embryonic Initiation of Shoot Meristem Stem Cells. Dev. Cell 2017, 40, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Floyd, S.K. Patterning and polarity in seed plant shoots. Annu. Rev. Plant. Biol. 2008, 59, 67–88. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Matsuda, Y.; Ozawa, K.; Nishimura, T.; Koshiba, T.; Fraaije, M.W.; Sekiguchi, H. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 2008, 279, 499–507. [Google Scholar] [CrossRef]
- Kong, X.; Huang, G.; Xiong, Y.; Zhao, C.; Wang, J.; Song, X.; Zuo, K. IBR5 Regulates Leaf Serrations Development via Modulation of the Expression of PIN1. Int. J. Mol. Sci. 2019, 20, 4429. [Google Scholar] [CrossRef]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef]
- Maugarny-Calès, A.; Cortizo, M.; Adroher, B.; Borrega, N.; Gonçalves, B.; Brunoud, G.; Laufs, P. Dissecting the pathways coordinating patterning and growth by plant boundary domains. PLoS Genet. 2019, 15, e1007913. [Google Scholar] [CrossRef]
- Hasson, A.; Plessis, A.; Blein, T.; Adroher, B.; Grigg, S.; Tsiantis, M.; Laufs, P. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 2011, 110, 54–68. [Google Scholar] [CrossRef]
- Sharma, A.; Zheng, B. Molecular Responses during Plant Grafting and Its Regulation by Auxins, Cytokinins, and Gibberellins. Biomolecules 2019, 9, 397. [Google Scholar] [CrossRef]
- Rast-Somssich, M.I.; Žádníková, P.; Schmid, S.; Kieffer, M.; Kepinski, S.; Simon, R. The Arabidopsis JAGGED LATERAL ORGANS (JLO) gene sensitizes plants to auxin. J. Exp. Bot. 2017, 68, 2741–2755. [Google Scholar] [CrossRef]
- Rast, M.I.; Simon, R. Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell 2012, 24, 2917–2933. [Google Scholar] [CrossRef] [PubMed]
- Žádníková, P.; Simon, R. How boundaries control plant development. Curr. Opin. Plant Biol. 2014, 17, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, B.; Hasson, A.; Belcram, K.; Cortizo, M.; Morin, H.; Nikovics, K.; Vialette-Guiraud, A.; Takeda, S.; Aida, M.; Laufs, P.; et al. A conserved role for CUP-SHAPED COTYLEDON genes during ovule development. Plant J. 2015, 83, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Engelhorn, J.; Reimer, J.J.; Leuz, I.; Göbel, U.; Huettel, B.; Farrona, S.; Turck, F. DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 controls leaf margin architecture in Arabidopsis thaliana. Development 2012, 139, 2566–2575. [Google Scholar] [CrossRef]
- Yang, Y.; Nicolas, M.; Zhang, J.; Yu, H.; Guo, D.; Yuan, R.; Qin, G. Correction: The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 2018, 14, e1007565. [Google Scholar] [CrossRef]
- Tao, Q.; Guo, D.; Wei, B.; Zhang, F.; Pang, C.; Jiang, H.; Zhang, J.; Wei, T.; Gu, H.; Qu, L.J.; et al. The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/ TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 2013, 25, 421–437. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, B.; Yuan, R.; Wang, J.; Ding, M.; Chen, Z.; Yu, H.; Qin, G. The Arabidopsis RING-type E3 ligase TEAR1 controls leaf development by targeting the TIE1 transcrip-tional repressor for degradation. Plant Cell 2017, 29, 243–259. [Google Scholar] [CrossRef]
- Tameshige, T.; Okamoto, S.; Lee, J.S.; Aida, M.; Tasaka, M.; Torii, K.U.; Uchida, N. A secreted peptide and its receptors shape the auxin response pattern and leaf margin morphogenesis. Curr. Biol. 2016, 26, 2478–2485. [Google Scholar] [CrossRef]
- Toriba, T.; Tokunaga, H.; Shiga, T.; Nie, F.; Naramoto, S.; Honda, E.; Kyozuka, J. BLADE-ON-PETIOLE genes temporally and developmentally regulate the sheath to blade ratio of rice leaves. Nat. Commun. 2019, 10, 619. [Google Scholar] [CrossRef]
- Norberg, M.; Holmlund, M.; Nilsson, O. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 2005, 132, 2203–2213. [Google Scholar] [CrossRef]
- Sakamoto, T.; Sotta, N.; Suzuki, T.; Fujiwara, T.; Matsunaga, S. The 26S proteasome is required for the maintenance of root apical meristem by modulating auxin and cytokinin responses under high-boron stress. Front. Plant Sci. 2019, 10, 590. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, A.M.; Nodine, M.D.; Gaj, M.D. miR160 and miR166/165 Contribute to the LEC2-Mediated Auxin Response Involved in the Somatic Embryogenesis Induction in Arabidopsis. Front. Plant Sci. 2017, 8, 2024. [Google Scholar] [CrossRef] [PubMed]
