MicroRNAs as Guardians of the Prostate: Those Who Stand before Cancer. What Do We Really Know about the Role of microRNAs in Prostate Biology?
Abstract
1. Introduction
2. Implications of Low Molecular Weight (LMW)- and High Molecular Weight (HMW)-RISCs and the Conundrum of the Let-7 miRNA Family
3. MiRNA Processing Machinery Genes Dicer and Dgcr8 in Prostate Biology and Their Surprising Loss of Function Phenotypes
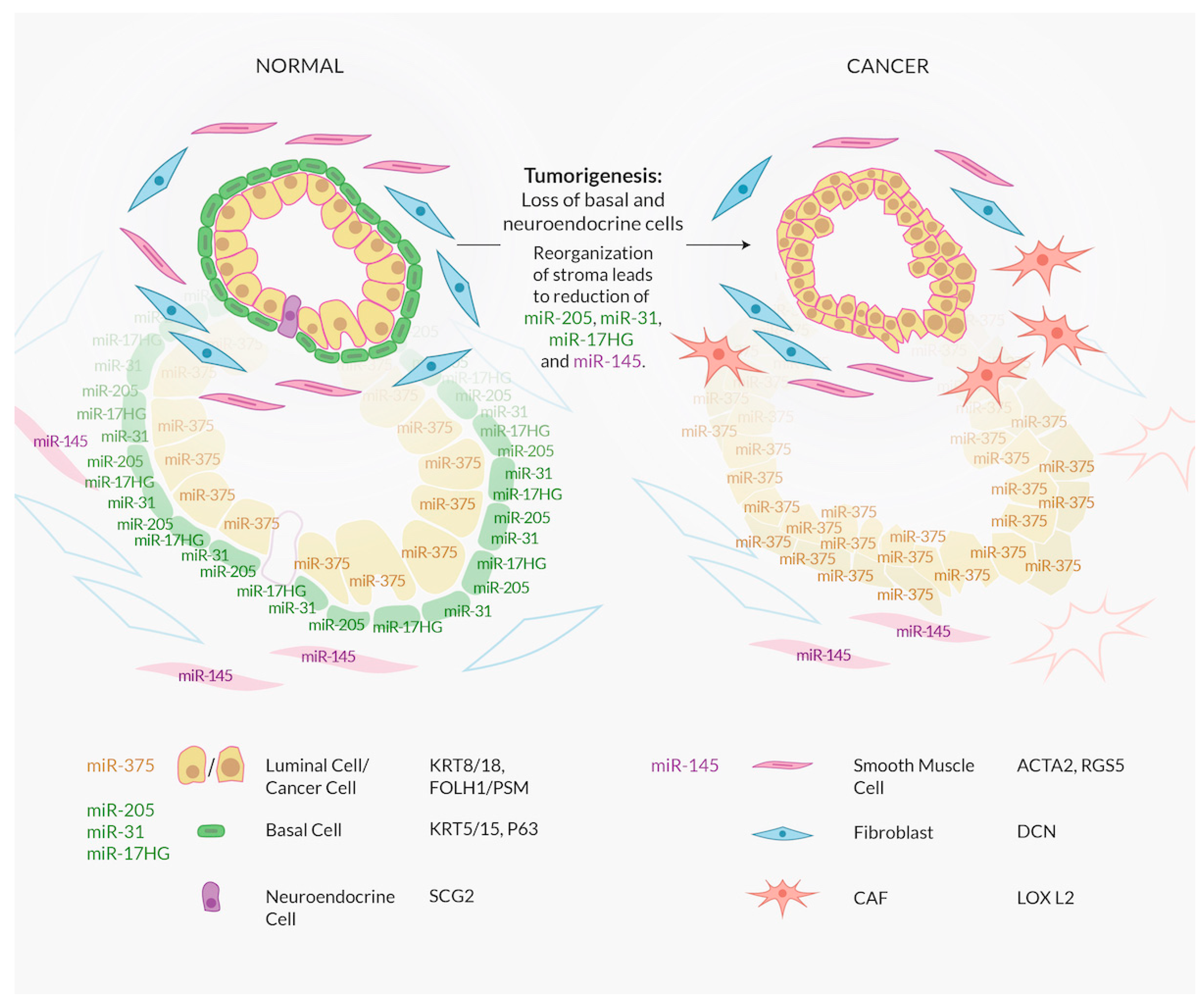
4. A Major Confounding Factor in Interpreting miRNA Expression Data: Cell Type and the Lack of Comprehensive Characterization of Luminal Cells
5. Characterizing miRNA Function through Rigorous Establishment of Relationship with Targets
6. In the miRNA World, Quantity and Numbers Matter
7. How to Evaluate miRNA Expression Data: A Cautionary Note
8. MiR-143/145: Highly Expressed vs. Highly Important
8.1. The miR-143/145 Cluster Is a Stromal Cell Marker
8.2. The Fallacy of Overexpressing a Stromal microRNA in Prostate Cancer Cells
8.3. For Few miRNAs in Addition to miR143/145 the Cell Type of Expression Is Known
9. MiR-375: An Indeterminant Oncogene, a Mirage of a Tumor Suppressor Gene
9.1. miR-375 a Luminal Epithelial Phenotype Gatekeeper?
9.2. The Molecular Mechanisms How miR-375 May Function as A Gatekeeper Are Unclear
9.3. No Conclusive Data on miR-375 Categorization as OncomiR or Tumor Suppressor
10. MiR-22: Possibly an Androgen-Regulated Tumor Suppressor miRNA: Few Data, Big Ideas
10.1. MiR-22 and Its Relationship with Androgen Receptor (AR) Signaling
10.2. Confusing Experiments Putting miRNAs in Regulatory Axes Which May Not Exist
11. MiR-148a: A Preferential oncomiR in Tumor Cells Amidst Proposed AR Signaling
12. The miR-182/-183/-96 Cluster: Overexpression Associated with Zinc Homeostasis, Angiogenesis, Hypoxia and Androgen Receptor Signaling
13. MiR-141/-200/-429: Controlling Epithelial Plasticity and Metastasis
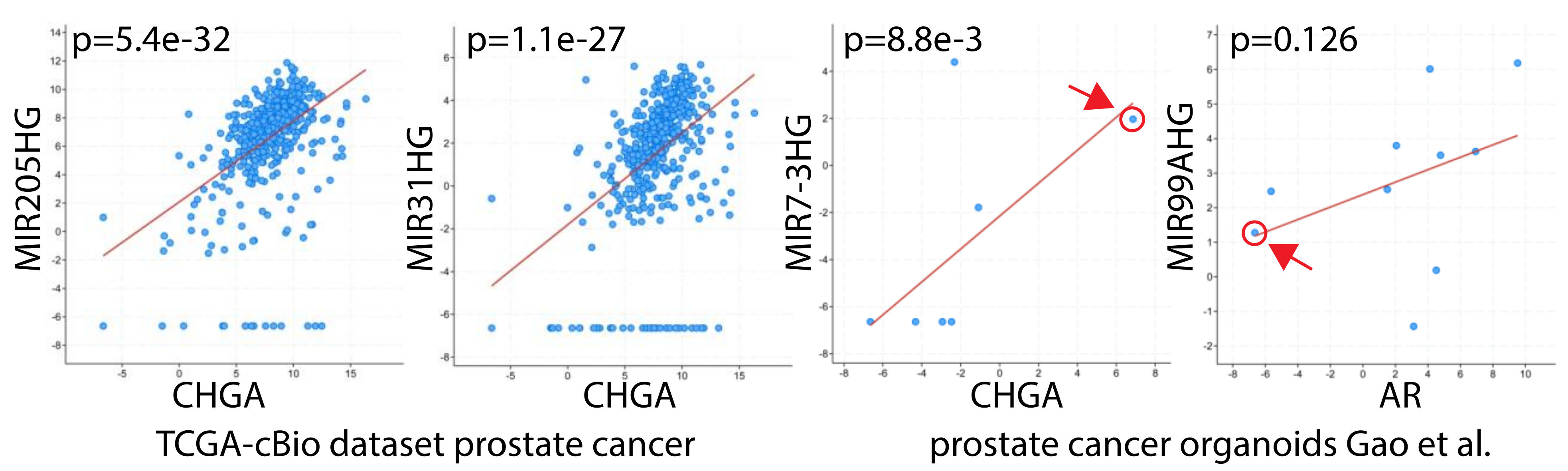
14. The Link Between High Grade Prostate Cancer, Neuroendocrine Differentiation, and miRNAs: miR-30
15. MiR-378: A Link Between Tumor Cachexia and Tumor Progression
16. MiRNA Relationships with Androgen Receptor (AR) Signaling
17. Serum/Plasma miRNA Levels and Relation to Diagnosis and Prognosis
18. Therapeutic Applications on the Horizon
19. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3’UTR | 3’untranslated region of the mRNA |
| Ago | argonaute |
| AR | androgen receptor |
| BPH | benign prostatic hyperplasia |
| ceRNA | competing endogenous RNA |
| CRPC | castration-resistant prostate cancer |
| Ct | threshold cycle |
| dsRNA | double-stranded RNA |
| EMT | epithelial-mesenchymal transition |
| ENCODE | Encyclopedia of DNA Elements |
| FDA | Food and Drug Administration |
| GEO | Gene Expression Omnibus |
| HMW | high molecular weight |
| ISH | in situ hybridization |
| KO | Knock-out |
| lacZ | β-galactosidase |
| LCM | laser capture microscopy |
| LMW | low molecular weight |
| miRNA | microRNA |
| Pca | prostate cancer |
| PSA | prostate specific antigen |
| qRT-PCR | quantitative reverse transcription-polymerase chain reaction |
| RISC | RNA-induced silencing complex |
| RNAi | RNA interference |
| RNAseq | high-thoughput RNA sequencing |
| siRNA | small interfering RNA |
| TCGA | The Cancer Genome Atlas |
| TGF-β | tumor growth factor beta |
References
- Bethune, J.; Artus-Revel, C.G.; Filipowicz, W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. Embo Rep. 2012, 13, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Vanacore, D.; Boccellino, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Di Franco, R.; Romano, F.J.; Montanari, M.; La Mantia, E.; Piscitelli, R.; et al. Micrornas in prostate cancer: An overview. Oncotarget 2017, 8, 50240–50251. [Google Scholar] [CrossRef] [PubMed]
- La Rocca, G.; Olejniczak, S.H.; González, A.J.; Briskin, D.; Vidigal, J.A.; Spraggon, L.; DeMatteo, R.G.; Radler, M.R.; Lindsten, T.; Ventura, A.; et al. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc. Natl. Acad. Sci. USA 2015, 112, 767–772. [Google Scholar] [CrossRef]
- Belair, C.D.; Paikari, A.; Moltzahn, F.; Shenoy, A.; Yau, C.; Dall’Era, M.; Simko, J.; Benz, C.; Blelloch, R. DGCR8 is essential for tumor progression following PTEN loss in the prostate. Embo Rep. 2015, 16, 1219–1232. [Google Scholar] [CrossRef]
- Dong, Q.; Meng, P.; Wang, T.; Qin, W.; Qin, W.; Wang, F.; Yuan, J.; Chen, Z.; Yang, A.; Wang, H. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS ONE 2010, 5, e10147. [Google Scholar] [CrossRef] [PubMed]
- Martens-Uzunova, E.S.; Jalava, S.E.; Dits, N.F.; van Leenders, G.J.; Moller, S.; Trapman, J.; Bangma, C.H.; Litman, T.; Visakorpi, T.; Jenster, G. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene 2012, 31, 978–991. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiss, J.L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270–284.e13. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Ngezahayo, A.; Murua Escobar, H.; Nolte, I. Role of miRNA let-7 and its major targets in prostate cancer. Biomed. Res. Int. 2014, 2014, 376326. [Google Scholar] [CrossRef]
- Albino, D.; Civenni, G.; Dallavalle, C.; Roos, M.; Jahns, H.; Curti, L.; Rossi, S.; Pinton, S.; D’Ambrosio, G.; Sessa, F.; et al. Activation of the Lin28/let-7 Axis by Loss of ESE3/EHF Promotes a Tumorigenic and Stem-like Phenotype in Prostate Cancer. Cancer Res. 2016, 76, 3629–3643. [Google Scholar] [CrossRef]
- Zhong, X.; Li, N.; Liang, S.; Huang, Q.; Coukos, G.; Zhang, L. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J. Biol. Chem. 2010, 285, 41961–41971. [Google Scholar] [CrossRef]
- Tummala, R.; Nadiminty, N.; Lou, W.; Zhu, Y.; Gandour-Edwards, R.; Chen, H.W.; Evans, C.P.; Gao, A.C. Lin28 promotes growth of prostate cancer cells and activates the androgen receptor. Am. J. Pathol. 2013, 183, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Lee, K.H.; Lee, H.S.; Jeong, C.W.; Ku, J.H.; Kim, H.H.; Kwak, C. Concurrent treatment with simvastatin and NF-kappaB inhibitor in human castration-resistant prostate cancer cells exerts synergistic anti-cancer effects via control of the NF-kappaB/LIN28/let-7 miRNA signaling pathway. PLoS ONE 2017, 12, e0184644. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Powers, J.T.; Einhorn, W.; Hoshida, Y.; Ng, T.L.; Toffanin, S.; O’Sullivan, M.; Lu, J.; Phillips, L.A.; Lockhart, V.L.; et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009, 41, 843–848. [Google Scholar] [CrossRef]
- Jang, H.S.; Shah, N.M.; Du, A.Y.; Dailey, Z.Z.; Pehrsson, E.C.; Godoy, P.M.; Zhang, D.; Li, D.; Xing, X.; Kim, S.; et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 2019, 51, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Song, C.J.; Chen, H.; Chen, L.Z.; Ru, G.M.; Guo, J.J.; Ding, Q.N. The potential of microRNAs as human prostate cancer biomarkers: A meta-analysis of related studies. J. Cell Biochem. 2018, 119, 2763–2786. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, B.; Valdez, J.M.; Wang, F.; Ittmann, M.; Xin, L. Dicer Ablation Impairs Prostate Stem Cell Activity and Causes Prostate Atrophy. Stem Cells 2010, 28, 1260–1269. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Macias, S.; Cordiner, R.; Gautier, P.; Plass, M.; Cáceres, J. DGCR8 Acts as an Adaptor for the Exosome Complex to Degrade Double-Stranded Structured RNAs. Mol. Cell 2015, 60, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Macias, S.; Plass, M.; Stajuda, A.; Michlewski, G.; Eyras, E.; Cáceres, J.F. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat. Struct. Mol. Biol. 2012, 19, 760–766. [Google Scholar] [CrossRef]
