Distal-Less Homeobox 5 Is a Therapeutic Target for Attenuating Hypertrophy and Apoptosis of Mesenchymal Progenitor Cells
Abstract
1. Introduction
2. Results
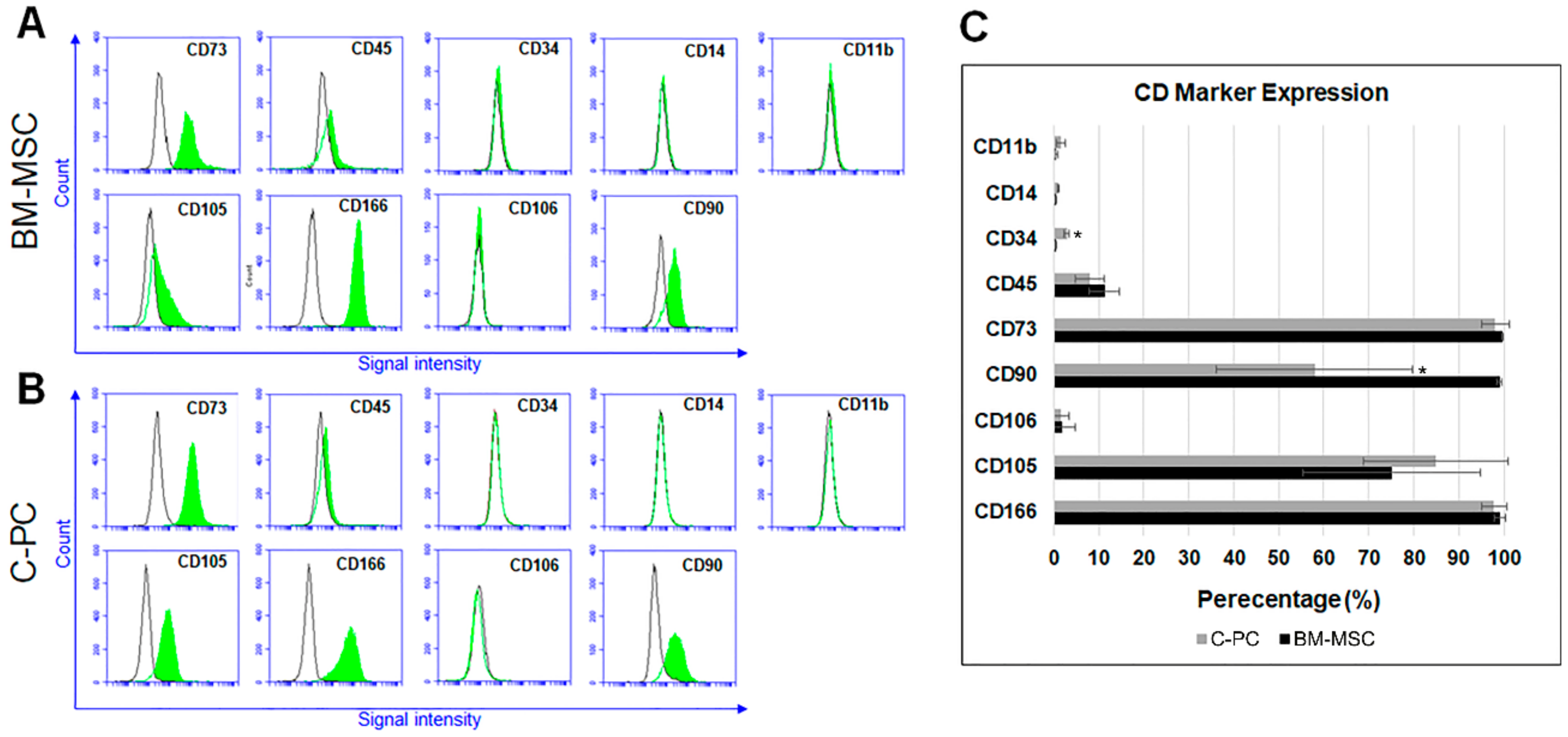
2.1. C-PCs Are Mesenchymal Progenitor Cells that Resist Cell Hypertrophy
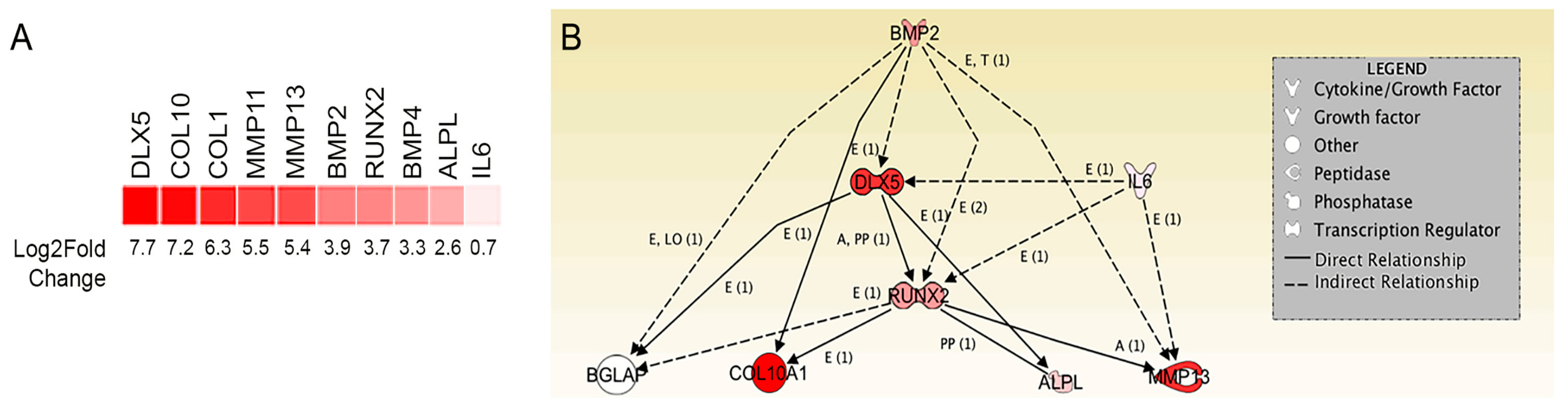
2.2. Transcriptome Analysis Identifies DLX5 as a Regulator of Terminal Chondrocyte Differentiation and Hypertrophy that is Elevated in Human BM-MSCs
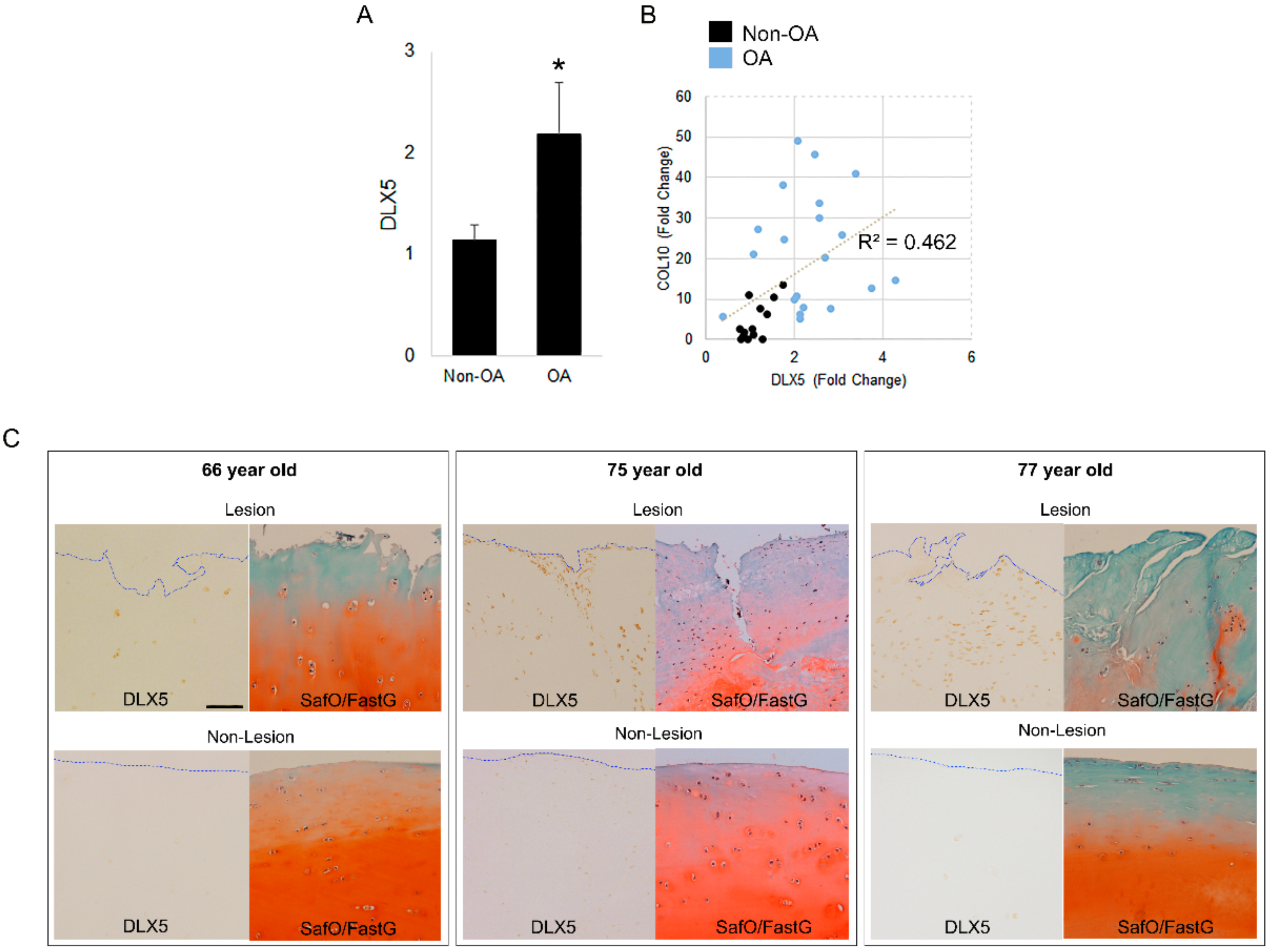
2.3. DLX5 Is Elevated in OA Cartilage Cells and Correlates Positively with COL10 Expression
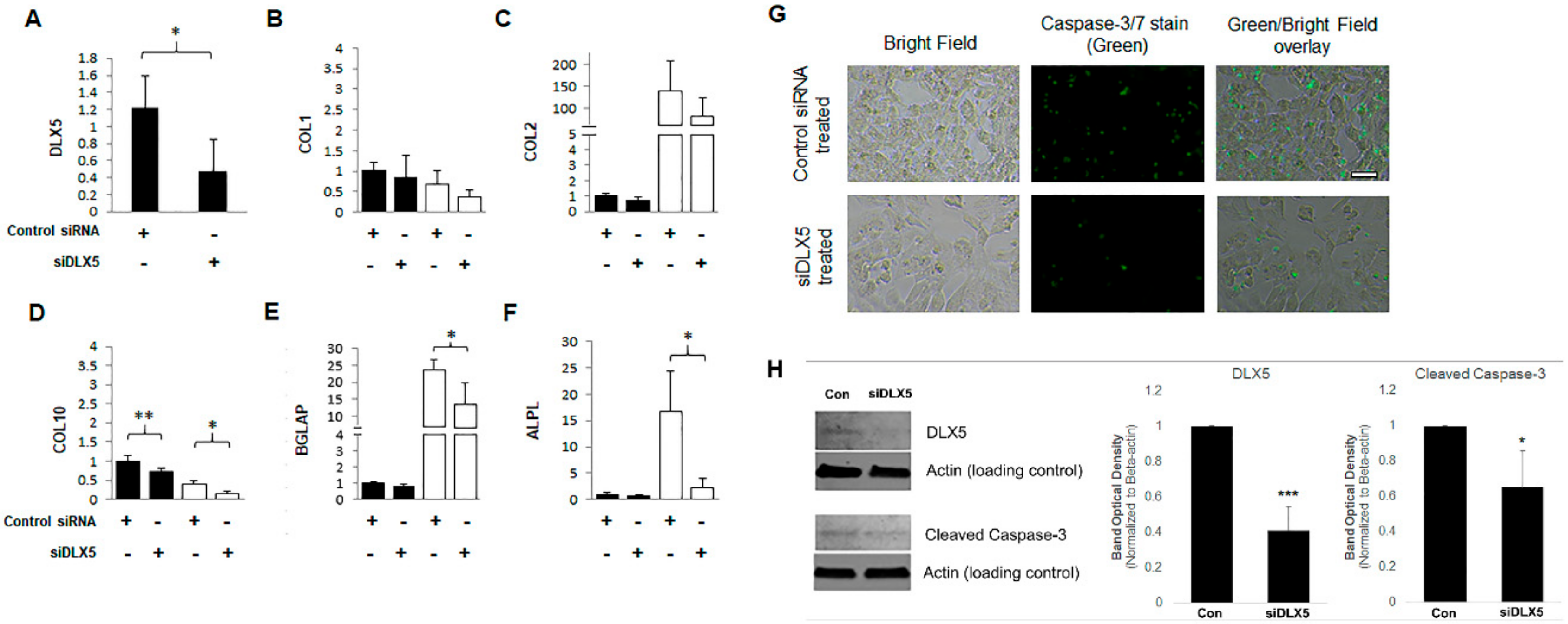
2.4. DLX5 Knockdown Attenuates Hypertrophy and Decreases Apoptotic Cell Fate without Deterring Chondrogenesis in BM-MSCs
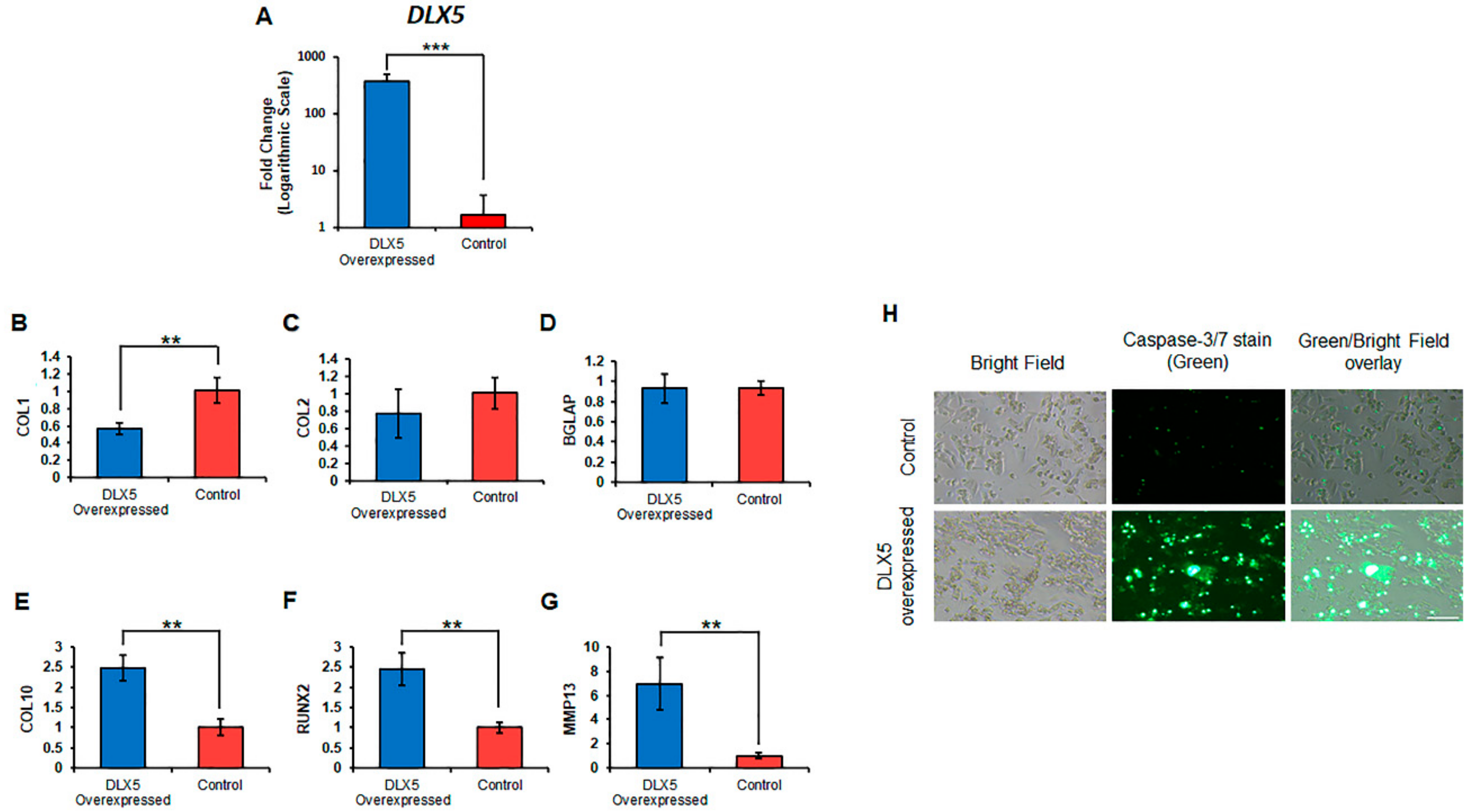
2.5. Elevated DLX5 Expression Stimulates Cell Hypertrophy and Apoptosis in C-PCs
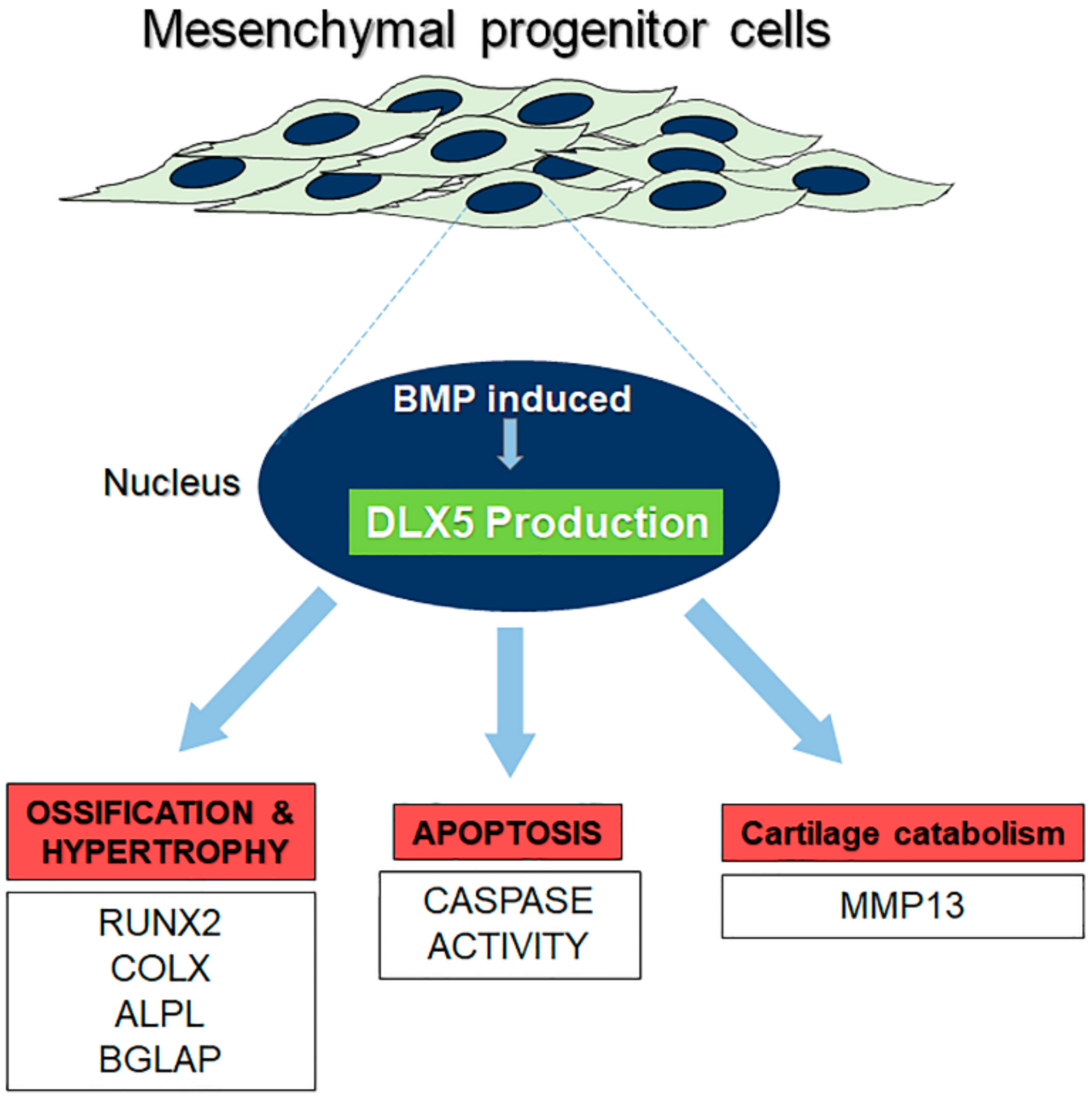
3. Discussion
4. Methods
4.1. Human Cell and Cell Line Culture
4.2. mRNA Expression Analysis via RT-qPCR
4.3. Immunophenotyping of BM-MSC and C-PC Cultures
4.4. RNA Sequencing and Transcriptome Analysis
4.5. DLX5 Knockdown and Overexpression Experiments
4.6. Staining of Patient Cartilage
4.7. Apoptotic Cell Detection Using Active Caspase-3/7 Immunostaining
4.8. Western Blotting
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Felson, D.; Lawrence, R.C.; Dieppe, P.A.; Hirsch, R.; Helmick, C.G.; Jordan, J.M.; Kington, R.S.; Lane, N.E.; Nevitt, M.C.; Zhang, Y.; et al. Osteoarthritis: New insights. Part 1: The disease and its risk factors. Ann. Intern. Med. 2000, 133, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Mandl, L.A. Osteoarthritis year in review 2018: Clinical. Osteoarthr. Cartil. 2019, 27, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Monaco, M.L.; Merckx, G.; Ratajczak, J.; Gervois, P.; Hilkens, P.; Clegg, P.D.; Bronckaers, A.; Vandeweerd, J.