Environmental DNA and RNA as Records of Human Exposome, Including Biotic/Abiotic Exposures and Its Implications in the Assessment of the Role of Environment in Chronic Diseases
Abstract
1. Introduction
2. eDNA and Environmental Impact Assessment
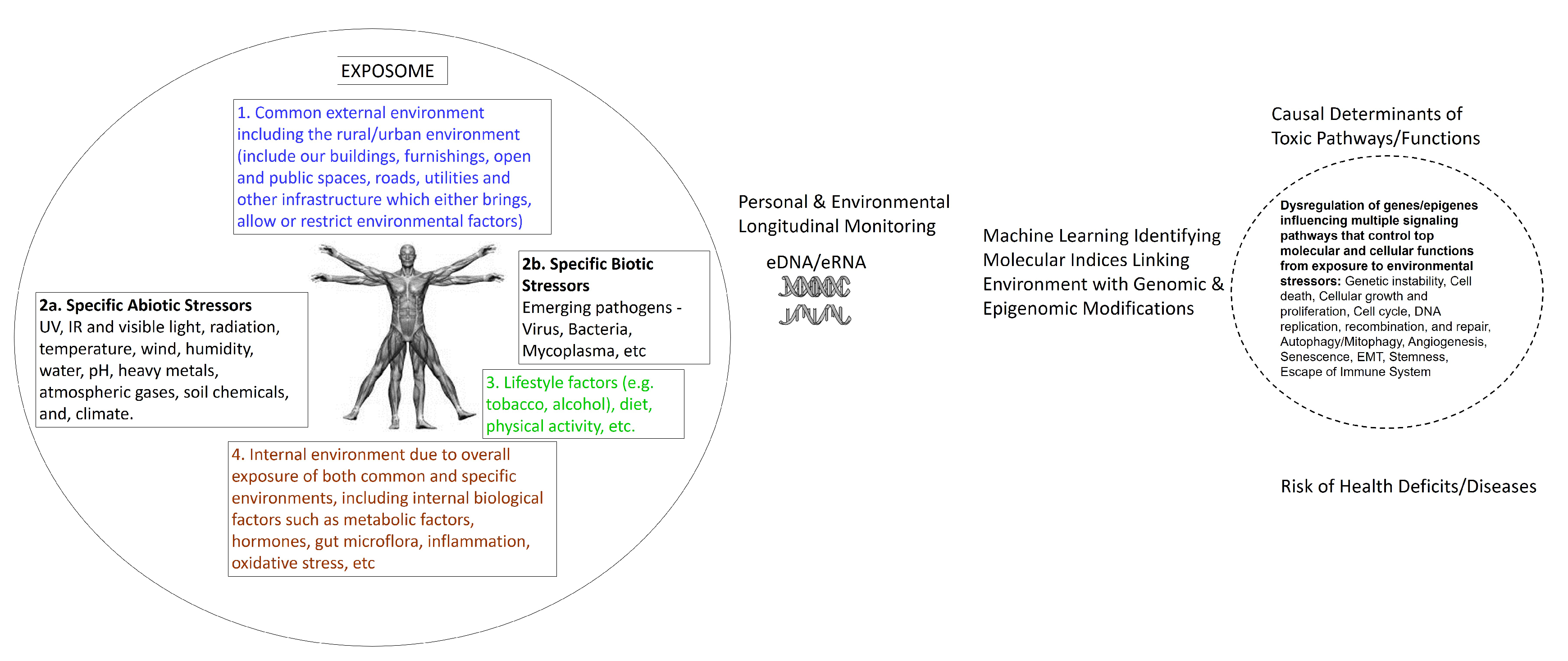
3. Can eDNA/eRNA Biomonitor the Exposome?
4. Biomonitoring Exposome of Terrestrial, Fresh Water, and Marine Life
5. Human Genome–Exposome Interface: Environmental Functional Genomics
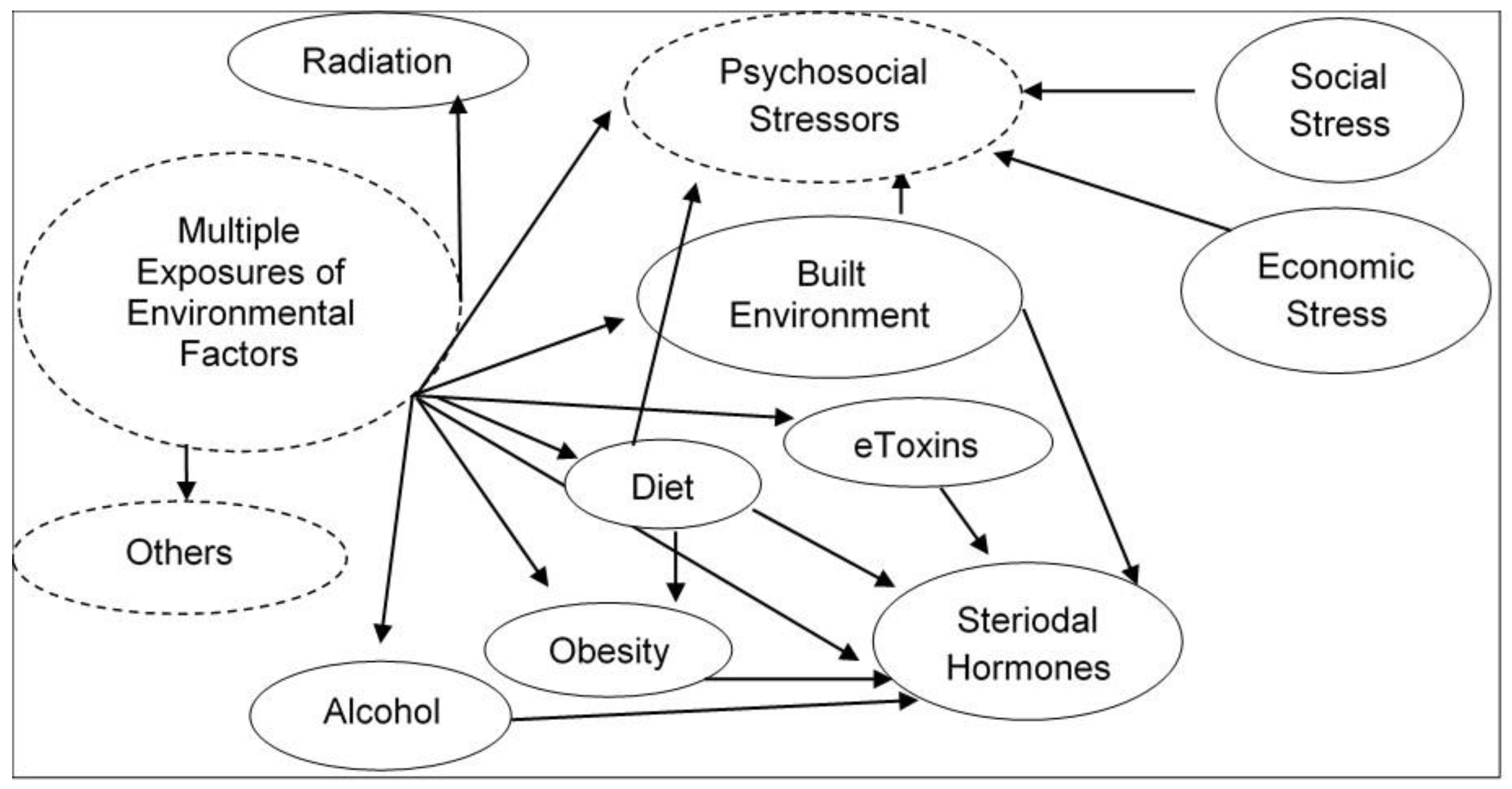
6. Epigenome Modifications from Exposure to Environmental Stressors that May Increase Susceptibility of Chronic Diseases
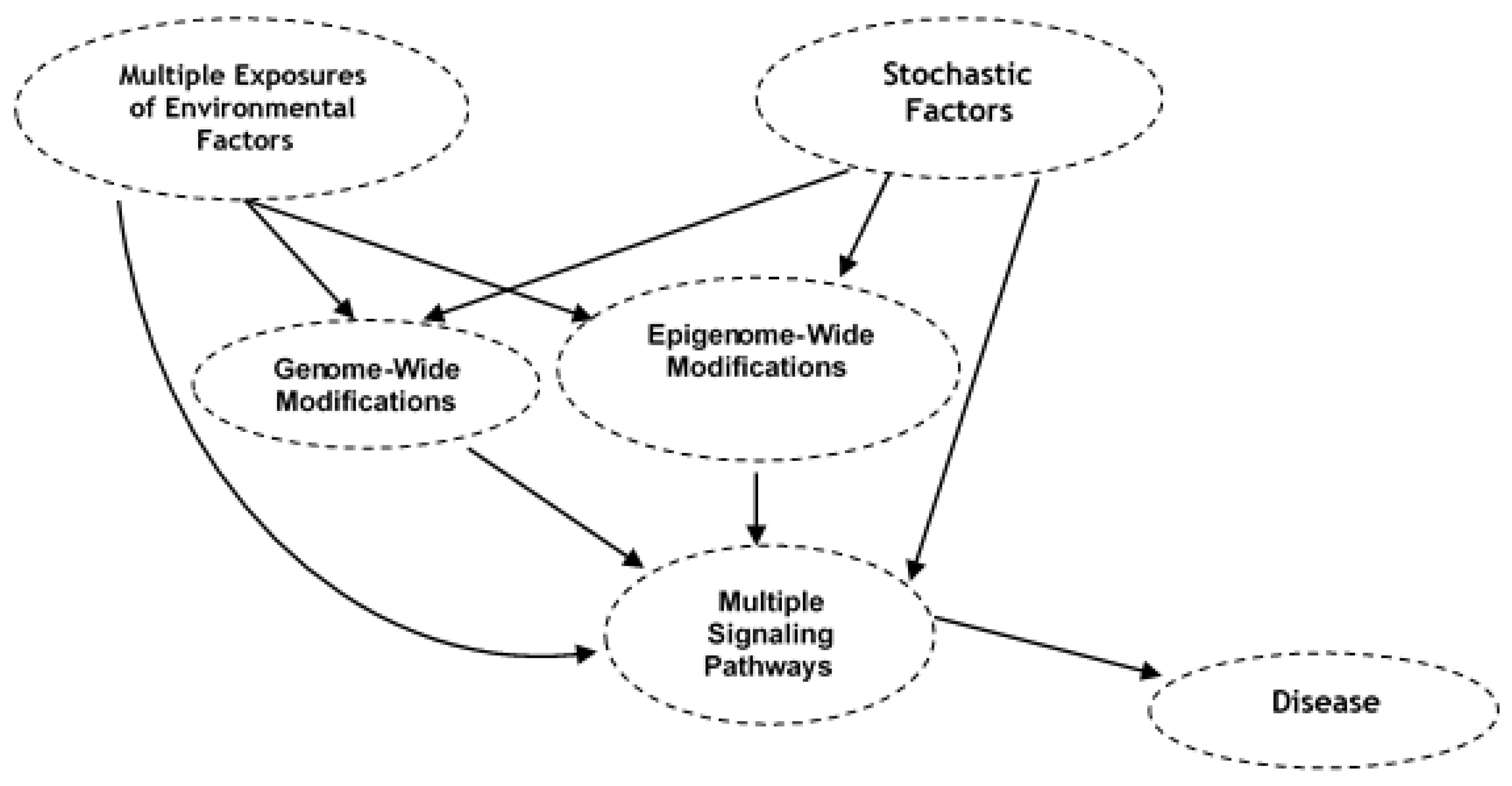
7. Bioinformatics to Assess Causal Association between Environmental Exposure, Genome-Wide and Epigenome-Wide Modifications, and Disease Phenotype
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wild, C.P. The exposome: From concept to utility. Int. J. Epidemiol. 2012, 41, 24–32. [Google Scholar] [CrossRef]