- Teta, M.; Choi, Y.S.; Okegbe, T.; Wong, G.; Tam, O.H.; Chong, M.M.W.; Seykora, J.T.; Nagy, A.; Littman, D.R.; Andl, T.; et al. Inducible deletion of epidermal Dicer and Drosha reveals multiple functions for miRNAs in postnatal skin. Development 2012, 139, 1405–1416. [Google Scholar] [CrossRef]
- Garber, K. A tale of two cells: Discovering the origin of prostate cancer. J. Natl. Cancer Inst. 2010, 102, 1528–1529, 1535. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.A.; Mitrofanova, A.; Bergren, S.K.; Abate-Shen, C.; Cardiff, R.D.; Califano, A.; Shen, M.M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell of origin model for prostate cancer heterogeneity. Nat. Cell Biol. 2013, 15, 274–283. [Google Scholar] [CrossRef]
- Gandellini, P.; Folini, M.; Longoni, N.; Pennati, M.; Binda, M.; Colecchia, M.; Salvioni, R.; Supino, R.; Moretti, R.; Limonta, P.; et al. miR-205 Exerts Tumor-Suppressive Functions in Human Prostate through Down-regulation of Protein Kinase Cε. Cell Tumor Stem Cell Biol. 2009. [Google Scholar] [CrossRef]
- Fan, X.; Bjerke, G.A.; Riemondy, K.; Wang, L.; Yi, R. A basal-enriched microRNA is required for prostate tumorigenesis in a Pten knockout mouse model. Mol. Carcinog. 2019, 58, 2241–2253. [Google Scholar] [CrossRef] [PubMed]
- Nordby, Y.; Richardsen, E.; Ness, N.; Donnem, T.; Patel, H.R.H.; Busund, L.T.; Bremnes, R.M.; Andersen, S. High miR-205 expression in normal epithelium is associated with biochemical failure - an argument for epithelial crosstalk in prostate cancer? Sci. Rep. 2017, 7, 16308. [Google Scholar] [CrossRef]
- Zedan, A.H.; Blavnsfeldt, S.G.; Hansen, T.F.; Nielsen, B.S.; Marcussen, N.; Pleckaitis, M.; Osther, P.J.S.; Sorensen, F.B. Heterogeneity of miRNA expression in localized prostate cancer with clinicopathological correlations. PLoS ONE 2017, 12, e0179113. [Google Scholar] [CrossRef]
- Eckstein, M.; Sailer, V.; Nielsen, B.S.; Wittenberg, T.; Wiesmann, V.; Lieb, V.; Nolte, E.; Hartmann, A.; Kristiansen, G.; Wernert, N.; et al. Co-staining of microRNAs and their target proteins by miRNA in situ hybridization and immunohistofluorescence on prostate cancer tissue microarrays. Lab. Investig. 2019. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liao, K.; Xiao, C. MicroRNA says no to mass production. Nat. Immunol. 2018, 19, 1040–1042. [Google Scholar] [CrossRef] [PubMed]
- Schmiedel, J.M.; Klemm, S.L.; Zheng, Y.; Sahay, A.; Bluthgen, N.; Marks, D.S.; van Oudenaarden, A. Gene expression. MicroRNA control of protein expression noise. Science 2015, 348, 128–132. [Google Scholar] [CrossRef]
- Siciliano, V.; Garzilli, I.; Fracassi, C.; Criscuolo, S.; Ventre, S.; di Bernardo, D. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat. Commun. 2013, 4, 2364. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, F.; Lee, J.A.; Gao, F.B. MicroRNA-9a ensures the precise specification of sensory organ precursors inDrosophila. Genes Dev. 2006, 20, 2793–2805. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.; Brennecke, J.; Bushati, N.; Russell, R.B.; Cohen, S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell 2005, 123, 1133–1146. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Halushka, M.K. Toward the promise of microRNAs - Enhancing reproducibility and rigor in microRNA research. RNA Biol. 2016, 13, 1103–1116. [Google Scholar] [CrossRef]
- Kuhn, D.E.; Martin, M.M.; Feldman, D.S.; Terry, A.V., Jr.; Nuovo, G.J.; Elton, T.S. Experimental validation of miRNA targets. Methods 2008, 44, 47–54. [Google Scholar] [CrossRef]
- Denzler, R.; McGeary, S.E.; Title, A.C.; Agarwal, V.; Bartel, D.P.; Stoffel, M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol. Cell 2016, 64, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Broderick, J.A.; Zamore, P.D. Competitive endogenous RNAs cannot alter microRNA function in vivo. Mol. Cell 2014, 54, 711–713. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Ridzon, D.; Wong, L.; Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007, 8, 166. [Google Scholar] [CrossRef] [PubMed]
- Minami, K.; Uehara, T.; Morikawa, Y.; Omura, K.; Kanki, M.; Horinouchi, A.; Ono, A.; Yamada, H.; Ohno, Y.; Urushidani, T. miRNA expression atlas in male rat. Sci. Data 2014, 1, 140005. [Google Scholar] [CrossRef]
- Mullokandov, G.; Baccarini, A.; Ruzo, A.; Jayaprakash, A.D.; Tung, N.; Israelow, B.; Evans, M.J.; Sachidanandam, R.; Brown, B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 2012, 9, 840–846. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, H.; Li, Y.; Jiang, L. MicroRNA-30a functions as tumor suppressor and inhibits the proliferation and invasion of prostate cancer cells by down-regulation of SIX1. Hum. Cell 2017, 30, 290–299. [Google Scholar] [CrossRef]
- Kozomara, A.; Hunt, S.; Ninova, M.; Griffiths-Jones, S.; Ronshaugen, M. Target repression induced by endogenous microRNAs: Large differences, small effects. PLoS ONE 2014, 9, e104286. [Google Scholar] [CrossRef]
- Mayya, V.K.; Duchaine, T.F. On the availability of microRNA-induced silencing complexes, saturation of microRNA-binding sites and stoichiometry. Nucleic Acids Res. 2015, 43, 7556–7565. [Google Scholar] [CrossRef] [PubMed]
- Bissels, U.; Wild, S.; Tomiuk, S.; Holste, A.; Hafner, M.; Tuschl, T.; Bosio, A. Absolute quantification of microRNAs by using a universal reference. Rna 2009, 15, 2375–2384. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Z.; O’Loughlin, E.; Lee, T.; Houel, S.; O’Carroll, D.; Tarakhovsky, A.; Ahn, N.G.; Yi, R. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 2012, 26, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.D.; Gentner, B.; Cantore, A.; Colleoni, S.; Amendola, M.; Zingale, A.; Baccarini, A.; Lazzari, G.; Galli, C.; Naldini, L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007, 25, 1457–1467. [Google Scholar] [CrossRef]
- Yang, J.S.; Phillips, M.D.; Betel, D.; Mu, P.; Ventura, A.; Siepel, A.C.; Chen, K.C.; Lai, E.C. Widespread regulatory activity of vertebrate microRNA* species. Rna 2011, 17, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Baccarini, A.; Chauhan, H.; Gardner, T.J.; Jayaprakash, A.D.; Sachidanandam, R.; Brown, B.D. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr. Biol. 2011, 21, 369–376. [Google Scholar] [CrossRef]
- Hsin, J.P.; Lu, Y.; Loeb, G.B.; Leslie, C.S.; Rudensky, A.Y. The effect of cellular context on miR-155-mediated gene regulation in four major immune cell types. Nat. Immunol. 2018, 19, 1137–1145. [Google Scholar] [CrossRef]
- Shan, J.; Al-Rumaihi, K.; Chouchane, K.; Al-Bozom, I.; Rabah, D.; Farhat, K.; Chouchane, L. Prostate cancer small non-coding RNA transcriptome in Arabs. J. Transl. Med. 2017, 15, 260. [Google Scholar] [CrossRef]
- Hart, M.; Nolte, E.; Wach, S.; Szczyrba, J.; Taubert, H.; Rau, T.T.; Hartmann, A.; Grasser, F.A.; Wullich, B. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol. Cancer Res. 2014, 12, 250–263. [Google Scholar] [CrossRef]