- Saini, K.; Markakis, M.N.; Zdanio, M.; Balcerowicz, D.M.; Beeckman, T.; De Veylder, L.; Vissenberg, K. Alteration in auxin homeostasis and signaling by overexpression of PINOID kinase causes leaf growth defects in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1009. [Google Scholar] [CrossRef] [PubMed]
- Kasprzewska, A.; Carter, R.; Swarup, R.; Bennett, M.; Monk, N.; Hobbs, J.K.; Fleming, A. Auxin influx importers modulate serration along the leaf margin. Plant J. 2015, 83, 705–718. [Google Scholar] [CrossRef]
- Stefano, P.; Ivana, M.; Arianna, G.; Letizia, Z.; Angelo, G.; Rosella, C.; Andrea, G. Identification of microRNAs and relative target genes in Moringa oleifera leaf and callus. Sci. Rep. 2019, 9, 1–14. [Google Scholar]
- Wang, J.; Mei, J.; Ren, G. Plant microRNAs: Biogenesis, homeostasis and degradation. Front. Plant Sci. 2019, 10, 360. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Du, T.; Zamore, P.D. microPrimer: The biogenesis and function of microRNA. Development 2005, 132, 4645–4652. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant MicroRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef] [PubMed]
- Bologna, N.G.; Mateos, J.L.; Bresso, E.G.; Palatnik, J.F. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009, 28, 3646–3656. [Google Scholar] [CrossRef]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Shao, C.; Chen, M. Toward microRNA-mediated gene regulatory networks in plants. Brief Bioinform. 2011, 12, 645–659. [Google Scholar] [CrossRef]
- Yu, Y.; Ji, L.; Le, B.H.; Zhai, J.; Chen, J.; Luscher, E.; Gao, L.; Liu, C.; Cao, X.; Mo, B.; et al. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 2017, 15, e2001272. [Google Scholar] [CrossRef]
- Armenta-Medina, A.; Lepe-Soltero, D.; Xiang, D.Q.; Datla, R.; Abreu-Goodger, C.; Gillmor, C.S. Arabidopsis thaliana miRNAs promote embryo pattern formation beginning in the zygote. Dev. Biol. 2017, 431, 145–151. [Google Scholar] [CrossRef]
- Knauer, S.; Holt, A.L.; Rubio-Somoza, I.; Tucker, E.J.; Hinze, A.; Pisch, M.; Javelle, M.; Timmermans, M.C.; Tucker, M.R.; Laux, T. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 2013, 24, 125–132. [Google Scholar] [CrossRef]
- Pulido, A.; Laufs, P. Co-ordination of developmental processes by small RNAs during leaf development. J. Exp. Bot. 2010, 61, 1277–1291. [Google Scholar] [CrossRef]
- Yang, T.; Wang, Y.; Teotia, S.; Wang, Z.; Shi, C.; Sun, H.; Tang, G. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci. Rep. 2019, 9, 2832. [Google Scholar] [CrossRef]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Chen, J.; Zhou, J.; Yu, H.; Ge, C.; Zhang, M.; Zhao, Y. An essential role for miRNA167 in maternal control of embryonic and seed development. Plant Physiol. 2019, 180, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Ding, N.; Fan, W.; Yan, J.; Gu, Y.; Tang, X.; Li, R.; Tang, G. Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. 2015, 233, 11–21. [Google Scholar] [CrossRef]
- Fahlgren, N.; Montgomery, T.A.; Howell, M.D.; Allen, E.; Dvorak, S.K.; Alexander, A.L.; Carrington, J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006, 16, 939–944. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Liu, Y.; Li, D.; Zhao, Y.; Li, Z.; Liu, Z. Overexpression of OsAGO1b Induces Adaxially Rolled Leaves by Affecting Leaf Abaxial Sclerenchymatous Cell Development in Rice. Rice 2019, 12, 60. [Google Scholar] [CrossRef]
- Kidner, C.A.; Martienssen, R.A. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 2004, 428, 81–84. [Google Scholar] [CrossRef]
- Nagasaki, H.; Itoh, J.I.; Hayashi, K.; Hibara, K.I.; Satoh-Nagasawa, N.; Nosaka, M.; Nagato, Y. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 14867–14871. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Scanlon, M.J.; Schneeberger, R.G.; Freeling, M. The maize mutant narrow sheath fails to establish leaf margin identity in a meristematic domain. Development 1996, 122, 1683–1691. [Google Scholar]
- Merelo, P.; Ram, H.; Caggiano, M.P.; Ohno, C.; Ott, F.; Straub, D.; Heisler, M.G. Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc. Natl. Acad. Sci. USA 2016, 113, 11973–11978. [Google Scholar] [CrossRef] [PubMed]
- Waites, R.; Hudson, A. phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 1995, 121, 2143–2154. [Google Scholar]