- Deoraj, A.; Yoo, C.; Roy, D. Integrated bioinformatics, biostatistics and molecular epidemiologic approaches to study how the environment and genes work together to influence the development of complex chronic diseases. In Gene Environment Interaction Studies; Anno, S., Ed.; Pan Stanford Publishing: Singapore, 2016; pp. 160–200. [Google Scholar]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome—From metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Ho, S.Y.W.; Heupink, T.H.; Rambaut, A.; Shapiro, B. Bayesian Estimation of Sequence Damage in Ancient DNA. Mol. Biol. Evol. 2007, 24, 1416–1422. [Google Scholar] [CrossRef]
- Orlando, L.; Cooper, A. Using ancient DNA to understand evolutionary and ecological processes. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 573–598. [Google Scholar] [CrossRef]
- Beng, K.C.; Corlett, R.T. Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Zaiko, A.; Pochon, X.; Garcia-Vazquez, E.; Olenin, S.; Wood, S.A. Advantages and limitations of environmental DNA/RNA tools for marine biosecurity: Management and surveillance of non-indigenous species. Front. Mar. Sci. 2018, 5, 322. [Google Scholar] [CrossRef]
- Harrison Jori, B.; Sunday Jennifer, M.; Rogers Sean, M. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B 2019, 286, 20191409. [Google Scholar] [CrossRef]
- Seymour, M. Rapid progression and future of environmental DNA research. Commun Biol. 2019, 2, 80. [Google Scholar] [CrossRef]
- Winding, A.; Bang-Andreasen, T.; Hansen, L.H.; Panitz, F.; Krogh, P.H.; Krause-Jensen, D.; Stæhr, P.; Nicolaisen, M.; Hendriksen, N.B.; Sapkota, R.; et al. eDNA in Environmental Monitoring. Aarhus University, DCE—Danish Centre for Environment and Energy. Technical Report No. 133. 2019. Available online: http://dce2.au.dk/pub/TR133.pdf (accessed on 10 July 2020).