- Xu, G.; Wu, J.; Zhou, L.; Chen, B.; Sun, Z.; Zhao, F.; Tao, Z. Characterization of the small RNA transcriptomes of androgen dependent and independent prostate cancer cell line by deep sequencing. PLoS ONE 2010, 5, e15519. [Google Scholar] [CrossRef]
- Sun, D.; Lee, Y.S.; Malhotra, A.; Kim, H.K.; Matecic, M.; Evans, C.; Jensen, R.V.; Moskaluk, C.A.; Dutta, A. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011, 71, 1313–1324. [Google Scholar] [CrossRef]
- Hamilton, M.P.; Rajapakshe, K.I.; Bader, D.A.; Cerne, J.Z.; Smith, E.A.; Coarfa, C.; Hartig, S.M.; McGuire, S.E. The Landscape of microRNA Targeting in Prostate Cancer Defined by AGO-PAR-CLIP. Neoplasia 2016, 18, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Brenner, J.L.; Jasiewicz, K.L.; Fahley, A.F.; Kemp, B.J.; Abbott, A.L. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr. Biol. 2010, 20, 1321–1325. [Google Scholar] [CrossRef]
- Li, X.; Cassidy, J.J.; Reinke, C.A.; Fischboeck, S.; Carthew, R.W. A microRNA imparts robustness against environmental fluctuation during development. Cell 2009, 137, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Hill, J.; Olson, E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Gordanpour, A.; Nam, R.K.; Sugar, L.; Seth, A. MicroRNAs in prostate cancer: From biomarkers to molecularly-based therapeutics. Prostate Cancer Prostatic Dis. 2012, 15, 314–319. [Google Scholar] [CrossRef]
- Fang, Y.X.; Gao, W.Q. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene 2014, 33, 135–147. [Google Scholar] [CrossRef]
- Wang, Y.L.; Wu, S.; Jiang, B.; Yin, F.F.; Zheng, S.S.; Hou, S.C. Role of MicroRNAs in Prostate Cancer Pathogenesis. Clin. Genitourin. Cancer 2015, 13, 261–270. [Google Scholar] [CrossRef]
- Patil, P.A.; Magi-Galluzzi, C. MicroRNA in prostate cancer: Practical aspects. Histol. Histopathol. 2015, 30, 1379–1396. [Google Scholar] [CrossRef]
- Goto, Y.; Kurozumi, A.; Arai, T.; Nohata, N.; Kojima, S.; Okato, A.; Kato, M.; Yamazaki, K.; Ishida, Y.; Naya, Y.; et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br. J. Cancer 2017, 117, 409–420. [Google Scholar] [CrossRef]
- Boettger, T.; Beetz, N.; Kostin, S.; Schneider, J.; Krüger, M.; Hein, L.; Braun, T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Investig. 2009. [Google Scholar] [CrossRef]
- Taboga, S.R.; Scortegagna, E.; Siviero, M.P.; Carvalho, H.F. Anatomy of smooth muscle cells in nonmalignant and malignant human prostate tissue. Anat. Record 2008, 291, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Larne, O.; Hagman, Z.; Lilja, H.; Bjartell, A.; Edsjo, A.; Ceder, Y. miR-145 suppress the androgen receptor in prostate cancer cells and correlates to prostate cancer prognosis. Carcinogenesis 2015, 36, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Wach, S.; Nolte, E.; Szczyrba, J.; Stohr, R.; Hartmann, A.; Orntoft, T.; Dyrskjot, L.; Eltze, E.; Wieland, W.; Keck, B.; et al. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int. J. Cancer 2012, 130, 611–621. [Google Scholar] [CrossRef]
- Medrano, S.; Sequeira-Lopez, M.L.; Gomez, R.A. Deletion of the miR-143/145 cluster leads to hydronephrosis in mice. Am. J. Pathol. 2014, 184, 3226–3238. [Google Scholar] [CrossRef] [PubMed]
- Chivukula, R.R.; Shi, G.; Acharya, A.; Mills, E.W.; Zeitels, L.R.; Anandam, J.L.; Abdelnaby, A.A.; Balch, G.C.; Mansour, J.C.; Yopp, A.C.; et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell 2014, 157, 1104–1116. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, S.; Baker, A.; Moller, S.; Nielsen, B.S. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods 2010, 52, 375–381. [Google Scholar] [CrossRef]
- Chiyomaru, T.; Enokida, H.; Tatarano, S.; Kawahara, K.; Uchida, Y.; Nishiyama, K.; Fujimura, L.; Kikkawa, N.; Seki, N.; Nakagawa, M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br. J. Cancer 2010, 102, 883–891. [Google Scholar] [CrossRef]
- Naito, Y.; Sakamoto, N.; Oue, N.; Yashiro, M.; Sentani, K.; Yanagihara, K.; Hirakawa, K.; Yasui, W. MicroRNA-143 regulates collagen type III expression in stromal fibroblasts of scirrhous type gastric cancer. Cancer Sci. 2014, 105, 228–235. [Google Scholar] [CrossRef]
- Kumar, B.; Rosenberg, A.Z.; Choi, S.M.; Fox-Talbot, K.; De Marzo, A.M.; Nonn, L.; Brennen, W.N.; Marchionni, L.; Halushka, M.K.; Lupold, S.E. Cell-type specific expression of oncogenic and tumor suppressive microRNAs in the human prostate and prostate cancer. Sci. Rep. 2018, 8, 7189. [Google Scholar] [CrossRef]
- Szczyrba, J.; Loprich, E.; Wach, S.; Jung, V.; Unteregger, G.; Barth, S.; Grobholz, R.; Wieland, W.; Stohr, R.; Hartmann, A.; et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol. Cancer Res. 2010, 8, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Rane, J.K.; Scaravilli, M.; Ylipää, A.; Pellacani, D.; Mann, V.M.; Simms, M.S.; Nykter, M.; Collins, A.T.; Visakorpi, T.; Maitland, N.J. MicroRNA Expression Profile of Primary Prostate Cancer Stem Cells as a Source of Biomarkers and Therapeutic Targets. Eur. Urol. 2017, 67, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.F.; Liang, Y.M.; Lau, P.N.; Shen, W.; Wang, D.K.; Cheung, W.T.; Xue, C.J.; Poon, L.M.; Lam, Y.W. Dynamic localisation of mature microRNAs in Human nucleoli is influenced by exogenous genetic materials. PLoS ONE 2013, 8, e70869. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 64. [Google Scholar] [CrossRef]
- Kent, O.A.; McCall, M.N.; Cornish, T.C.; Halushka, M.K. Lessons from miR-143/145: The importance of cell-type localization of miRNAs. Nucleic Acids Res. 2014, 42, 7528–7538. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Liang, J.; Wei, J.; Basturk, O.; Wang, J.; Daniels, G.; Gellert, L.L.; Li, Y.; Shen, Y.; Osman, I.; et al. Epithelial and stromal expression of miRNAs during prostate cancer progression. Am. J. Transl. Res. 2014, 6, 329–339. [Google Scholar] [PubMed]
- Melbø-Jørgensen, C.; Ness, N.; Andersen, S.; Valkov, A.; Dønnem, T.; Al-Saad, S.; Kiselev, Y.; Berg, T.; Nordby, Y.; Bremnes, R.M.; et al. Stromal Expression of MiR-21 Predicts Biochemical Failure in Prostate Cancer Patients with Gleason Score 6. PLoS ONE 2014, 9, e113039. [Google Scholar] [CrossRef] [PubMed]
- Iscaife, A.; Reis, S.T.; Morais, D.R.; Viana, N.I.; da Silva, I.A.; Pimenta, R.; Bordini, A.; Dip, N.; Srougi, M.; Leite, K.R.M. Treating metastatic prostate cancer with microRNA-145. Apoptosis 2018, 23, 388–395. [Google Scholar] [CrossRef]
- Costa-Pinheiro, P.; Ramalho-Carvalho, J.; Vieira, F.Q.; Torres-Ferreira, J.; Oliveira, J.; Goncalves, C.S.; Costa, B.M.; Henrique, R.; Jeronimo, C. MicroRNA-375 plays a dual role in prostate carcinogenesis. Clin. Epigenetics 2015, 7, 42. [Google Scholar] [CrossRef]
- Selth, L.A.; Das, R.; Townley, S.L.; Coutinho, I.; Hanson, A.R.; Centenera, M.M.; Stylianou, N.; Sweeney, K.; Soekmadji, C.; Jovanovic, L.; et al. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene 2017, 36, 24–34. [Google Scholar] [CrossRef]
