Protein–Protein Interactions Efficiently Modeled by Residue Cluster Classes
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Datasets
4.2. Machine Learning and Statistical Testing
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.; Lewis, S.E.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Wang, J.P.; Liu, B.; Sun, Y.; Chiang, V.L.; Sederoff, R.R. Enzyme-enzyme interactions in monolignol biosynthesis. Front Plant Sci. 2019, 9, 1942. [Google Scholar] [CrossRef] [PubMed]
- Freilich, R.; Arhar, T.; Abrams, J.L.; Gestwicki, J.E. Protein-Protein Interactions in the Molecular Chaperone Network. Acc. Chem. Res. 2018, 51, 940–949. [Google Scholar] [CrossRef]
- Zahiri, J.; Emamjomeh, A.; Bagheri, S.; Ivazeh, A.; Mahdevar, G.; Sepasi Tehrani, H.; Mirzaie, M.; Fakheri, B.A.; Mohammad-Noori, M. Protein complex prediction: A survey. Genomics 2020, 112, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, C.; Deng, L. Machine learning approaches for protein-protein interaction hot spot prediction: Progress and comparative assessment. Molecules 2018, 23, 2535. [Google Scholar] [CrossRef]
- Kotlyar, M.; Rossos, A.E.M.; Jurisica, I. Prediction of Protein-Protein Interactions. Curr. Protoc. Bioinform. 2017, 60, 8.2.1–8.2.14. [Google Scholar] [CrossRef]
- Bzdok, D.; Krzywinski, M.; Altman, N. Points of significance: Machine learning: Supervised methods. Nat. Methods 2018, 15, 5–6. [Google Scholar] [CrossRef]
- Ruiz-Blanco, Y.B.; Paz, W.; Green, J.; Marrero-Ponce, Y. ProtDCal: A program to compute general-purpose-numerical descriptors for sequences and 3D-structures of proteins. BMC Bioinform. 2015, 16, 162. Available online: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-015-0586-0 (accessed on 3 February 2020). [CrossRef]
- Shen, H.B.; Chou, K.C. PseAAC: A flexible web server for generating various kinds of protein pseudo amino acid composition. Anal. Biochem. 2008, 373, 386–388. [Google Scholar] [CrossRef]
- Li, Z.R.; Lin, H.H.; Han, L.Y.; Jiang, L.; Chen, X.; Chen, Y.Z. PROFEAT: A web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence. Nucleic Acids Res. 2006, 34, w32–w37. [Google Scholar] [CrossRef]
- Sarkar, D.; Saha, S. Machine-learning techniques for the prediction of protein–protein interactions. J. Biosci. 2019, 44, 104. [Google Scholar] [CrossRef]
- Romero-Molina, S.; Ruiz-Blanco, Y.B.; Green, J.R.; Sanchez-Garcia, E. ProtDCal-Suite: A web server for the numerical codification and functional analysis of proteins. Protein Sci. 2019, 28, 1734–1743. Available online: http://www.ncbi.nlm.nih.gov/pubmed/31271472 (accessed on 3 February 2020). [CrossRef] [PubMed]