- Zhang, P.; Arora, M.; Chaleckis, R.; Isobe, T.; Jain, M.; Meister, I.; Melén, E.; Perzanowski, M.; Torta, F.; Wenk, M.R.; et al. Tackling the complexity of the exposome: Considerations from the Gunma University Initiative for Advanced Research (GIAR) Exposome Symposium. Metabolites 2019, 9, 106. [Google Scholar] [CrossRef]
- Zinger, L.; Donald, J.; Brosse, S.; Gonzalez, M.A.; Iribar, A.; Leroy, C.; Murienne, J.; Orivel, J.; Schimann, H.; Taberlet, P.; et al. Advances and prospects of environmental DNA in neotropical rainforests. Adv. Ecol. Res. 2020. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Mishra, A.; Jha, G.; Thakur, I.S. Draft genome sequence of Zhihengliuella sp. ISTPL4, a psychrotolerant and halotolerant bacterium isolated from Pangong Lake, India. Genome Announc. 2018, 6, e01533-17. [Google Scholar] [CrossRef] [PubMed]
- Rathour, R.; Gupta, J.; Kumar, M.; Hiloidhari, M.; Mehrotra, A.K.; Thakur, I.S. Metagenomic sequencing of microbial communities from a brackish water of pangong lake of north west Indian Himalayas. Genome Announc. 2017, 5, e01029-17. [Google Scholar] [CrossRef]
- Gupta, J.; Rathour, R.; Kumar, M.; Hiloidhari, M.; Mehrotra, A.K.; Thakur, I.S. Metagenomic analysis of microbial diversity in landfill lysimeter soil of Ghazipur landfill site, New Delhi, India. Genome Announc. 2017, 5, e01104. [Google Scholar] [CrossRef]
- Rathour, R.; Gupta, J.; Mishra, A.; Rajeev, A.C.; Dupont, C.L.; Thakur, I.S. A comparative metagenomic study reveals microbial diversity and their role in the biogeochemical cycling of Pangong lake. Sci. Total Environ. 2020. [Google Scholar] [CrossRef]
- Biggs, J.; Ewald, N.; Valentini, A.; Gaboriaud, C.; Dejean, T.; Griffiths, R.A.; Foster, J.; Wilkinson, W.K.; Arnell, A.; Brotherton, P.; et al. Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol. Conserv. 2015, 183, 19–28. [Google Scholar] [CrossRef]
- Klymus, K.E.; Richter, C.A.; Chapman, D.C.; Paukert, C. Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 2014, 183, 77–84. [Google Scholar] [CrossRef]
- Brown, E.A.; Chain, F.J.J.; Zhan, A.; MacIsaac, H.J.; Cristescu, M.E. Early detection of aquatic invaders using metabarcoding reveals a high number of non-indigenous species in Canadian ports. Divers. Distrib. 2016, 22, 1045–1059. [Google Scholar] [CrossRef]
- Zaiko, A.; Martinez, J.L.; Schmidt-Petersen, J.; Ribicic, D.; Samuiloviene, A.; Garcia-Vazquez, E. Metabarcoding approach for the ballast water surveillance an advantageous solution or an awkward challenge? Mar. Pollut. Bull. 2015, 92, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Zaiko, A.; Samuiloviene, A.; Ardura, A.; Garcia-Vazquez, E. Metabarcoding approach for nonindigenous species surveillance in marine coastal waters. Mar. Pollut. Bull. 2015, 100, 53–59. [Google Scholar] [CrossRef]
- Zaiko, A.; Schimanski, K.; Pochon, X.; Hopkins, G.A.; Goldstien, S.; Floerl, O.; Wood, S.A. Metabarcoding improves detection of eukaryotes from early biofouling communities: Implications for pest monitoring and pathway management. Biofouling J. Bioadh. Biofilm Res. 2016, 32, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Lanzen, A.; Lekang, K.; Jonassen, I.; Thompson, E.M.; Troedsson, C. High-throughput metabarcoding of eukaryotic diversity for environmental monitoring of offshore oil-drilling activities. Mol. Ecol. 2016, 25, 4392–4406. [Google Scholar] [CrossRef] [PubMed]