-M.; Lambrichts, I. Stem Cells for Cartilage Repair: Preclinical Studies and Insights in Translational Animal Models and Outcome Measures. Stem Cells Int. 2018, 2018, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Lei, G.; Lin, Z.; Yang, Y.; Lin, H.; Tuan, R.S. Engineering hyaline cartilage from mesenchymal stem cells with low hypertrophy potential via modulation of culture conditions and Wnt/beta-catenin pathway. Biomaterials 2019, 192, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Magnussen, R.A.; Dunn, W.R.; Carey, J.L.; Spindler, K.P. Treatment of Focal Articular Cartilage Defects in the Knee. Clin. Orthop. Relat. Res. 2008, 466, 952–962. [Google Scholar] [CrossRef]
- Paschos, N.; Sennett, M.L. Update on mesenchymal stem cell therapies for cartilage disorders. World J. Orthop. 2017, 8, 853–860. [Google Scholar] [CrossRef]
- Dzobo, K.; Thomford, N.E.; Senthebane, D.A.; Shipanga, H.; Rowe, A.; Dandara, C.; Pillay, M.; Motaung, K.S.C.M. Advances in Regenerative Medicine and Tissue Engineering: Innovation and Transformation of Medicine. Stem Cells Int. 2018, 2018, 1–24. [Google Scholar] [CrossRef]
- Kuo, C.K.; Li, W.-J.; Mauck, R.L.; Tuan, R.S. Cartilage tissue engineering: Its potential and uses. Curr. Opin. Rheumatol. 2006, 18, 64–73. [Google Scholar] [CrossRef]
- Vinatier, C.; Guicheux, J. Cartilage tissue engineering: From biomaterials and stem cells to osteoarthritis treatments. Ann. Phys. Rehabil. Med. 2016, 59, 139–144. [Google Scholar] [CrossRef]
- Goldberg, A.; Mitchell, K.; Soans, J.; Kim, L.; Zaidi, R. The use of mesenchymal stem cells for cartilage repair and regeneration: A systematic review. J. Orthop. Surg. Res. 2017, 12, 39. [Google Scholar] [CrossRef]
- Mueller, M.B.; Tuan, R.S. Functional characterization of hypertrophy in chondrogenesis of human mesenchymal stem cells. Arthritis Rheum. 2008, 58, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, I.; Vuoristo, J.T.; Larson, B.L.; Prockop, D.J. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 4397–4402. [Google Scholar] [CrossRef]