- Chen, M.; Ju, C.J.T.; Zhou, G.; Chen, X.; Zhang, T.; Chang, K.W.; Zaniolo, C.W.; Wang, W. Multifaceted Protein-Protein Interaction Prediction Based on Siamese Residual RCNN. Bioinformatics 2019, 35, i305–i314. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Chan, K.C.C. Extracting Coevolutionary Features from Protein Sequences for Predicting Protein-Protein Interactions. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Ardakani, F.B.; Schmidt, F.; Schulz, M.H. Predicting transcription factor binding using ensemble random forest models [version 2; peer review: 2 approved]. F1000Research 2019, 7, 1603. [Google Scholar] [CrossRef]
- Hue, M.; Riffle, M.; Vert, J.P.; Noble, W.S. Large-scale prediction of protein-protein interactions from structures. BMC Bioinform. 2010, 11, 144. [Google Scholar] [CrossRef]
- Chang, J.W.; Zhou, Y.Q.; Ul Qamar, M.T.; Chen, L.L.; Ding, Y.D. Prediction of protein–protein interactions by evidence combining methods. Int. J. Mol. Sci. 2016, 17, 1946. [Google Scholar] [CrossRef]
- Ding, Z.; Kihara, D. Computational Methods for Predicting Protein-Protein Interactions Using Various Protein Features. Curr. Protoc. Protein Sci. 2018, 93, e62. Available online: http://doi.wiley.com/10.1002/cpps.62 (accessed on 3 February 2020). [CrossRef]
- Zhang, S.B.; Tang, Q.R. Protein-protein interaction inference based on semantic similarity of Gene Ontology terms. J. Theor. Biol. 2016, 401, 30–37. [Google Scholar] [CrossRef]
- Corral-Corral, R.; Chavez, E.; Del Rio, G. Machine Learnable Fold Space Representation based on Residue Cluster Classes. Comput. Biol. Chem. 2015, 59, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mosca, R.; Céol, A.; Stein, A.; Olivella, R.; Aloy, P. 3did: A catalog of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2014, 42, D374. [Google Scholar] [CrossRef] [PubMed]
- Blohm, P.; Frishman, G.; Smialowski, P.; Goebels, F.; Wachinger, B.; Ruepp, A.; Frishman, D. Negatome 2.0: A database of non-interacting proteins derived by literature mining, manual annotation and protein structure analysis. Nucleic Acids Res. 2014, 42, D396–D400. Available online: http://www.ncbi.nlm.nih.gov/pubmed/24214996 (accessed on 16 February 2020). [CrossRef] [PubMed]
- Atkeson, C.G.; Moore, A.W.; Schaal, S. Locally Weighted Learning. Artif. Intell. Rev. 1997, 11, 11–73. [Google Scholar] [CrossRef]
- Fontove, F.; Del Rio, G. Residue cluster classes: A unified protein representation for efficient structural and functional classification. Entropy 2020, 22, 472. [Google Scholar] [CrossRef]
- Zhang, Q.C.; Petrey, D.; Deng, L.; Qiang, L.; Shi, Y.; Thu, C.A.; Bisikirska, B.; Lefebvre, C.; Accili, D.; Hunter, T.; et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature 2012, 490, 556–560. [Google Scholar] [CrossRef]
- Elefsinioti, A.; Saraç, Ö.S.; Hegele, A.; Plake, C.; Hubner, N.C.; Poser, I.; Sarov, M.; Hyman, A.; Mann, M.; Schroeder, M.; et al. Large-scale de novo prediction of physical protein-protein association. Mol. Cell. Proteomics 2011, 10, M111.010629. [Google Scholar] [CrossRef]
- Petschnigg, J.; Groisman, B.; Kotlyar, M.; Taipale, M.; Zheng, Y.; Kurat, C.F.; Sayad, A.; Sierra, J.R.; Usaj, M.M.; Snider, J.; et al. The mammalian-membrane two-hybrid assay (MaMTH) for probing membrane-protein interactions in human cells. Nat. Methods 2014, 11, 585–592. [Google Scholar] [CrossRef]