- Chuck, G.; Whipple, C.; Jackson, D.; Hake, S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 2010, 137, 1243–1250. [Google Scholar] [CrossRef]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef]
- Damodharan, S.; Zhao, D.; Arazi, T. A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 2016, 86, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Kasschau, K.D.; Carrington, J.C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003, 13, 784–789. [Google Scholar] [CrossRef]
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76. [Google Scholar] [CrossRef]
- Busch, B.L.; Schmitz, G.; Rossmann, S.; Piron, F.; Ding, J.; Bendahmane, A.; Theres, K. Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell 2011, 23, 3595–3609. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yao, X.; Pi, L.; Wang, H.; Cui, X.; Huang, H. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 2009, 58, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2- like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Cartolano, M.; Castillo, R.; Efremova, N.; Kuckenberg, M.; Zethof, J.; Gerats, T.; Vandenbussche, M. A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat. Genet. 2007, 39, 901. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef]
- Prigge, M.J.; Otsuga, D.; Alonso, J.M.; Ecker, J.R.; Drews, G.N.; Clark, S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005, 17, 61–76. [Google Scholar] [CrossRef]
- Vaucheret, H. AGO1 Homeostasis Involves Differential Production of 21-nt and 22-nt miR168 Species by MIR168a and MIR168b. PLoS ONE 2009, 4, e6442. [Google Scholar] [CrossRef]
- Ji, L.; Liu, X.; Yan, J.; Wang, W.; Yumul, R.E.; Kim, Y.J.; Dinh, T.T.; Liu, J.; Cui, X.; Zheng, B.; et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011, 7, e1001358. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, S.; Yun, J.; Lee, M.; Park, C.M. The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci. 2014, 215, 29–38. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Wollmann, H.; Schommer, C.; Schwab, R.; Boisbouvier, J.; Rodriguez, R.; Warthmann, N.; Allen, E.; Dezulian, T.; Huson, D.; et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 2007, 13, 115–125. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7–miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Laufs, P.; Peaucelle, A.; Morin, H.; Traas, J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 2004, 131, 4311–4322. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.S.; Li, J.; Stahle, M.I.; Dubroué, A.; Gubler, F.; Millar, A.A. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. USA 2007, 104, 16371–16376. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.K.; Xu, Y.M.; Shi, M.; Lai, Y.M.; Wu, X.; Wang, H.S.; Zhu, Z.; Poethig, R.S.; Wu, G. Repression of miR156 by miR159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell 2017, 29, 1293–1304. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef]
- Wollmann, H.; Mica, E.; Todesco, M.; Long, J.A.; Weigel, D. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 2010, 137, 3633–3642. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ping, X.; Cao, Y.; Jian, H.; Gao, Y.; Wang, J.; Liu, L. Genome-wide exploration and characterization of miR172/euAP2 genes in Brassica napus L. for likely role in flower organ development. BMC Plant Biol. 2019, 19, 336. [Google Scholar] [CrossRef]
- Koyama, T.; Furutani, M.; Tasaka, M.; Ohme-Takagi, M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 2007, 19, 473–484. [Google Scholar] [CrossRef]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. Ethylene-insensitive3 is a senescence associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef] [PubMed]
- Parry, G.; Calderon-Villalobos, L.I.; Prigge, M.; Peret, B.; Dharmasiri, S.; Itoh, H.; Lechner, E.; Gray, W.M.; Bennett, M.; Estelle, M. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. USA 2009, 106, 22540–22545. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Chen, R. PHANTASTICA regulates leaf polarity and petiole identity in Medicago truncatula. Plant Signal. Behav. 2014, 9, e28121. [Google Scholar] [CrossRef] [PubMed]


| Gene | Description | Biological Function | Species | References |
|---|---|---|---|---|
| ASYMMETRIC LEAVES1/ROUGH SHEATH 2/PHANTASTICA (ARP) | MYB domain protein | Stem cell differentiation | Arabidopsis, Zea mays, Antirrhinum majus | [31,32,36,37,38] |