- Studer, D.; Millan, C.; Maniura-Weber, K.; Zenobi-Wong, M.; Öztürk, E. Molecular and biophysical mechanisms regulating hypertrophic differentiation in chondrocytes and mesenchymal stem cells. Eur. Cell Mater. 2012, 24, 118–135. [Google Scholar] [CrossRef]
- Derfus, B.; Kranendonk, S.; Camacho, N.; Mandel, N.; Kushnaryov, V.; Lynch, K.; Ryan, L. Human osteoarthritic cartilage matrix vesicles generate both calcium pyrophosphate dihydrate and apatite in vitro. Calcif. Tissue Int. 1998, 63, 258–262. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, H.E.; Bara, J.J.; Brakspear, K.; Singhrao, S.K.; Archer, C. The comparison of equine articular cartilage progenitor cells and bone marrow-derived stromal cells as potential cell sources for cartilage repair in the horse. Veter. J. 2012, 192, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.; Khan, I.; Richardson, K.; Nelson, L.; McCarthy, H.E.; Analbelsi, T.; Singhrao, S.K.; Dowthwaite, G.P.; Jones, R.E.; Baird, D.M.; et al. Identification and Clonal Characterisation of a Progenitor Cell Sub-Population in Normal Human Articular Cartilage. PLoS ONE 2010, 5, e13246. [Google Scholar] [CrossRef]
- Jayasuriya, C.T.; Twomey-Kozak, J.; Newberry, J.; Desai, S.; Feltman, P.; Franco, J.R.; Li, N.; Terek, R.; Ehrlich, M.G.; Owens, B.D. Human Cartilage-Derived Progenitors Resist Terminal Differentiation and Require CXCR4 Activation to Successfully Bridge Meniscus Tissue Tears. Stem Cells 2018, 37, 102–114. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Bendall, A.J.; Hu, G.; Levi, G.; Abate-Shen, C. Dlx5 regulates chondrocyte differentiation at multiple stages. Int. J. Dev. Biol. 2003, 47, 335–344. [Google Scholar]