- Lanzen, A.; Lekang, K.; Jonassen, I.; Thompson, E.M.; Troedsson, C. DNA extraction replicates improve diversity and compositional dissimilarity y in metabarcoding of eukaryotes in marine sediments. PLoS ONE 2017, 12, e0179443. [Google Scholar] [CrossRef] [PubMed]
- Laroche, O.; Wood, S.A.; Tremblay, L.A.; Ellis, J.I.; Lear, G.; Pochon, X. A cross-taxa study using environmental DNA/RNA metabarcoding to measure biological impacts of offshore oil and gas drilling and production operations. Mar. Pollut. Bull. 2018, 127, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Jiang, W.; Wang, B.; Fang, J.; Lang, J.; Tian, G.; Jiang, J.; Zhu, T.F. Inhalable microorganisms in Beijing’s PM2.5 and PM10 pollutants during a severe smog event. Environ. Sci. Technol. 2014, 48, 1499–14507. [Google Scholar] [CrossRef]
- Tong, X.; Xu, H.; Zou, L.; Cai, M.; Xu, X.; Zhao, Z.; Xiao, F.; Li, Y. High diversity of airborne fungi in the hospital environment as revealed by metasequencing-based microbiome analysis. Sci. Rep. 2017, 7, 39606. [Google Scholar] [CrossRef]
- Dennis, K.K.; Marder, E.; Balshaw, D.M.; Cui, Y.; Lynes, A.M.; Patti, J.G.; Rappaport, M.S.; Shaughnessy, T.D.; Vrijheid, M.; Barr, B.D. Biomonitoring in the Era of the Exposome. Environ. Health Perspect. 2017, 125, 502–510. [Google Scholar] [CrossRef]
- Allwood, J.S.; Fierer, N.; Dunn, R.R. The Future of Environmental DNA in Forensic Science. Appl. Environ. Microbiol. 2020, 86, e01504-19. [Google Scholar] [CrossRef]
- Fløjgaard, C.; Frøslev, T.G.; Brunbjerg, A.K.; Bruun, H.H.; Moeslund, J.; Hansen, A.J.; Ejrnæs, R. Predicting provenance of forensic soil samples: Linking soil to ecological habitats by metabarcoding and supervised classification. PLoS ONE 2019, 14, e0202844. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Johnson, M.D.; Cox, R.D.; Barnes, M.A. Analyzing airborne environmental DNA: A comparison of extraction methods, primer type, and trap type on the ability to detect airborne eDNA from terrestrial plant communities. Environ. DNA 2019, 1, 176–185. [Google Scholar] [CrossRef]
- Johnson, M.D.; Cox, R.D.; Barnes, M.A. The detection of a non-anemophilous plant species using airborne eDNA. PLoS ONE 2019, 14, e0225262. [Google Scholar] [CrossRef] [PubMed]
- Sansom, B.J.; Sassoubre, L.M. Environmental DNA (eDNA) shedding and decay rates to model freshwater mussel eDNA transport in a river. Environ. Sci. Technol. 2017, 51, 14244–14253. [Google Scholar] [CrossRef]
- Shogren, A.J.; Tank, J.L.; Andruszkiewicz, E.A.; Olds, B.; Jerde, C.; Bolster, D. Modelling the transport of environmental DNA through a porous substrate using continuous flow-through column experiments. J. R. Soc. Interface 2016, 13, 20160290. [Google Scholar] [CrossRef]
- Pedersen, M.W.; Overballe-Petersen, S.; Ermini, L.; Sarkissian, D.C.; Haile, J.; Hellstrom, M.; Spens, J.; Thomsen, F.P.; Bohmann, K.; Cappellini, E.; et al. Ancient and modern environmental DNA. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015, 370, 20130383. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. Ancient-human genomes plucked from cave dirt. Nature, 27 April 2017; 21910. [Google Scholar]
- Cristescu, M.E. Can environmental RNA revolutionize biodiversity science? Trends Ecol. Evol. 2019, 34, 694–697. [Google Scholar] [CrossRef]
- Von Ammon, U.; Wood, S.A.; Laroche, O.; Zaiko, A.; Lavery, S.D.; Inglis, G.J.; Pochon, X. Linking environmental DNA and RNA for improved detection of the marine invasive Fanworm Sabella spallanzanii. Front. Mar. Sci. 2019, 6, 621. [Google Scholar] [CrossRef]