- Schwartz, A.S.; Yu, J.; Gardenour, K.R.; Finley, R.L.; Ideker, T. Cost-effective strategies for completing the interactome. Nat. Methods 2009, 6, 55–61. [Google Scholar] [CrossRef]
- Ben-Hur, A.; Noble, W.S. Choosing negative examples for the prediction of protein-protein interactions. BMC Bioinform. 2006, 7, S2. [Google Scholar] [CrossRef]
- Hamp, T.; Rost, B. More challenges for machine-learning protein interactions. Bioinformatics 2015, 31, 1521–1525. Available online: https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btu857 (accessed on 19 February 2020). [CrossRef] [PubMed]
- Park, Y.; Marcotte, E.M. Flaws in evaluation schemes for pair-input computational predictions. Nat. Methods 2012, 9, 1134–1136. [Google Scholar] [CrossRef] [PubMed]
- Basile, W.; Sachenkova, O.; Light, S.; Elofsson, A. High GC content causes orphan proteins to be intrinsically disordered. PLoS Comput. Biol. 2017, 13, e1005375. Available online: https://dx.plos.org/10.1371/journal.pcbi.1005375 (accessed on 1 April 2020). [CrossRef] [PubMed]
- Kotlyar, M.; Pastrello, C.; Sheahan, N.; Jurisica, I. Integrated interactions database: Tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 2016, 44, D536–D541. Available online: https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkv1115 (accessed on 19 February 2020). [CrossRef] [PubMed]
- Snider, J.; Kotlyar, M.; Saraon, P.; Yao, Z.; Jurisica, I.; Stagljar, I. Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 2015, 11, 848. Available online: https://onlinelibrary.wiley.com/doi/abs/10.15252/msb.20156351 (accessed on 19 February 2020). [CrossRef] [PubMed]
- Wang, Z.; Clark, N.R.; Ma’ayan, A. Dynamics of the discovery process of protein-protein interactions from low content studies. BMC Syst. Biol. 2015, 9, 26. Available online: https://bmcsystbiol.biomedcentral.com/articles/10.1186/s12918-015-0173-z (accessed on 19 February 2020). [CrossRef] [PubMed]
- Fischer, H.; Polikarpov, I.; Craievich, A.F. Average protein density is a molecular-weight-dependent function. Protein Sci. 2009, 13, 2825–2828. [Google Scholar] [CrossRef]
- Kaddis, C.S.; Lomeli, S.H.; Yin, S.; Berhane, B.; Apostol, M.I.; Kickhoefer, V.A.; Rome, L.H.; Loo, J.A. Sizing Large Proteins and Protein Complexes by Electrospray Ionization Mass Spectrometry and Ion Mobility. J. Am. Soc. Mass Spectrom. 2007, 18, 1206–1216. [Google Scholar] [CrossRef]
- Alquraishi, M.; Valencia, A. AlphaFold at CASP13. Bioinformatics 2019, 35, 4862–4865. Available online: http://www.ncbi.nlm.nih.gov/pubmed/31116374 (accessed on 19 February 2020). [CrossRef]