- Ferrari, D.; Kosher, R.A. Dlx5 is a positive regulator of chondrocyte differentiation during endochondral ossification. Dev. Biol. 2002, 252, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-H.C.; Noamani, B.; Abernethy, D.E.; Zhu, H.; Levi, G.; Bendall, A.J. Dlx5- and Dlx6-mediated chondrogenesis: Differential domain requirements for a conserved function. Mech. Dev. 2006, 123, 819–830. [Google Scholar] [CrossRef]
- Zhu, H.; Bendall, A.J. Dlx5 Is a Cell Autonomous Regulator of Chondrocyte Hypertrophy in Mice and Functionally Substitutes for Dlx6 during Endochondral Ossification. PLoS ONE 2009, 4, e8097. [Google Scholar] [CrossRef] [PubMed]
- Reppel, L.; Schiavi, J.; Charif, N.; Leger, L.; Yu, H.; Pinzano, A.; Henrionnet, C.; Stoltz, J.-F.; Bensoussan, D.; Huselstein, C. Chondrogenic induction of mesenchymal stromal/stem cells from Wharton’s jelly embedded in alginate hydrogel and without added growth factor: An alternative stem cell source for cartilage tissue engineering. Stem Cell Res. Ther. 2015, 6, 260. [Google Scholar] [CrossRef] [PubMed]
- Schmid, T.M.; Popp, R.G.; Linsenmayer, T.F. Hypertrophic Cartilage Matrix. Ann. N. Y. Acad. Sci. 1990, 580, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.-G.; Zhao, X.; Yang, Q.; Li, Y.; Ge, C.; Zhao, G.; Franceschi, R.T. Physical and functional interactions between Runx2 and HIF-1α induce vascular endothelial growth factor gene expression. J. Cell. Biochem. 2011, 112, 3582–3593. [Google Scholar] [CrossRef]
- Shibakawa, A.; Yudoh, K.; Masuko, K.; Kato, T.; Nishioka, K.; Nakamura, H. The role of subchondral bone resorption pits in osteoarthritis: MMP production by cells derived from bone marrow. Osteoarthr. Cartil. 2005, 13, 679–687. [Google Scholar] [CrossRef]
- Stanton, L.-A.; Sabari, S.; Sampaio, A.V.; Underhill, T.M.; Beier, F. p38 MAP kinase signalling is required for hypertrophic chondrocyte differentiation. Biochem. J. 2004, 378, 53–62. [Google Scholar] [CrossRef]