- Chu, M.; Chang, Y.; Li, P.; Guo, Y.; Zhang, K.; Gao, W. Androgen receptor is negatively correlated with the methylation-mediated transcriptional repression of miR-375 in human prostate cancer cells. Oncol. Rep. 2014, 31, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Choi, N.; Park, J.; Lee, J.S.; Yoe, J.; Park, G.Y.; Kim, E.; Jeon, H.; Cho, Y.M.; Roh, T.Y.; Lee, Y. miR-93/miR-106b/miR-375-CIC-CRABP1: A novel regulatory axis in prostate cancer progression. Oncotarget 2015, 6, 23533–23547. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Bjorling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lieberman, R.; Pan, J.; Zhang, Q.; Du, M.; Zhang, P.; Nevalainen, M.; Kohli, M.; Shenoy, N.K.; Meng, H.; et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol. Cancer 2016, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Pickl, J.M.; Tichy, D.; Kuryshev, V.Y.; Tolstov, Y.; Falkenstein, M.; Schuler, J.; Reidenbach, D.; Hotz-Wagenblatt, A.; Kristiansen, G.; Roth, W.; et al. Ago-RIP-Seq identifies Polycomb repressive complex I member CBX7 as a major target of miR-375 in prostate cancer progression. Oncotarget 2016, 7, 59589–59603. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef]
- Hu, X.; Jia, Y.; Yu, J.; Chen, J.; Fu, Q. Loss of YAP protein in prostate cancer is associated with Gleason score increase. Tumori 2015, 101, 189–193. [Google Scholar] [CrossRef]
- Pillman, K.A.; Phillips, C.A.; Roslan, S.; Toubia, J.; Dredge, B.K.; Bert, A.G.; Lumb, R.; Neumann, D.P.; Li, X.; Conn, S.J.; et al. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. Embo J. 2018, 37. [Google Scholar] [CrossRef]
- Renwick, N.; Cekan, P.; Masry, P.A.; McGeary, S.E.; Miller, J.B.; Hafner, M.; Li, Z.; Mihailovic, A.; Morozov, P.; Brown, M.; et al. Multicolor microRNA FISH effectively differentiates tumor types. J. Clin. Investig. 2013, 123, 2694–2702. [Google Scholar] [CrossRef]
- Jung, H.M.; Patel, R.S.; Phillips, B.L.; Wang, H.; Cohen, D.M.; Reinhold, W.C.; Chang, L.J.; Yang, L.J.; Chan, E.K. Tumor suppressor miR-375 regulates MYC expression via repression of CIP2A coding sequence through multiple miRNA-mRNA interactions. Mol. Biol. Cell 2013, 24, 1638–1648. [Google Scholar] [CrossRef]
- Falzone, L.; Lupo, G.; La Rosa, G.R.M.; Crimi, S.; Anfuso, C.D.; Salemi, R.; Rapisarda, E.; Libra, M.; Candido, S. Identification of Novel MicroRNAs and Their Diagnostic and Prognostic Significance in Oral Cancer. Cancers 2019, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Hanazawa, T.; Nohata, N.; Okamoto, Y.; Seki, N. The functional significance of microRNA-375 in human squamous cell carcinoma: Aberrant expression and effects on cancer pathways. J. Hum. Genet. 2012, 57, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Shi, X.; Wang, N.; Liu, C.; Wang, J. YAP1, targeted by miR-375, enhanced the pro-angiogenesis of airway smooth muscle cells in asthma via STAT3 activation. Cell Cycle 2020, 19, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Liu, Q.; Jiang, Y.; Wang, B.; Ji, Y.; Liu, H.; Xie, Y. Long non-coding RNA LNC_000898 alleviates cardiomyocyte apoptosis and promotes cardiac repair after myocardial infarction via modulating miR-375/PDK1 axis. J. Cardiovasc. Pharm. 2020. [Google Scholar] [CrossRef]
- Wang, C.; Luo, J.; Chen, Z.; Ye, M.; Hong, Y.; Liu, J.; Nie, J.; Zhao, Q.; Chang, Y. MiR-375 Impairs the Invasive Capabilities of Hepatoma Cells by Targeting HIF1alpha Under Hypoxia. Dig. Dis. Sci. 2020. [Google Scholar] [CrossRef]
- Li, H.; Li, Z.; Pi, Y.; Chen, Y.; Mei, L.; Luo, Y.; Xie, J.; Mao, X. MicroRNA-375 exacerbates knee osteoarthritis through repressing chondrocyte autophagy by targeting ATG2B. Aging 2020, 12, 7248–7261. [Google Scholar] [CrossRef]
- Cheng, S.; Di, Z.; Hirman, A.R.; Zheng, H.; Duo, L.; Zhai, Q.; Xu, J. MiR-375-3p alleviates the severity of inflammation through targeting YAP1/LEKTI pathway in HaCaT cells. Biosci. Biotechnol. Biochem. 2020, 1–9. [Google Scholar] [CrossRef]
- Xu, F.; Ye, M.L.; Zhang, Y.P.; Li, W.J.; Li, M.T.; Wang, H.Z.; Qiu, X.; Xu, Y.; Yin, J.W.; Hu, Q.; et al. MicroRNA-375-3p enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Cancer Sci. 2020, 111, 1528–1541. [Google Scholar] [CrossRef]
- Fan, K.; Zebisch, A.; Horny, K.; Schrama, D.; Becker, J.C. Highly Expressed miR-375 is not an Intracellular Oncogene in Merkel Cell Polyomavirus-Associated Merkel Cell Carcinoma. Cancers 2020, 12, 529. [Google Scholar] [CrossRef]
- Liu, A.S.; Yu, H.Y.; Yang, Y.L.; Xue, F.Y.; Chen, X.; Zhang, Y.; Zhou, Z.Y.; Zhang, B.; Li, L.; Sun, C.Z.; et al. A Chemotherapy-Driven Increase in Mcl-1 Mediates the Effect of miR-375 on Cisplatin Resistance in Osteosarcoma Cells. Onco Targets 2019, 12, 11667–11677. [Google Scholar] [CrossRef]
- Tang, W.; Li, G.S.; Li, J.D.; Pan, W.Y.; Shi, Q.; Xiong, D.D.; Mo, C.H.; Zeng, J.J.; Chen, G.; Feng, Z.B.; et al. The role of upregulated miR-375 expression in breast cancer: An in vitro and in silico study. Pathol. Res. Pr. 2020, 216, 152754. [Google Scholar] [CrossRef] [PubMed]
- Jaafarpour, Z.; Soleimani, M.; Hosseinkhani, S.; Geramizadeh, B.; Yaghmaei, P.; Mobarra, N.; Karimi, M.H. Overexpression of microRNA-375 and microRNA-122 promotes the differentiation of human induced pluripotent stem cells into hepatocyte-like cells. Biologicals 2020, 63, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.; Balwierz, A.; Zhang, J.D.; Kublbeck, M.; Pawitan, Y.; Hielscher, T.; Wiemann, S.; Sahin, O. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene 2013, 32, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.L.; Kwong, D.L.; Chan, T.H.; Law, S.Y.; Chen, L.; Li, Y.; Qin, Y.R.; Guan, X.Y. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut 2012, 61, 33–42. [Google Scholar] [CrossRef]
- He, X.X.; Chang, Y.; Meng, F.Y.; Wang, M.Y.; Xie, Q.H.; Tang, F.; Li, P.Y.; Song, Y.H.; Lin, J.S. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene 2012, 31, 3357–3369. [Google Scholar] [CrossRef]
- Chang, Y.; Yan, W.; He, X.; Zhang, L.; Li, C.; Huang, H.; Nace, G.; Geller, D.A.; Lin, J.; Tsung, A. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology 2012, 143, 177–187.e8. [Google Scholar] [CrossRef]
- Tsukamoto, Y.; Nakada, C.; Noguchi, T.; Tanigawa, M.; Nguyen, L.T.; Uchida, T.; Hijiya, N.; Matsuura, K.; Fujioka, T.; Seto, M.; et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010, 70, 2339–2349. [Google Scholar] [CrossRef]
- Liu, A.M.; Poon, R.T.; Luk, J.M. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem. Biophys. Res. Commun. 2010, 394, 623–627. [Google Scholar] [CrossRef]
- Ding, L.; Xu, Y.; Zhang, W.; Deng, Y.; Si, M.; Du, Y.; Yao, H.; Liu, X.; Ke, Y.; Si, J.; et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010, 20, 784–793. [Google Scholar] [CrossRef]
- De Souza Rocha Simonini, P.; Breiling, A.; Gupta, N.; Malekpour, M.; Youns, M.; Omranipour, R.; Malekpour, F.; Volinia, S.; Croce, C.M.; Najmabadi, H.; et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010, 70, 9175–9184. [Google Scholar] [CrossRef]
- Poy, M.N.; Hausser, J.; Trajkovski, M.; Braun, M.; Collins, S.; Rorsman, P.; Zavolan, M.; Stoffel, M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. USA 2009, 106, 5813–5818. [Google Scholar] [CrossRef] [PubMed]
- El Ouaamari, A.; Baroukh, N.; Martens, G.A.; Lebrun, P.; Pipeleers, D.; van Obberghen, E. miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 2008, 57, 2708–2717. [Google Scholar] [CrossRef]
- Ioannidis, J.P. Why most published research findings are false. PLoS Med. 2005, 2, e124. [Google Scholar] [CrossRef] [PubMed]