- Wood, S.A.; Biessy, L.; Latchford, J.L.; Zaiko, A.; Ammon, V.U.; Audrezet, F.; Cristescu, E.M.; Pochon, X. Release and degradation of environmental DNA and RNA in a marine system. Sci. Total. Environ. 2020, 704, 135314. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, Y.; Wu, H.; Liu, F.; Peng, W.; Zhang, X.; Chang, F.; Xie, P.; Zhang, H. A review and perspective of eDNA application to eutrophication and HAB control in freshwater and marine ecosystems. Microorganisms 2020, 8, 417. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Howland, K.; Normandeau, E.; Grey, K.E.; Archambault, P.; Deiner, K.; Lodge, M.D.; Hernandez, C.; Leduc, N.; Bernatchez, L. eDNA metabarcoding as a new surveillance approach for coastal Arctic biodiversity. Ecol. Evol. 2018, 8, 7763–7777. [Google Scholar] [CrossRef]
- Djurhuus, A.; Closek, C.J.; Kelly, R.P.; Pitz, J.K.; Michisaki, P.R.; Starks, A.H.; Walz, R.K.; Andruskiewicz, A.E.; Olesin, E.; Hubbrad, K.; et al. Environmental DNA reveals seasonal shifts and potential interactions in a marine community. Nat. Commun. 2020, 11, 254. [Google Scholar] [CrossRef] [PubMed]
- Kontaş, S.; Bostancı, D. Genotoxic effects of environmental pollutant heavy metals on Alburnus chalcoides (Pisces: Cyprinidae) inhabiting lower Melet river (Ordu, Turkey). Bull. Environ. Contam. Toxicol. 2020, 104, 763–769. [Google Scholar] [CrossRef]
- Fathallah, S.; Beltifa, A.; Fekih, O.; Mansour, H.B. Bio monitoring of heavy metals genotoxicity in Tunisian coastal using the comet assay in the European calm Ruditapes decussatus. J. Env. Toxicol. Anal. Res. 2019, 1, 105. [Google Scholar]
- Sebbio, C.; Carere, C.; Nascetti, G.; Bellisario, B.; Mosesso, P.; Cimmaruta, R.; Angeletti, D. Interspecies variation in DNA damage induced by pollution. Curr. Zool. 2014, 60, 308–321. [Google Scholar] [CrossRef]
- Nik-Zainal, S.; Kucab, J.E.; Morganella, S.; Glodzik, D.; Alexandrov, B.L.; Arlt, M.V.; Weninger, A.; Hollstein, M.; Stratton, R.M.; Phillips, H.D. The genome as a record of environmental exposure. Mutagenesis 2015, 30, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Murray, V.; Hardie, M.E.; Gautam, S.D. Comparison of different methods to determine the DNA sequence preference of ionising radiation-induced DNA damage. Genes 2020, 11, 8. [Google Scholar] [CrossRef]
- Zatopek, K.M.; Potapov, V.; Maduzia, L.L.; Alpaslan, E.; Chen, L.; Evans, T.C.; Ong, J.L.; Ettwiller, L.M.; Gardner, A.F. RADAR-seq: A RAre DAmage and Repair sequencing method for detecting DNA damage on a genome-wide scale. DNA Repair (Amst.) 2019, 80, 36–44. [Google Scholar] [CrossRef]
- De Luca, F.; Rotunno, G.; Salvianti, F.; Galardi, F.; Pestrin, M.; Gabellini, S.; Simi, L.; Mancini, I.; Vannucchi, A.M.; Pazzagli, M.; et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget 2016, 7, 26107–26119. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.I.; Knapp, M.; Gemmell, N.J.; Jeunen, G.-J.; Bunce, M.; Lamare, M.D.; Taylor, H.R. Beyond Biodiversity: Can Environmental DNA (eDNA) Cut It as a Population Genetics Tool? Genes 2019, 10, 192. [Google Scholar] [CrossRef] [PubMed]
- McCord, B.R.; Opel, K.L.; Funes, M.; Zoppis, S.; Jantz, L.M. An Investigation of the Effect of DNA Degradation and Inhibition on PCR Amplification of Single Source and Mixed Forensic Samples; The U.S. Department of Justice grant final report; The U.S. Department of Justice: Washington, DC, USA, 2011.