- Chao, P.-H.G.; West, A.C.; Hung, C.T. Chondrocyte intracellular calcium, cytoskeletal organization, and gene expression responses to dynamic osmotic loading. Am. J. Physiol. Physiol. 2006, 291, C718–C725. [Google Scholar] [CrossRef]
- Clark, A.L.; Votta, B.J.; Kumar, S.; Liedtke, W.; Guilak, F. Chondroprotective role of the osmotically sensitive ion channel transient receptor potential vanilloid 4: Age- and sex-dependent progression of osteoarthritis in Trpv4-deficient mice. Arthritis Rheum. 2010, 62, 2973–2983. [Google Scholar] [CrossRef]
- Negoro, K.; Kobayashi, S.; Takeno, K.; Uchida, K.; Baba, H. Effect of osmolarity on glycosaminoglycan production and cell metabolism of articular chondrocyte under three-dimensional culture system. Clin. Exp. Rheumatol. 2008, 26, 534–541. [Google Scholar] [PubMed]
- Terkeltaub, R.; Lotz, M.; Johnson, K.; Deng, D.; Hashimoto, S.; Goldring, M.B.; Burton, D.; Deftos, L.J. Parathyroid hormone-related proteins is abundant in osteoarthritic cartilage, and the parathyroid hormone-related protein 1–173 isoform is selectively induced by transforming growth factor beta in articular chondrocytes and suppresses generation of extracellular inorganic pyrophosphate. Arthritis Rheum. 1998, 41, 2152–2164. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, T.; Swoboda, B.; Nah, H.-D. Activation of annexin II and V expression, terminal differentiation, mineralization and apoptosis in human osteoarthritic cartilage. Osteoarthr. Cartil. 2000, 8, 294–302. [Google Scholar] [CrossRef]
- Pfander, D.; Swoboda, B.; Kirsch, T. Expression of Early and Late Differentiation Markers (Proliferating Cell Nuclear Antigen, Syndecan-3, Annexin VI, and Alkaline Phosphatase) by Human Osteoarthritic Chondrocytes. Am. J. Pathol. 2001, 159, 1777–1783. [Google Scholar] [CrossRef]
- Tchetina, E.V.; Squires, G.; Poole, A.R. Increased type II collagen degradation and very early focal cartilage degeneration is associated with upregulation of chondrocyte differentiation related genes in early human articular cartilage lesions. J. Rheumatol. 2005, 32, 876–886. [Google Scholar]