- Li, S.L.; An, N.; Liu, B.; Wang, S.Y.; Wang, J.J.; Ye, Y. Exosomes from LNCaP cells promote osteoblast activity through miR-375 transfer. Oncol. Lett. 2019, 17, 4463–4473. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.P.; Chen, J.; Seok, H.Y.; Zhang, Z.; Kataoka, M.; Hu, X.; Wang, D.Z. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ. Res. 2013, 112, 1234–1243. [Google Scholar] [CrossRef]
- Nam, J.W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell 2014, 53, 1031–1043. [Google Scholar] [CrossRef]
- Song, S.J.; Pandolfi, P.P. miR-22 in tumorigenesis. Cell Cycle 2014, 13, 11–12. [Google Scholar] [CrossRef]
- Pasqualini, L.; Bu, H.; Puhr, M.; Narisu, N.; Rainer, J.; Schlick, B.; Schafer, G.; Angelova, M.; Trajanoski, Z.; Borno, S.T.; et al. miR-22 and miR-29a Are Members of the Androgen Receptor Cistrome Modulating LAMC1 and Mcl-1 in Prostate Cancer. Mol. Endocrinol. 2015, 29, 1037–1054. [Google Scholar] [CrossRef]
- Wang, W.L.; Chatterjee, N.; Chittur, S.V.; Welsh, J.; Tenniswood, M.P. Effects of 1alpha,25 dihydroxyvitamin D3 and testosterone on miRNA and mRNA expression in LNCaP cells. Mol. Cancer 2011, 10, 58. [Google Scholar] [CrossRef]
- Delic, D.; Grosser, C.; Dkhil, M.; Al-Quraishy, S.; Wunderlich, F. Testosterone-induced upregulation of miRNAs in the female mouse liver. Steroids 2010, 75, 998–1004. [Google Scholar] [CrossRef]
- Ottman, R.; Nguyen, C.; Lorch, R.; Chakrabarti, R. MicroRNA expressions associated with progression of prostate cancer cells to antiandrogen therapy resistance. Mol. Cancer 2014, 13, 1. [Google Scholar] [CrossRef]
- Budd, K.E.; McCoy, F.; Monecke, S.; Cormican, P.; Mitchell, J.; Keane, O.M. Extensive Genomic Diversity among Bovine-Adapted Staphylococcus aureus: Evidence for a Genomic Rearrangement within CC97. PLoS ONE 2015, 10, e0134592. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ladd, A.; Dragoescu, E.; Budd, W.T.; Ware, J.L.; Zehner, Z.E. MicroRNA-17-3p is a prostate tumor suppressor in vitro and in vivo, and is decreased in high grade prostate tumors analyzed by laser capture microdissection. Clin. Exp. Metastasis 2009, 26, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Henry, G.H.; Malewska, A.; Joseph, D.B.; Malladi, V.S.; Lee, J.; Torrealba, J.; Mauck, R.J.; Gahan, J.C.; Raj, G.V.; Roehrborn, C.G.; et al. A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra. Cell Rep. 2018, 25, 3530–3542.e5. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Ibeagha-Awemu, E.M.; Liang, G.; Beaudoin, F.; Zhao, X.; Guan le, L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genom. 2014, 15, 181. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Kumar, A.; Gomez, C.R.; Akhtar, I.; Hancock, J.C.; Lage, J.M.; Pound, C.R.; Levenson, A.S. MTA1-activated Epi-microRNA-22 regulates E-cadherin and prostate cancer invasiveness. FEBS Lett. 2017, 591, 924–933. [Google Scholar] [CrossRef]
- Zhang, Z.; Lanz, R.B.; Xiao, L.; Wang, L.; Hartig, S.M.; Ittmann, M.M.; Feng, Q.; He, B. The tumor suppressive miR-200b subfamily is an ERG target gene in human prostate tumors. Oncotarget 2016, 7, 37993–38003. [Google Scholar] [CrossRef]
- Maina, P.K.; Shao, P.; Liu, Q.; Fazli, L.; Tyler, S.; Nasir, M.; Dong, X.; Qi, H.H. c-MYC drives histone demethylase PHF8 during neuroendocrine differentiation and in castration-resistant prostate cancer. Oncotarget 2016, 7, 75585–75602. [Google Scholar] [CrossRef]
- Budd, W.T.; Seashols-Williams, S.J.; Clark, G.C.; Weaver, D.; Calvert, V.; Petricoin, E.; Dragoescu, E.A.; O’Hanlon, K.; Zehner, Z.E. Dual Action of miR-125b As a Tumor Suppressor and OncomiR-22 Promotes Prostate Cancer Tumorigenesis. PLoS ONE 2015, 10, e0142373. [Google Scholar] [CrossRef]
- Chen, Z.; Qi, Y.; Gao, C. Cardiac myocyte-protective effect of microRNA-22 during ischemia and reperfusion through disrupting the caveolin-3/eNOS signaling. Int. J. Clin. Exp. Pathol. 2015, 8, 4614–4626. [Google Scholar]
- Gurha, P.; Abreu-Goodger, C.; Wang, T.; Ramirez, M.O.; Drumond, A.L.; van Dongen, S.; Chen, Y.; Bartonicek, N.; Enright, A.J.; Lee, B.; et al. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation 2012, 125, 2751–2761. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wu, M.; Zhao, P.; Huang, Y.; Wang, W.; Yin, W. Neuroprotective effects of viral overexpression of microRNA-22 in rat and cell models of cerebral ischemia-reperfusion injury. J. Cell. Biochem. 2015, 116, 233–241. [Google Scholar] [CrossRef]
- Tan, G.; Shi, Y.; Wu, Z.H. MicroRNA-22 promotes cell survival upon UV radiation by repressing PTEN. Biochem. Biophys. Res. Commun. 2012, 417, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Li, F.; Meng, Q.; Zhao, Y.; Chen, L.; Zhang, H.; Xue, L.; Zhang, X.; Lengner, C.; Yu, Z. Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by miR-22. PLoS Genet. 2015, 11, e1005253. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, J.; Li, C.; Kong, J.; Wang, J.; Wu, Y.; Xu, E.; Lai, M. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015, 356, 781–790. [Google Scholar] [CrossRef]
- Watahiki, A.; Wang, Y.; Morris, J.; Dennis, K.; O’Dwyer, H.M.; Gleave, M.; Gout, P.W. MicroRNAs associated with metastatic prostate cancer. PLoS ONE 2011, 6, e24950. [Google Scholar] [CrossRef]
- Wang, P.; Phan, T.; Gordon, D.; Chung, S.; Henning, S.M.; Vadgama, J.V. Arctigenin in combination with quercetin synergistically enhances the antiproliferative effect in prostate cancer cells. Mol. Nutr. Food Res. 2015, 59, 250–261. [Google Scholar] [CrossRef]
- Walter, B.A.; Valera, V.A.; Pinto, P.A.; Merino, M.J. Comprehensive microRNA Profiling of Prostate Cancer. J. Cancer 2013, 4, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Jalava, S.E.; Urbanucci, A.; Latonen, L.; Waltering, K.K.; Sahu, B.; Janne, O.A.; Seppala, J.; Lahdesmaki, H.; Tammela, T.L.; Visakorpi, T. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene 2012, 31, 4460–4471. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Takayama, K.; Katayama, S.; Urano, T.; Horie-Inoue, K.; Ikeda, K.; Takahashi, S.; Kawazu, C.; Hasegawa, A.; Ouchi, Y.; et al. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 2010, 13, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Aakula, A.; Kohonen, P.; Leivonen, S.K.; Makela, R.; Hintsanen, P.; Mpindi, J.P.; Martens-Uzunova, E.; Aittokallio, T.; Jenster, G.; Perala, M.; et al. Systematic Identification of MicroRNAs That Impact on Proliferation of Prostate Cancer Cells and Display Changed Expression in Tumor Tissue. Eur. Urol. 2016, 69, 1120–1128. [Google Scholar] [CrossRef]
- Lichner, Z.; Ding, Q.; Samaan, S.; Saleh, C.; Nasser, A.; Al-Haddad, S.; Samuel, J.N.; Fleshner, N.E.; Stephan, C.; Jung, K.; et al. miRNAs dysregulated in association with Gleason grade regulate extracellular matrix, cytoskeleton and androgen receptor pathways. J. Pathol. 2015, 237, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, Y.X.; Wang, Z.N. The microRNA-148/152 family: Multi-faceted players. Mol. Cancer 2013, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Cimino, D.; De Pitta, C.; Orso, F.; Zampini, M.; Casara, S.; Penna, E.; Quaglino, E.; Forni, M.; Damasco, C.; Pinatel, E.; et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 2013, 27, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Xu, C.; Fang, Z.; Li, Y.; Liu, H.; Wang, Y.; Xu, C.; Sun, Y. Androgen receptor regulated microRNA miR-182-5p promotes prostate cancer progression by targeting the ARRDC3/ITGB4 pathway. Biochem. Biophys. Res. Commun. 2016, 474, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiyama, K.; Ito, H.; Taga, M.; Naganuma, S.; Oshinoya, Y.; Nagano, K.; Yokoyama, O.; Itoh, H. Expression of microRNAs associated with Gleason grading system in prostate cancer: miR-182-5p is a useful marker for high grade prostate cancer. Prostate 2013, 73, 827–834. [Google Scholar] [CrossRef]
