Characterization of PtAOS1 Promoter and Three Novel Interacting Proteins Responding to Drought in Poncirus trifoliata
Abstract
1. Introduction
2. Results
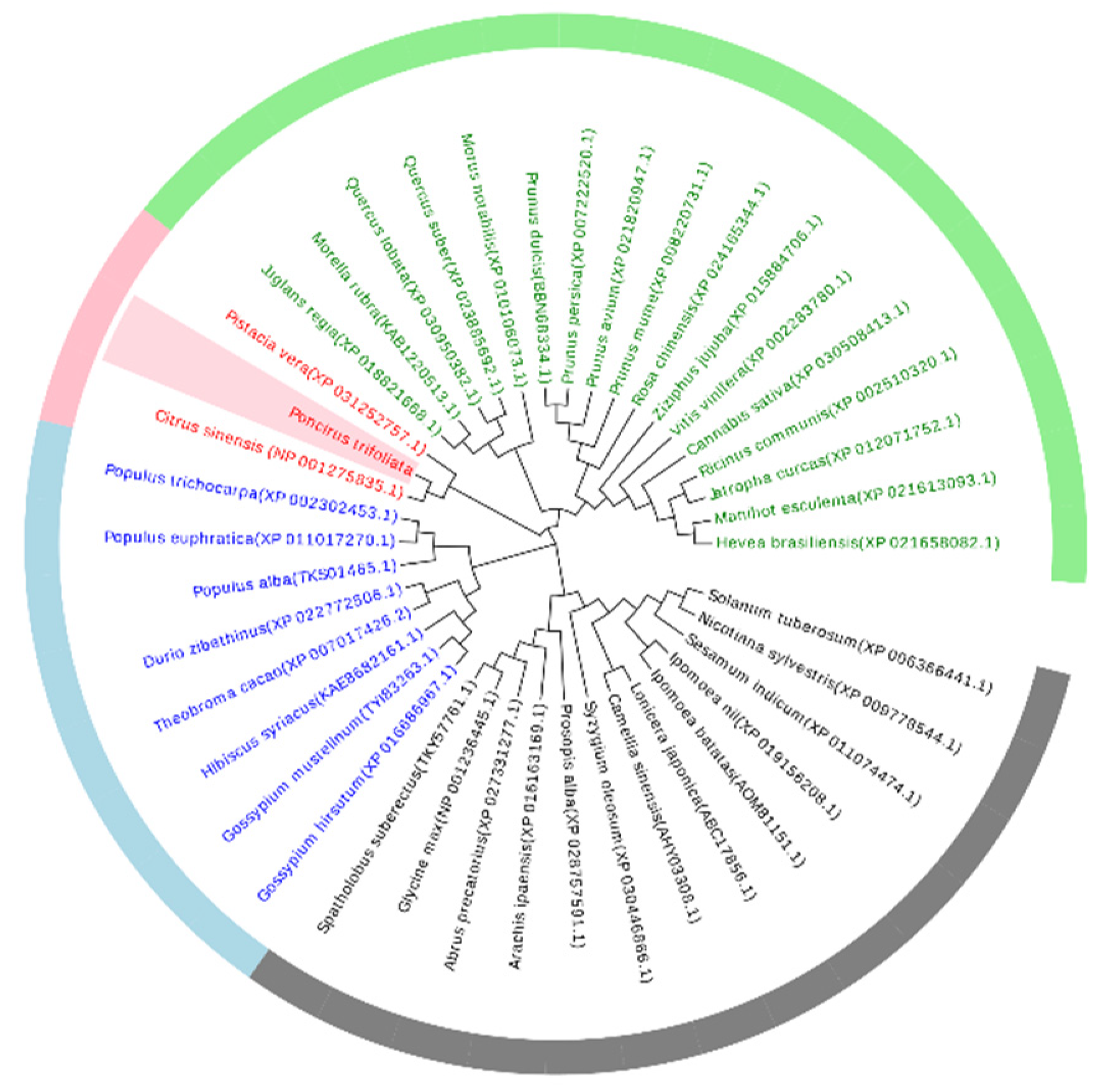