- Roche, D.B.; McGuffin, L.J. Toolbox for protein structure prediction. Methods in Molecular Biology 2016, 1369, 363–377. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019, 47, D464–D474. Available online: https://academic.oup.com/nar/article/47/D1/D464/5144139 (accessed on 16 February 2020). [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA Data Mining Software: An Update. ACM SIGKDD Explor. Newsl. 2009, 11, 1. Available online: https://www.kdd.org/exploration_files/p2V11n1.pdf (accessed on 12 February 2020). [CrossRef]
- Kotthoff, L.; Thornton, C.; Hoos, H.H.; Hutter, F.; Leyton-Brown, K. Auto-WEKA 2.0: Automatic model selection and hyperparameter optimization in WEKA. J. Mach. Learn. Res. 2017, 18, 1–5. Available online: http://automl.org/autoweka (accessed on 12 February 2020).
- Mckinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference (SCIPY 2010), Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Van Der Walt, S.; Colbert, S.C.; Varoquaux, G. The NumPy array: A structure for efficient numerical computation. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar] [CrossRef]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and Statistical Modeling with Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 92–96. Available online: http://statsmodels.sourceforge.net/ (accessed on 5 April 2020).
- Waskom, M. Seaborn: Statistical Data Visualization—Seaborn 0.10.0 Documentation. 2012. Available online: https://seaborn.pydata.org/ (accessed on 9 April 2020).
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 99–104. [Google Scholar] [CrossRef]








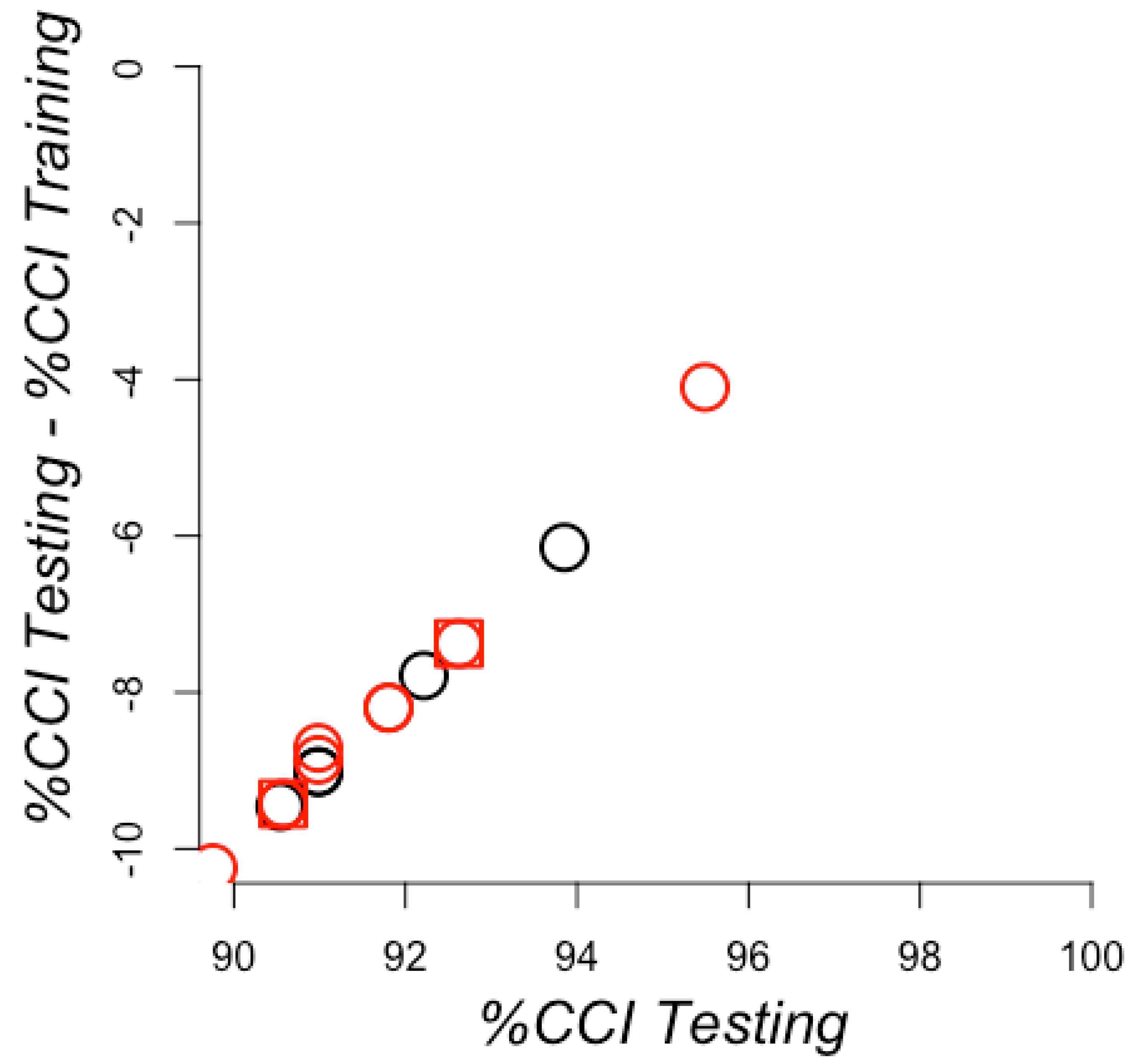
| P:N | Training | Testing | ||
|---|---|---|---|---|
| P | N | P | N | |
| 1:1 | 692 | 692 | 4819 | 692 |
| 2:1 | 1384 | 692 | 4819 | 692 |
| 3:1 | 2076 | 692 | 4819 | 692 |
| 1:2 | 692 | 1384 | 4819 | 692 |
| 1:3 | 692 | 2076 | 4819 | 692 |
| Training | Testing | ||||
|---|---|---|---|---|---|
| Positives | Negatives | Positives | Negatives | ||
| Concatenation | 1:1 | 489 | 489 | 122 | 122 |
| 2:1 | 978 | 489 | 122 | 122 | |
| 3:1 | 1467 | 489 | 122 | 122 | |
| Sum | 1:1 | 448 | 448 | 111 | 111 |
| 2:1 | 896 | 448 | 111 | 111 | |
| 3:1 | 1344 | 448 | 111 | 111 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poot Velez, A.H.; Fontove, F.; Del Rio, G. Protein–Protein Interactions Efficiently Modeled by Residue Cluster Classes. Int. J. Mol. Sci. 2020, 21, 4787. https://doi.org/10.3390/ijms21134787
Poot Velez AH, Fontove F, Del Rio G. Protein–Protein Interactions Efficiently Modeled by Residue Cluster Classes. International Journal of Molecular Sciences. 2020; 21(13):4787. https://doi.org/10.3390/ijms21134787
Chicago/Turabian StylePoot Velez, Albros Hermes, Fernando Fontove, and Gabriel Del Rio. 2020. "Protein–Protein Interactions Efficiently Modeled by Residue Cluster Classes" International Journal of Molecular Sciences 21, no. 13: 4787. https://doi.org/10.3390/ijms21134787
APA StylePoot Velez, A. H., Fontove, F., & Del Rio, G. (2020). Protein–Protein Interactions Efficiently Modeled by Residue Cluster Classes. International Journal of Molecular Sciences, 21(13), 4787. https://doi.org/10.3390/ijms21134787