| AUXIN RESPONSE FACTORS (ARF) | Protein with N-terminal DNA binding domain, activator/repressor | Leaf polarity | Arabidopsis | [39] |
| ANGUSTIFOLIA(AN3)/GRF-INTERACTING FACTOR1(GIF1) | Transcription coactivators | Cell proliferation | Arabidopsis | [40,41] |
| CLAVATA (CLV) | CLV1 (receptor kinase); CLV2 (transmembrane protein); CLV3 (extracellular protein) | Maintain stem cell size | Arabidopsis | [42,43] |
| Class-1 KNOTTED-like homeobox (KNOX1) | Homeodomain protein | Maintain stem cell identity | Arabidopsis, Zea mays | [28,44] |
| CUP-SHAPED COTYLEDON2 (CUC2) | Protein containing the NAC DNA binding domain | Shoot meristem formation; organ boundary specification; and leaf margin development | Arabidopsis | [45] |
| DEVELOPMENT-RELATE PcG TARGET IN THE APEX 4 (DPA4) | RAV transcription repressor | Organ initiation and development; leaf margin development | Arabidopsis | [46] |
| GROWTH-REGULATING FACTOR5 (GRF5) | Transcription activators containing N-terminal QLQ or WRC domain | Cell proliferation | Arabidopsis | [47] |
| Narrow sheath (ns)/PRESSED FLOWER (PRS) | Homeodomain protein | Recruitment of leaf founder cells and leaf expansion/Marginal cell proliferation | Zea mays Arabidopsis | [44,48,49] |
| PIN-FORMED1 (PIN1) | Transmembrane protein | Auxin efflux | Arabidopsis | [34] |
| PHANTASTICA (PHAN) | MYB domain protein | Stem cell differentiation | Antirrhinum majus | [50] |
| WUSCHEL (WUS) | Homeodomain protein | Maintain shoot and floral meristem identity | Arabidopsis | [51] |
| YABBY (YAB) | Protein with zinc-finger and helix-loop-helix domains | Specification of Leaf polarity and lamina expansion | Arabidopsis | [52,53] |
| miRNAs Family | Target Families/Gene | Biological Function | Species | References |
|---|---|---|---|---|
| miR156 | SPL | Promote vegetative phase change and floral transition | Arabidopsis, Zea mays | [239,240,241] |
| miR159 | MYB TFS:GAMYB, MYB33 | Control floral identity and flower development | Arabidopsis | [242] |
| miR160 | ARF | Leaf and root development, auxin response, floral organ identity | Arabidopsis | [229,243] |
| miR162 | DCL1 | Arabidopsis | [244] | |
| miR164 | NAC-TF: CUC1,CUC2 | Shoot and root development | Arabidopsis, Solanum, and Oryza | [45,192,238,245] |
| miR164a | NAC-TF: CUC1, CUC2 | Leaf development, patterning, and polarity | [26,246] | |
| miR164c | NAC-TF: CUC1,CUC2 | Floral identity and flower development | [247] | |
| miR165/166 | HD-ZIP, PHB | Meristem maintenance, vascular development and organ polarity | Arabidopsis | [160,248,249,250] |
| miR167 | ARF6 and 8 | Auxin response | Arabidopsis | [251] |
| miR168 | AGO1 | Arabidopsis | [252] | |
| miR172 | AP2 | Developmental timing and floral organ identity | Arabidopsis, Z. mays, S. tuberosum | [253,254,255,256] |
| miR319 | TCP | Leaf development | Arabidopsis and Solanum lycopersicum | [257] |
| miR390 | TAS3 | Auxin response, developmental timing, lateral organ polarity | Arabidopsis | [233,258] |
| miR393 | F-box protein: TIR1 | Hormone signaling for plant development | Arabidopsis, Oryza | [259] |
| miR396 | GRF faimly | Control cell proliferation | Arabidopsis, Medicago and Oryza | [127,260,261] |
| miR408 | Plantacyanin, Laccases | Stress response | Arabidopsis | [262] |
| TAS3 | ARF3 and (only mosses) AP2 like | Leaf polarity | All land plants | [177,233] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, S.; Khan, N.; Xie, L. Molecular and Hormonal Regulation of Leaf Morphogenesis in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 5132. https://doi.org/10.3390/ijms21145132
Ali S, Khan N, Xie L. Molecular and Hormonal Regulation of Leaf Morphogenesis in Arabidopsis. International Journal of Molecular Sciences. 2020; 21(14):5132. https://doi.org/10.3390/ijms21145132
Chicago/Turabian StyleAli, Shahid, Naeem Khan, and Linan Xie. 2020. "Molecular and Hormonal Regulation of Leaf Morphogenesis in Arabidopsis" International Journal of Molecular Sciences 21, no. 14: 5132. https://doi.org/10.3390/ijms21145132
APA StyleAli, S., Khan, N., & Xie, L. (2020). Molecular and Hormonal Regulation of Leaf Morphogenesis in Arabidopsis. International Journal of Molecular Sciences, 21(14), 5132. https://doi.org/10.3390/ijms21145132