- Walker, G.D.; Fischer, M.; Gannon, J.; Thompson, R.C.; Oegema, T.R. Expression of type-X collagen in osteoarthritis. J. Orthop. Res. 1995, 13, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Kozak, T. Department of Orthopaedics, Warren Alpert Medical School of Brown University and the Rhode Island Hospital, Providence, RI. RUNX2 mRNA expression in DLX5 KD C-PCs. Unpublished work. 2019. [Google Scholar]
- Miura, M.; Chen, X.-D.; Allen, M.R.; Bi, Y.; Gronthos, S.; Seo, B.-M.; Lakhani, S.; Flavell, R.A.; Feng, X.-H.; Robey, P.G.; et al. A crucial role of caspase-3 in osteogenic differentiation of bone marrow stromal stem cells. J. Clin. Investig. 2004, 114, 1704–1713. [Google Scholar] [CrossRef]
- Svandova, E.; Lesot, H.; Berghe, T.V.; Tucker, A.S.; Sharpe, P.T.; Vandenabeele, P.; Matalova, E. Non-apoptotic functions of caspase-7 during osteogenesis. Cell Death Dis. 2014, 5, e1366. [Google Scholar] [CrossRef]
- Janečková, E.; Bíliková, P.; Matalová, E. Osteogenic Potential of Caspases Related to Endochondral Ossification. J. Histochem. Cytochem. 2017, 66, 47–58. [Google Scholar] [CrossRef]
- Adams, C.S.; Horton, W.E., Jr. Chondrocyte apoptosis increases with age in the articular cartilage of adult animals. Anat. Rec. 1998, 250, 418–425. [Google Scholar] [CrossRef]
- Hwang, H.S.; Kim, H.A. Chondrocyte Apoptosis in the Pathogenesis of Osteoarthritis. Int. J. Mol. Sci. 2015, 16, 26035–26054. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.M.; Lai, Y.K.Y.; Rakoczy, P.E. Adenovirus and Adeno-Associated Virus Vectors. DNA Cell Biol. 2002, 21, 895–913. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Y.; Peng, P.D.; Ehrhardt, A.; Storm, T.A.; Kay, M.A. Comparison of Adenoviral and Adeno-Associated Viral Vectors for Pancreatic Gene DeliveryIn Vivo. Hum. Gene Ther. 2004, 15, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Maleki, M.; Ghanbarvand, F.; Behvarz, M.R.; Ejtemaei, M.; Ghadirkhomi, E. Comparison of Mesenchymal Stem Cell Markers in Multiple Human Adult Stem Cells. Int. J. Stem Cells 2014, 7, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Sidney, L.E.; Branch, M.J.; Dunphy, S.; Dua, H.S.; Hopkinson, A. Concise Review: Evidence for CD34 as a Common Marker for Diverse Progenitors. Stem Cells 2014, 32, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Hackett, C.H.; Flaminio, M.J.B.; Fortier, L.A. Analysis of CD14 Expression Levels in Putative Mesenchymal Progenitor Cells Isolated from Equine Bone Marrow. Stem Cells Dev. 2010, 20, 721–735. [Google Scholar] [CrossRef]
- Ohradanova-Repic, A.; Machacek, C.; Fischer, M.B.; Stockinger, H. Differentiation of human monocytes and derived subsets of macrophages and dendritic cells by the HLDA10 monoclonal antibody panel. Clin. Transl. Immunol. 2016, 5, e55. [Google Scholar] [CrossRef]
- Mafi, P.; Hindocha, S.; Mafi, R.; Griffin, M.; Khan, W.S. Adult Mesenchymal Stem Cells and Cell Surface Characterization—A Systematic Review of the Literature. Open Orthop. J. 2011, 5, 253–260. [Google Scholar] [CrossRef]
| Gene | Forward Seq. | Reverse Seq. | Accession |
|---|---|---|---|
| ALPL | CTGGACGGACCCTCGCCAGTG | TGCAATCGACGTGGGTGGGAGG | NM_000478.5 |
| Beta-Actin | GGACCTGACTGACTACCTCAT | CGTAGCACAGCTTCTCCTTAAT | NM_001101.4 |
| BGLAP | AGCAAAGGTGCAGCCTTTGT | GGCTCCCAGCCATTGATACA | NM_199173.6 |
| COL1A1 | CAGGAGGCACGCGGAGTGTG | GGCAGGGCTCGGGTTTCCAC | NM_000088.3 |
| COL2A1 | CTCCCAGAACATCACCTACCACT | CGTGAACCTGCTATTGCCCT | NM_001844.4 |
| COL10A1 | GCCCACAGGCATAAAAGGCCC | GAAGGACCTGGGTGCCCTCGA | NM_000493.3 |
| DLX5 | ACAGCCATGTCTGCTTAGACC | AGACGGATGGTGCATAGCTG | NM_005221.6 |
| MMP13 | ATGCGGGGTTCCTGATGTGG | GGCCCAGGAGGAAAAGCATG | NM_002427.4 |
| RUNX2 | CTCTGACTTCTGCCTCTGGC | GGTGTGGTAGTGAGTGGTGG | NM_001024630.4 |
| Cell Surface Marker | Description of Marker |