- Dambal, S.; Baumann, B.; McCray, T.; Williams, L.; Richards, Z.; Deaton, R.; Prins, G.S.; Nonn, L. The miR-183 family cluster alters zinc homeostasis in benign prostate cells, organoids and prostate cancer xenografts. Sci. Rep. 2017, 7, 7704. [Google Scholar] [CrossRef]
- Mihelich, B.L.; Khramtsova, E.A.; Arva, N.; Vaishnav, A.; Johnson, D.N.; Giangreco, A.A.; Martens-Uzunova, E.; Bagasra, O.; Kajdacsy-Balla, A.; Nonn, L. miR-183-96-182 cluster is overexpressed in prostate tissue and regulates zinc homeostasis in prostate cells. J. Biol. Chem. 2011, 286, 44503–44511. [Google Scholar] [CrossRef]
- Siu, M.K.; Tsai, Y.C.; Chang, Y.S.; Yin, J.J.; Suau, F.; Chen, W.Y.; Liu, Y.N. Transforming growth factor-beta promotes prostate bone metastasis through induction of microRNA-96 and activation of the mTOR pathway. Oncogene 2015, 34, 4767–4776. [Google Scholar] [CrossRef]
- Kong, D.; Li, Y.; Wang, Z.; Banerjee, S.; Ahmad, A.; Kim, H.R.; Sarkar, F.H. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells 2009, 27, 1712–1721. [Google Scholar] [CrossRef]
- Puhr, M.; Hoefer, J.; Schafer, G.; Erb, H.H.; Oh, S.J.; Klocker, H.; Heidegger, I.; Neuwirt, H.; Culig, Z. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am. J. Pathol. 2012, 181, 2188–2201. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, R.; Zhang, D.; Deng, Q.; Liu, B.; Chao, H.P.; Rycaj, K.; Takata, Y.; Lin, K.; Lu, Y.; et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 2017, 8, 14270. [Google Scholar] [CrossRef]
- Li, J.Z.; Li, J.; Wang, H.Q.; Li, X.; Wen, B.; Wang, Y.J. MiR-141-3p promotes prostate cancer cell proliferation through inhibiting kruppel-like factor-9 expression. Biochem. Biophys. Res. Commun. 2017, 482, 1381–1386. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Gao, P.; Zhu, B.; Chen, X.; Lin, F.; Wang, X.; Wei, J.; Zhang, H. Downregulation of microRNA-429 inhibits cell proliferation by targeting p27Kip1 in human prostate cancer cells. Mol. Med. Rep. 2015, 11, 1435–1441. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Xiao, H.; Yang, T.; Chang, L.; Tian, Y.; Wu, B.; Xu, H. Effects of miR-200c on the migration and invasion abilities of human prostate cancer Du145 cells and the corresponding mechanism. Front. Med. 2014, 8, 456–463. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Liu, Y.; Deng, X.; Qi, S.; Sun, X.; Liu, G.; Liu, Y.; Liu, Y.; Zhao, M. Down-regulation of miR-200b-3p by low p73 contributes to the androgen-independence of prostate cancer cells. Prostate 2013, 73, 1048–1056. [Google Scholar] [CrossRef]
- Yu, J.; Lu, Y.; Cui, D.; Li, E.; Zhu, Y.; Zhao, Y.; Zhao, F.; Xia, S. miR-200b suppresses cell proliferation, migration and enhances chemosensitivity in prostate cancer by regulating Bmi-1. Oncol. Rep. 2014, 31, 910–918. [Google Scholar] [CrossRef]
- Williams, L.V.; Veliceasa, D.; Vinokour, E.; Volpert, O.V. miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS ONE 2013, 8, e83991. [Google Scholar] [CrossRef]
- Lynch, S.M.; O’Neill, K.M.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. Regulation of miR-200c and miR-141 by Methylation in Prostate Cancer. Prostate 2016, 76, 1146–1159. [Google Scholar] [CrossRef]
- Xu, S.; Ge, J.; Zhang, Z.; Zhou, W. miR-141 inhibits prostatic cancer cell proliferation and migration, and induces cell apoptosis via targeting of RUNX1. Oncol. Rep. 2018, 39, 1454–1460. [Google Scholar] [CrossRef]
- Hua, Y.; Liang, C.; Miao, C.; Wang, S.; Su, S.; Shao, P.; Liu, B.; Bao, M.; Zhu, J.; Xu, A.; et al. MicroRNA-126 inhibits proliferation and metastasis in prostate cancer via regulation of ADAM9. Oncol. Lett. 2018, 15, 9051–9060. [Google Scholar] [CrossRef]
- Ottman, R.; Levy, J.; Grizzle, W.E.; Chakrabarti, R. The other face of miR-17-92a cluster, exhibiting tumor suppressor effects in prostate cancer. Oncotarget 2016, 7, 73739–73753. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Imamura, Y.; Sadar, M.D. Androgen receptor targeted therapies in castration-resistant prostate cancer: Bench to clinic. Int. J. Urol. 2016, 23, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Nouri, M.; Ratther, E.; Stylianou, N.; Nelson, C.C.; Hollier, B.G.; Williams, E.D. Androgen-targeted therapy-induced epithelial mesenchymal plasticity and neuroendocrine transdifferentiation in prostate cancer: An opportunity for intervention. Front. Oncol. 2014, 4, 370. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Studach, L.; Hullinger, R.L.; Xie, J.; Andrisani, O.M. Down-regulation of RE-1 silencing transcription factor (REST) in advanced prostate cancer by hypoxia-induced miR-106b~25. Exp. Cell Res. 2014, 320, 188–199. [Google Scholar] [CrossRef]
- Kao, C.J.; Martiniez, A.; Shi, X.B.; Yang, J.; Evans, C.P.; Dobi, A.; deVere White, R.W.; Kung, H.J. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene 2014, 33, 2495–2503. [Google Scholar] [CrossRef]
- Kumar, B.; Khaleghzadegan, S.; Mears, B.; Hatano, K.; Kudrolli, T.A.; Chowdhury, W.H.; Yeater, D.B.; Ewing, C.M.; Luo, J.; Isaacs, W.B.; et al. Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening. Oncotarget 2016, 7, 72593–72607. [Google Scholar] [CrossRef]
- Lin, Z.Y.; Chen, G.; Zhang, Y.Q.; He, H.C.; Liang, Y.X.; Ye, J.H.; Liang, Y.K.; Mo, R.J.; Lu, J.M.; Zhuo, Y.J.; et al. MicroRNA-30d promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer. Mol. Cancer 2017, 16, 48. [Google Scholar] [CrossRef]
- Ostano, P.; Mello-Grand, M.; Sesia, D.; Gregnanin, I.; Peraldo-Neia, C.; Guana, F.; Jachetti, E.; Farsetti, A.; Chiorino, G. Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 1078. [Google Scholar] [CrossRef]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Devor, E.J.; Schickling, B.M.; Leslie, K.K. microRNA expression patterns across seven cancers are highly correlated and dominated by evolutionarily ancient families. Biomed. Rep. 2014, 2, 384–387. [Google Scholar] [CrossRef] [PubMed]
- McCall, M.N.; Kim, M.S.; Adil, M.; Patil, A.H.; Lu, Y.; Mitchell, C.J.; Leal-Rojas, P.; Xu, J.; Kumar, M.; Dawson, V.L.; et al. Toward the human cellular microRNAome. Genome Res. 2017, 27, 1769–1781. [Google Scholar] [CrossRef] [PubMed]
- Carrer, M.; Liu, N.; Grueter, C.E.; Williams, A.H.; Frisard, M.I.; Hulver, M.W.; Bassel-Duby, R.; Olson, E.N. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc. Natl. Acad. Sci. USA 2012, 109, 15330–15335. [Google Scholar] [CrossRef] [PubMed]
- Denham, J.; Marques, F.Z.; O’Brien, B.J.; Charchar, F.J. Exercise: Putting action into our epigenome. Sports Med. 2014, 44, 189–209. [Google Scholar] [CrossRef]