- Rizzi, E.; Lari, M.; Gigli, E.; Bellis, D.G.; Caramelli, D. Ancient DNA studies: New perspectives on old samples. Genet. Sel. Evol. 2012, 44, 21. [Google Scholar] [CrossRef] [PubMed]
- Rohland, N.; Glocke, I.; Aximu-Petri, A.; Meyer, M. Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat. Protoc. 2018, 13, 2447–2461. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T. Epigenetic alterations induced by environmental stress associated with metabolic and neurodevelopmental disorders. Environ. Epigenet. 2016, 2, dvw017. [Google Scholar] [CrossRef] [PubMed]
- Meehan, R.R.; Thomson, J.P.; Lentini, A.; Nestor, E.C.; Pennings, S. DNA methylation as a genomic marker of exposure to chemical and environmental agents. Curr. Opin. Chem. Biol. 2018, 45, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Gluckman, P.D.; Seng, C.Y.; Fukuoka, H.; Beedle, S.A.; Hanson, A.M. Low birthweight and subsequent obesity in Japan. Lancet 2007, 369, 1081–1082. [Google Scholar] [CrossRef]
- Painter, R.C.; de Rooij, S.R.; Bossuyt, P.M.; Simmers, A.T.; Osmond, C.; Barker, J.D.; Bleker, P.O.; Roseboom, J.T. Early onset of coronary artery disease after prenatal exposure to the Dutch famine. Am. J. Clin. Nutr. 2006, 84, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Tobi, E.W.; Lumey, L.H.; Talens, R.P.; Kremer, D.; Putter, H.; Stein, D.A.; Slagboom, E.P.; Heijmans, T.B. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum. Mol. Genet. 2009, 18, 4046–4053. [Google Scholar] [CrossRef] [PubMed]
- Cooper, W.; Khulan, B.; Owens, S.; Elks, E.C.; Seidel, V.; Prentice, M.A.; Gusztav, B.; Ong, K.K.; Affara, A.N.; Constancia, M.; et al. DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: Results of a pilot randomized controlled trial. FASEB J. 2012, 26, 1782–1790. [Google Scholar] [CrossRef] [PubMed]
- Yaoi, T.; Itoh, K.; Nakamura, K.; Ogi, H.; Fujiwara, Y.; Fushiki, S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem. Biophys. Res. Commun. 2008, 376, 563–567. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.; Dolinoy, D.C.; Mancuso, P. Perinatal bisphenol A exposures increase production of pro-inflammatory mediators in bone marrow-derived mast cells of adult mice. J. Immunotoxicol. 2014, 11, 205–212. [Google Scholar] [CrossRef]
- Weinhouse, C.; Bergin, I.L.; Harris, C.; Dolinoy, C.D. Stat3 is a candidate epigenetic biomarker of perinatal Bisphenol A exposure associated with murine hepatic tumors with implications for human health. Epigenetics 2015, 10, 1099–1110. [Google Scholar] [CrossRef]
- Dao, T.; Hong, X.; Wang, X.; Tang, W.-Y. Aberrant 5′-CpG methylation of cord blood TNFα associated with maternal exposure to polybrominated diphenyl ethers. PLoS ONE 2015, 10, e0138815. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Jones, M.J.; Sava, F.; Kobor, M.C.; Carlsten, C. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part. Fibre Toxicol. 2014, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Richmond, R.C.; Simpkin, A.J.; Woodward, G.; Gaunt, R.T.; Lyttleton, O.; McArdle, L.W.; Ring, M.S.; Smith, D.A.C.A.; Timpson, J.N.; Tilling, K.; et al. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: Findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum. Mol. Genet. 2015, 24, 2201–2217. [Google Scholar] [CrossRef]
- Miousse, I.R.; Chalbot, M.C.; Lumen, A.; Ferguson, A.; Kavouras, G.I.; Koturbash, I. Response of transposable elements to environmental stressors. Mutation research. Rev. Mutat. Res. 2015, 765, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Casier, K.; Boivin, A.; Carré, C.; Teysset, L. Environmentally-induced transgenerational epigenetic inheritance: Implication of PIWI interacting RNAs. Cells 2019, 8, 1108. [Google Scholar] [CrossRef] [PubMed]
- Dorazio, R.M. eDNAoccupancy: An R Package for Multi-scale Occupancy Modeling of Environmental DNA Data. Mol. Ecol. Resour. 2017, 18, 368–380. [Google Scholar] [CrossRef]
- Dufresne, Y.; Lejzerowicz, F.; Perret-Gentil, L.A.; Pawlowski, J.; Cordier, T. SLIM: A flexible web application for the reproducible processing of environmental DNA metabarcoding data. BMC Bioinform. 2019, 20, 88. [Google Scholar] [CrossRef]
- Jeunen, G.-J. Bioinformatics for eDNA Metabarcoding. 2019. Available online: https://otagomohio.github.io/workshops/eDNA_Metabarcoding.html (accessed on 10 July 2020).