|---|---|
| CD166 | Mesenchymal stem cell/progenitor marker [45] |
| CD73 | Multipotent mesenchymal stromal cell marker [19] |
| CD90 | Multipotent mesenchymal stromal cell marker [19] |
| CD105 | Multipotent mesenchymal stromal cell marker [19] |
| CD106/VCAM1 | Vascular cell adhesion marker [45] |
| CD45 | Hematopoietic cell marker [45] |
| CD34 | Hematopoietic stem/progenitor cell and endothelial marker [46] |
| CD14 | Monocyte/macrophage and hematopoietic marker [47,48] |
| CD11b | Macrophage/monocyte cell marker [49] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Twomey-Kozak, J.; Desai, S.; Liu, W.; Li, N.Y.; Lemme, N.; Chen, Q.; Owens, B.D.; Jayasuriya, C.T. Distal-Less Homeobox 5 Is a Therapeutic Target for Attenuating Hypertrophy and Apoptosis of Mesenchymal Progenitor Cells. Int. J. Mol. Sci. 2020, 21, 4823. https://doi.org/10.3390/ijms21144823
Twomey-Kozak J, Desai S, Liu W, Li NY, Lemme N, Chen Q, Owens BD, Jayasuriya CT. Distal-Less Homeobox 5 Is a Therapeutic Target for Attenuating Hypertrophy and Apoptosis of Mesenchymal Progenitor Cells. International Journal of Molecular Sciences. 2020; 21(14):4823. https://doi.org/10.3390/ijms21144823
Chicago/Turabian StyleTwomey-Kozak, John, Salomi Desai, Wenguang Liu, Neill Y. Li, Nicholas Lemme, Qian Chen, Brett D. Owens, and Chathuraka T. Jayasuriya. 2020. "Distal-Less Homeobox 5 Is a Therapeutic Target for Attenuating Hypertrophy and Apoptosis of Mesenchymal Progenitor Cells" International Journal of Molecular Sciences 21, no. 14: 4823. https://doi.org/10.3390/ijms21144823
APA StyleTwomey-Kozak, J., Desai, S., Liu, W., Li, N. Y., Lemme, N., Chen, Q., Owens, B. D., & Jayasuriya, C. T. (2020). Distal-Less Homeobox 5 Is a Therapeutic Target for Attenuating Hypertrophy and Apoptosis of Mesenchymal Progenitor Cells. International Journal of Molecular Sciences, 21(14), 4823. https://doi.org/10.3390/ijms21144823