- Pan, D.; Mao, C.; Quattrochi, B.; Friedline, R.H.; Zhu, L.J.; Jung, D.Y.; Kim, J.K.; Lewis, B.; Wang, Y.X. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 2014, 5, 4725. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Xie, W.; Yang, M.; Hsieh, C.L.; Drouin, S.; Lee, G.S.; Kantoff, P.W. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate 2013, 73, 346–354. [Google Scholar] [CrossRef]
- Avgeris, M.; Stravodimos, K.; Scorilas, A. Loss of miR-378 in prostate cancer, a common regulator of KLK2 and KLK4, correlates with aggressive disease phenotype and predicts the short-term relapse of the patients. Biol. Chem. 2014, 395, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Kulyte, A.; Lorente-Cebrian, S.; Gao, H.; Mejhert, N.; Agustsson, T.; Arner, P.; Ryden, M.; Dahlman, I. MicroRNA profiling links miR-378 to enhanced adipocyte lipolysis in human cancer cachexia. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E267–E274. [Google Scholar] [CrossRef] [PubMed]
- Valentino, A.; Calarco, A.; Di Salle, A.; Finicelli, M.; Crispi, S.; Calogero, R.A.; Riccardo, F.; Sciarra, A.; Gentilucci, A.; Galderisi, U.; et al. Deregulation of MicroRNAs mediated control of carnitine cycle in prostate cancer: Molecular basis and pathophysiological consequences. Oncogene 2017, 36, 6030–6040. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.; Hickey, T.; Tilley, W.D.; Selth, L.A. Interplay between the androgen receptor signalling axis and microRNAs in prostate cancer. Endocr. Relat. Cancer 2019. [Google Scholar] [CrossRef] [PubMed]
- Ostling, P.; Leivonen, S.K.; Aakula, A.; Kohonen, P.; Makela, R.; Hagman, Z.; Edsjo, A.; Kangaspeska, S.; Edgren, H.; Nicorici, D.; et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011, 71, 1956–1967. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, C.E.; Sulpice, E.; Combe, S.; Shibakawa, A.; Leach, D.A.; Hamilton, M.P.; Chrysostomou, S.L.; Sharp, A.; Welti, J.; Yuan, W.; et al. Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene 2019, 38, 5700–5724. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.I.; Misawa, A.; Inoue, S. Significance of microRNAs in Androgen Signaling and Prostate Cancer Progression. Cancers 2017, 9, 102. [Google Scholar] [CrossRef]
- Fletcher, C.E.; Dart, D.A.; Sita-Lumsden, A.; Cheng, H.; Rennie, P.S.; Bevan, C.L. Androgen-regulated processing of the oncomir miR-27a, which targets Prohibitin in prostate cancer. Hum. Mol. Genet. 2012, 21, 3112–3127. [Google Scholar] [CrossRef]
- Mo, W.; Zhang, J.; Li, X.; Meng, D.; Gao, Y.; Yang, S.; Wan, X.; Zhou, C.; Guo, F.; Huang, Y.; et al. Identification of novel AR-targeted microRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate cancer. PLoS ONE 2013, 8, e56592. [Google Scholar] [CrossRef]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef]
- Goto, Y.; Kojima, S.; Nishikawa, R.; Kurozumi, A.; Kato, M.; Enokida, H.; Matsushita, R.; Yamazaki, K.; Ishida, Y.; Nakagawa, M.; et al. MicroRNA expression signature of castration-resistant prostate cancer: The microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br. J. Cancer 2015, 113, 1055–1065. [Google Scholar] [CrossRef]
- Goto, Y.; Kojima, S.; Nishikawa, R.; Enokida, H.; Chiyomaru, T.; Kinoshita, T.; Nakagawa, M.; Naya, Y.; Ichikawa, T.; Seki, N. The microRNA-23b/27b/24-1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget 2014, 5, 7748–7759. [Google Scholar] [CrossRef]
- Leidner, R.S.; Li, L.; Thompson, C.L. Dampening enthusiasm for circulating microRNA in breast cancer. PLoS ONE 2013, 8, e57841. [Google Scholar] [CrossRef]
- Djulbegovic, M.; Beyth, R.J.; Neuberger, M.M.; Stoffs, T.L.; Vieweg, J.; Djulbegovic, B.; Dahm, P. Screening for prostate cancer: Systematic review and meta-analysis of randomised controlled trials. BMJ 2010, 341, c4543. [Google Scholar] [CrossRef] [PubMed]
- McDonald, A.C.; Vira, M.; Shen, J.; Sanda, M.; Raman, J.D.; Liao, J.; Patil, D.; Taioli, E. Circulating microRNAs in plasma as potential biomarkers for the early detection of prostate cancer. Prostate 2018, 78, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Cui, Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef] [PubMed]
- Paiva, R.M.; Zauli, D.A.G.; Neto, B.S.; Brum, I.S. Urinary microRNAs expression in prostate cancer diagnosis: A systematic review. Clin. Transl. Oncol. 2020. [Google Scholar] [CrossRef]
- Mihelich, B.L.; Maranville, J.C.; Nolley, R.; Peehl, D.M.; Nonn, L. Elevated serum microRNA levels associate with absence of high-grade prostate cancer in a retrospective cohort. PLoS ONE 2015, 10, e0124245. [Google Scholar] [CrossRef]
- Haldrup, C.; Kosaka, N.; Ochiya, T.; Borre, M.; Hoyer, S.; Orntoft, T.F.; Sorensen, K.D. Profiling of circulating microRNAs for prostate cancer biomarker discovery. Drug Deliv. Transl. Res. 2014, 4, 19–30. [Google Scholar] [CrossRef]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar] [CrossRef]
- Brase, J.C.; Johannes, M.; Schlomm, T.; Falth, M.; Haese, A.; Steuber, T.; Beissbarth, T.; Kuner, R.; Sultmann, H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 2011, 128, 608–616. [Google Scholar] [CrossRef]
- Lodes, M.J.; Caraballo, M.; Suciu, D.; Munro, S.; Kumar, A.; Anderson, B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS ONE 2009, 4, e6229. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Al-Qatati, A.; Akrong, C.; Stevic, I.; Pantel, K.; Awe, J.; Saranchuk, J.; Drachenberg, D.; Mai, S.; Schwarzenbach, H. Plasma microRNA signature is associated with risk stratification in prostate cancer patients. Int. J. Cancer 2017, 141, 1231–1239. [Google Scholar] [CrossRef]
- Souza, M.F.; Kuasne, H.; Barros-Filho, M.C.; Ciliao, H.L.; Marchi, F.A.; Fuganti, P.E.; Paschoal, A.R.; Rogatto, S.R.; Colus, I.M.S. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS ONE 2017, 12, e0184094. [Google Scholar] [CrossRef] [PubMed]
- Sharova, E.; Grassi, A.; Marcer, A.; Ruggero, K.; Pinto, F.; Bassi, P.; Zanovello, P.; Zattoni, F.; D’Agostino, D.M.; Iafrate, M.; et al. A circulating miRNA assay as a first-line test for prostate cancer screening. Br. J. Cancer 2016, 114, 1362–1366. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs as Biomarkers for Diagnosis, Prognosis and Theranostics in Prostate Cancer. Int. J. Mol. Sci. 2016, 17, 421. [Google Scholar] [CrossRef] [PubMed]
- Mello-Grand, M.; Gregnanin, I.; Sacchetto, L.; Ostano, P.; Zitella, A.; Bottoni, G.; Oderda, M.; Marra, G.; Munegato, S.; Pardini, B.; et al. Circulating microRNAs combined with PSA for accurate and non-invasive prostate cancer detection. Carcinogenesis 2019, 40, 246–253. [Google Scholar] [CrossRef]
- Yang, M.; Liu, E.; Tang, L.; Lei, Y.; Sun, X.; Hu, J.; Dong, H.; Yang, S.M.; Gao, M.; Tang, B. Emerging roles and regulation of MiT/TFE transcriptional factors. Cell Commun. Signal. 2018, 16, 31. [Google Scholar] [CrossRef]
- Endzelins, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Sobolevska, K.; Abols, A.; Rodriguez, M.; Santare, D.; Rudnickiha, A.; Lietuvietis, V.; et al. Detection of circulating miRNAs: Comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017, 17, 730. [Google Scholar] [CrossRef]
- Urabe, F.K.N.; Sawa, Y.; Yamamoto, Y.; Ito, K.; Yamamoto, T.; Kimura, T.; Egawa, S.; Ochiya, T. miR-26a regulates extracellular vesicle secretion from prostate cancer cells via targeting SHC4, PFDN4, and CHORDC1. Sci. Adv. 2020, 6, eaay3051. [Google Scholar] [CrossRef]