- Wahlberg, E. FACEPAI: A script for fast and consistent environmental DNA processing and identification. BMC Ecol. 2019, 19, 51. [Google Scholar] [CrossRef]
- Cordier, T.; Forster, D.; Dufresne, Y.; Martins, C.I.; Stock, T.; Pawlowski, J. Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring. Mol. Ecol. Resour. 2018, 8, 1381–1391. [Google Scholar] [CrossRef]
- Ramos, J.; Felty, Q.; Roy, D. Integrated Chip-Seq and RNA-Seq data analysis coupled with bioinformatics approaches to investigate regulatory landscape of transcription modulators in breast cancer cells. Methods Mol. Biol. 2020, 2102, 35–59. [Google Scholar] [CrossRef]
- Kunkle, B.; Yoo, C.; Roy, D. Discovering gene–environment interactions in glioblastoma through a comprehensive data integration bioinformatics method. Neurotoxicology 2013, 35, 1–14. [Google Scholar] [CrossRef]
- Roy, D.; Morgan, M.; Yoo, C.; Deoraj, A.; Roy, S.; Yadav, V.K.; Garoub, M.; Assaggaf, H.; Doke, M. Integrated bioinformatics, environmental epidemiologic and genomic approaches to identify environmental and molecular links between endometriosis and breast cancer. Int. J. Mol. Sci. 2015, 16, 25285–25322. [Google Scholar] [CrossRef] [PubMed]
- Preciados, M.; Yoo, C.; Roy, D. Estrogenic Endocrine Disrupting Chemicals Influencing NRF1 Regulated Gene Networks in the Development of Complex Human Brain Diseases. Int. J. Mol. Sci. 2016, 17, 2086. [Google Scholar] [CrossRef]
- Luna, B.; Bhatia, S.; Yoo, C.; Felty, Q.; Sanberg, I.D.; Duchowny, M.; Khatib, Z.; Miller, I.; Ragheb, J.; Prasanna, J.; et al. Bayesian network and mechanistic hierarchical structure modeling of increased likelihood of developing intractable childhood epilepsy from the combined effect of mtDNA variants, oxidative damage, and copy number. J. Mol. Neurosci. 2014, 54, 752–766. [Google Scholar] [CrossRef] [PubMed]
- Luna, B.; Bhatia, S.; Yoo, C.; Felty, Q.; Sandberg, I.D.; Duchowny, M.; Khatib, Z.; Miller, I.; Ragheb, J.; Prasanna, J.; et al. Proteomic and mitochondrial genomic analyses of pediatric brain tumors. Mol. Neurobiol. 2015, 52, 1341–1363. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, T.M.; Obuse, C.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. [Google Scholar] [CrossRef]
- Budnik, L.T.; Adam, B.; Albin, M.; Banelli, B.; Baur, X.; Belpoggi, F.; Bolognesi, C.; Broberg, K.; Gustavsson, P.; Goen, T.; et al. Diagnosis, monitoring and prevention of exposure-related non-communicable diseases in the living and working environment: DiMoPEx-project is designed to determine the impacts of environmental exposure on human health. J. Occup. Med. Toxicol. 2018, 13, 6. [Google Scholar] [CrossRef]
- Lee, D.H.; Yoon, H.; Park, S.; Kim, S.J.; Ahn, Y.-H.; Kwon, K.; Lee, D.; Kim, H.K. Urinary exosomal and cell-free DNA detects somatic mutation and copy number alteration in urothelial carcinoma of bladder. Sci. Rep. 2018, 8, 14707. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, X.; Li, X.; Inlora, J.; Wang, T.; Liu, Q.; Snyder, M. Dynamic human environmental exposome revealed by longitudinal personal monitoring. Cell 2018, 175, 277–291.e31. [Google Scholar] [CrossRef] [PubMed]
- Carter Atkinson, USGS-PIERC. Environmental DNA (eDNA): A new tool for monitoring status and trends of ecosystems and taxa in Hawaii and Pacific Islands. Available online: https://www.usgs.gov/centers/pierc/science/environmental-dna-edna-new-tool-monitoring-status-and-trends-ecosystems-and?qt-science_center_objects=0# (accessed on 10 July 2020).
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thakur, I.S.; Roy, D. Environmental DNA and RNA as Records of Human Exposome, Including Biotic/Abiotic Exposures and Its Implications in the Assessment of the Role of Environment in Chronic Diseases. Int. J. Mol. Sci. 2020, 21, 4879. https://doi.org/10.3390/ijms21144879
Thakur IS, Roy D. Environmental DNA and RNA as Records of Human Exposome, Including Biotic/Abiotic Exposures and Its Implications in the Assessment of the Role of Environment in Chronic Diseases. International Journal of Molecular Sciences. 2020; 21(14):4879. https://doi.org/10.3390/ijms21144879
Chicago/Turabian StyleThakur, Indu Shekhar, and Deodutta Roy. 2020. "Environmental DNA and RNA as Records of Human Exposome, Including Biotic/Abiotic Exposures and Its Implications in the Assessment of the Role of Environment in Chronic Diseases" International Journal of Molecular Sciences 21, no. 14: 4879. https://doi.org/10.3390/ijms21144879
APA StyleThakur, I. S., & Roy, D. (2020). Environmental DNA and RNA as Records of Human Exposome, Including Biotic/Abiotic Exposures and Its Implications in the Assessment of the Role of Environment in Chronic Diseases. International Journal of Molecular Sciences, 21(14), 4879. https://doi.org/10.3390/ijms21144879