- Li, W.; Jiang, Y.; Wu, X.; Yang, F. Targeted Regulation of miR-26a on PTEN to Affect Proliferation and Apoptosis of Prostate Cancer Cells. Cancer Biother. Radiopharm. 2019, 34, 480–485. [Google Scholar] [CrossRef]
- Rizzo, M.; Berti, G.; Russo, F.; Fazio, S.; Evangelista, M.; D’Aurizio, R.; Pellegrini, M.; Rainaldi, G. Discovering the miR-26a-5p Targetome in Prostate Cancer Cells. J. Cancer 2017, 8, 2729–2739. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Falzone, L.; Candido, S.; Salemi, R.; Basile, M.S.; Scalisi, A.; McCubrey, J.A.; Torino, F.; Signorelli, S.S.; Montella, M.; Libra, M. Computational identification of microRNAs associated to both epithelial to mesenchymal transition and NGAL/MMP-9 pathways in bladder cancer. Oncotarget 2016, 7, 72758–72766. [Google Scholar] [CrossRef] [PubMed]
- Ghorbanmehr, N.; Gharbi, S.; Korsching, E.; Tavallaei, M.; Einollahi, B.; Mowla, S.J. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate 2019, 79, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating cancer with microRNA replacement therapy: A literature review. J. Cell Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef]
- Schmidt, M.F. miRNA Targeting Drugs: The Next Blockbusters? Methods Mol. Biol. 2017, 1517, 3–22. [Google Scholar] [CrossRef]
- Haussecker, D.; Kay, M.A. RNA interference. Drugging RNAi. Science 2015, 347, 1069–1070. [Google Scholar] [CrossRef]
- Krieg, A.M. Is RNAi dead? Mol. Ther. 2011, 19, 1001–1002. [Google Scholar] [CrossRef]
- Baek, J.; Kang, S.; Min, H. MicroRNA-targeting therapeutics for hepatitis C. Arch. Pharmacal. Res. 2014, 37, 299–305. [Google Scholar] [CrossRef]
- Adams, B.D.; Parsons, C.; Slack, F.J. The tumor-suppressive and potential therapeutic functions of miR-34a in epithelial carcinomas. Expert Opin. Ther. Targets 2016, 20, 737–753. [Google Scholar] [CrossRef]
- Reid, G.; Kao, S.C.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; van Zandwijk, N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 2016, 8, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.C.; Fulham, M.; Wong, K.; Cooper, W.; Brahmbhatt, H.; MacDiarmid, J.; Pattison, S.; Sagong, J.O.; Huynh, Y.; Leslie, F.; et al. A Significant Metabolic and Radiological Response after a Novel Targeted MicroRNA-based Treatment Approach in Malignant Pleural Mesothelioma. Am. J. Respir. Crit. Care Med. 2015, 191, 1467–1469. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.O.; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, F.; Zubovic, L.; Pavelitz, T.; Yang, W.; Godin, K.; Walker, M.; Zheng, S.; Macchi, P.; Varani, G. Targeted inhibition of oncogenic miR-21 maturation with designed RNA-binding proteins. Nat. Chem. Biol. 2016, 12, 717–723. [Google Scholar] [CrossRef]
- Vos, P.D.; Leedman, P.J.; Filipovska, A.; Rackham, O. Modulation of miRNA function by natural and synthetic RNA-binding proteins in cancer. Cell Mol. Life Sci. 2019. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, H.; Tan, Z.; Zhang, C.; Fu, X. Bottleneck limitations for microRNA-based therapeutics from bench to the bedside. Die Pharm. 2015, 70, 147–154. [Google Scholar]
- Diaz, M.R.; Vivas-Mejia, P.E. Nanoparticles as Drug Delivery Systems in Cancer Medicine: Emphasis on RNAi-Containing Nanoliposomes. Pharmaceuticals 2013, 6, 1361–1380. [Google Scholar] [CrossRef]
- Esposito, C.L.; Catuogno, S.; de Franciscis, V. Aptamer-MiRNA Conjugates for Cancer Cell-Targeted Delivery. Methods Mol. Biol. 2016, 1364, 197–208. [Google Scholar] [CrossRef]
- Bender, E. Nanoparticle theories slowly turn into practice. Cancer Discov. 2011, 1, 280. [Google Scholar]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Loeb, G.B.; Khan, A.A.; Canner, D.; Hiatt, J.B.; Shendure, J.; Darnell, R.B.; Leslie, C.S.; Rudensky, A.Y. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 2012, 48, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Rodosthenous, R.S.; Kashanchi, F.; Gingeras, T.; Gould, S.J.; Kuo, L.S.; Kurre, P.; Lee, H.; Leonard, J.N.; Liu, H.; et al. Advances, challenges, and opportunities in extracellular RNA biology: Insights from the NIH exRNA Strategic Workshop. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242. [Google Scholar] [CrossRef] [PubMed]
- Tarver, J.E.; Taylor, R.S.; Puttick, M.N.; Lloyd, G.T.; Pett, W.; Fromm, B.; Schirrmeister, B.E.; Pisani, D.; Peterson, K.J.; Donoghue, P.C.J. Well-Annotated microRNAomes Do Not Evidence Pervasive miRNA Loss. Genome Biol. Evol. 2018, 10, 1457–1470. [Google Scholar] [CrossRef]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. microRNA Therapeutics in Cancer - An Emerging Concept. EBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef]
| In Vivo Top 10 Expressed | In Vitro Top 10 Expressed | |||||||
|---|---|---|---|---|---|---|---|---|
| microRNA $ | TCGA 1 Rank | N 2,3 | PCa 2,3 | RWPE 4 | LNCaP 5,6,7,8 | Microarray 9 (RWPE/ LNCaP/ PC3/ DU145/ Stromal) | Clip RNAseq 10 (LNCaP/ LAPC4/ 22Rv1/ DU145/ PC3) | Altered in Cancer |
| miR143/145 | 1 | 1/1 | 1/1 | - | -/-/-/- | -/-/-/-/- | -/-/-/-/- | down |
| miR125/99/100/10 | 2 | 2/5 | 2/3 | - | -/3/-/- | -/1/10/-/- | 6/-/-/8/- | down |
| miR375 | 3 | -/- | 4/- | - | -/-/-/- | -/-/-/-/- | -/-/-/-/- | up |
| miR148/152 | 4 | -/- | 9/- | - | -/-/2/5 | -/-/-/-/- | 3/2/1/6/- | up |
| miR21 | 5 | -/10 | -/10 | 1 | -/4/6/1 | 8/-/7/2/- | 10/-/-/3/3 | |
| miR30 | 6 | -/- | -/- | - | -/-/-/3 | -/-/-/10/- | 9/-/-/-/- | |
| miR182/183/96 | 7 | -/- | -/- | 7 | -/-/5/6 | -/-/-/-/- | 2/1/4/4/1 | up |
| miR22 | 8 | 8/8 | -/9 | 4 | -/-/4/- | 9/-/-/8/6 | 5/8/-/7/6 | up |
| let7 | 9 | 3/3 | 3/4 | 3 | 1/1/1/2 | 3/2/1/4/1 | 1/3/2/5/2 | |
| miR141/200/429 | 10 | -/9 | -/7 | - | -/6/10/8 | 10/10/-/-/- | 7/-/-/-/- | up |
| Mir-23/24/27 | - | 5/2 | 7/2 | 5 | -/-/9/- | 1/-/3/1/2 | -/-/-/2/7 | down |
| miR-26 | - | 6/6 | 6/6 | 9 | -/-/8/- | -/7/-/-/- | -/-/9/-/- | |
| miR-191 | - | 7/- | 5/- | 10 | 2/-/3/- | -/-/-/-/- | -/-/-/-/- | |
| miR-15/16 | - | -/7 | -/8 | - | 5/8/-/- | 7/-/-/9/- | -/-/8/9/- | |
| miR-29 | - | -/4 | -/5 | - | 6/-/-/- | -/-/9/7/7 | -/-/-/-/8 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andl, T.; Ganapathy, K.; Bossan, A.; Chakrabarti, R. MicroRNAs as Guardians of the Prostate: Those Who Stand before Cancer. What Do We Really Know about the Role of microRNAs in Prostate Biology? Int. J. Mol. Sci. 2020, 21, 4796. https://doi.org/10.3390/ijms21134796
Andl T, Ganapathy K, Bossan A, Chakrabarti R. MicroRNAs as Guardians of the Prostate: Those Who Stand before Cancer. What Do We Really Know about the Role of microRNAs in Prostate Biology? International Journal of Molecular Sciences. 2020; 21(13):4796. https://doi.org/10.3390/ijms21134796
Chicago/Turabian StyleAndl, Thomas, Kavya Ganapathy, Alexia Bossan, and Ratna Chakrabarti. 2020. "MicroRNAs as Guardians of the Prostate: Those Who Stand before Cancer. What Do We Really Know about the Role of microRNAs in Prostate Biology?" International Journal of Molecular Sciences 21, no. 13: 4796. https://doi.org/10.3390/ijms21134796
APA StyleAndl, T., Ganapathy, K., Bossan, A., & Chakrabarti, R. (2020). MicroRNAs as Guardians of the Prostate: Those Who Stand before Cancer. What Do We Really Know about the Role of microRNAs in Prostate Biology? International Journal of Molecular Sciences, 21(13), 4796. https://doi.org/10.3390/ijms21134796