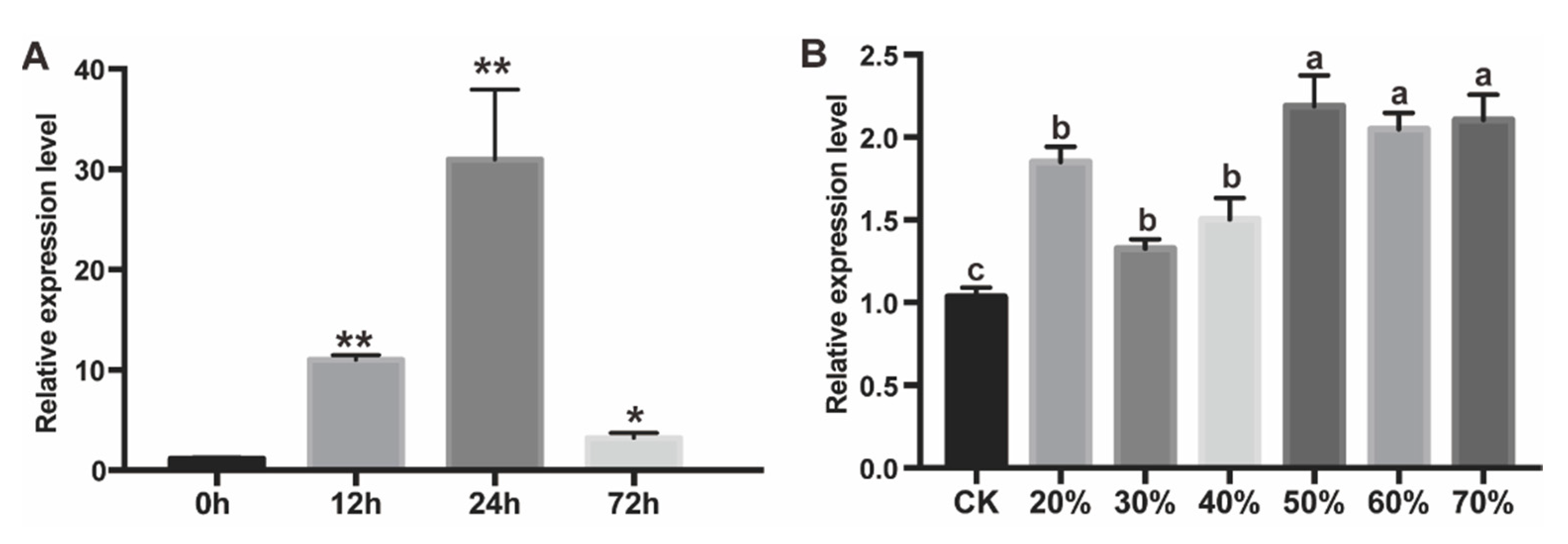
2.1. PtAOS1 Gene Expression and its Induction by Drought and JA Treatments
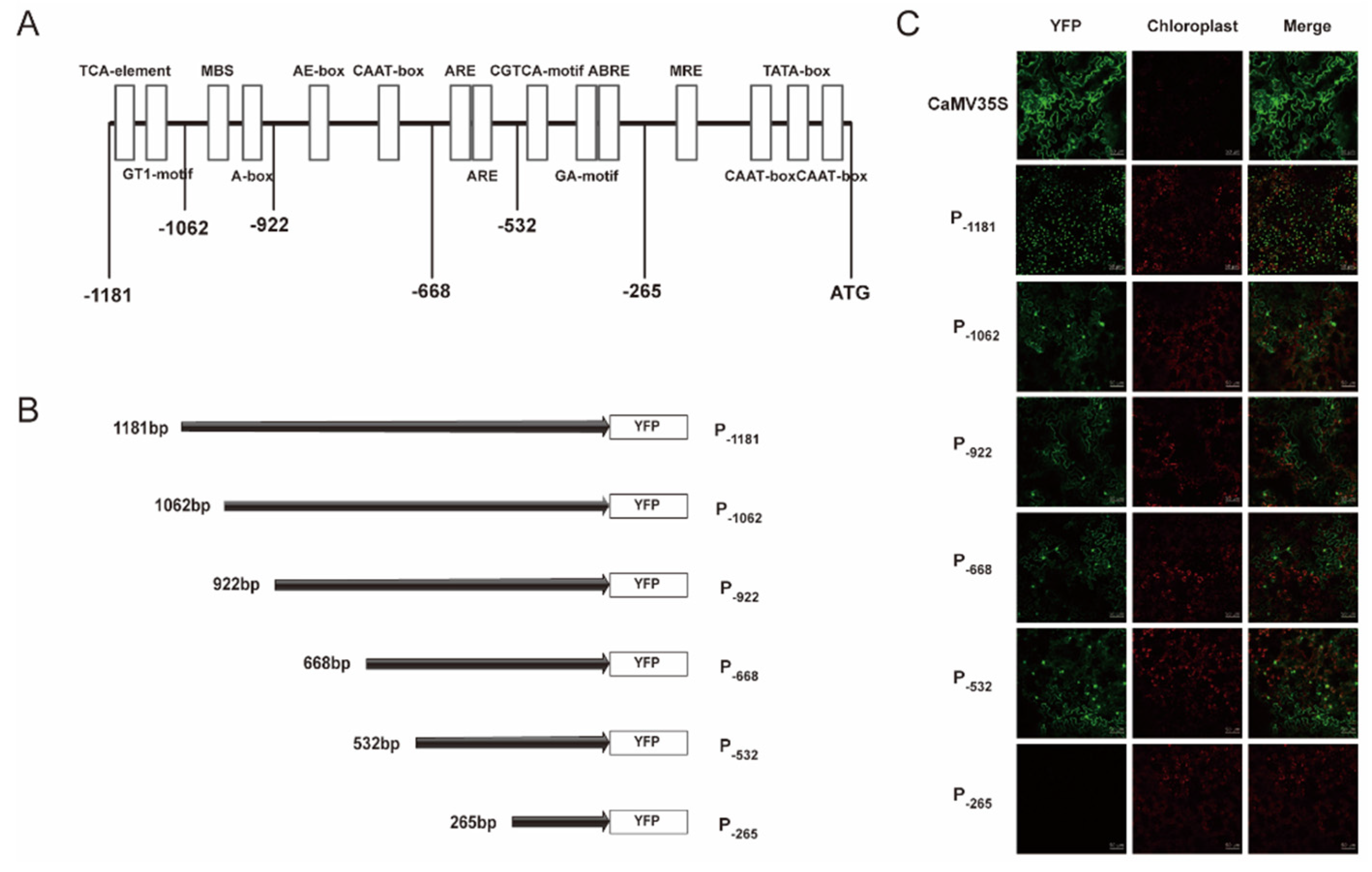
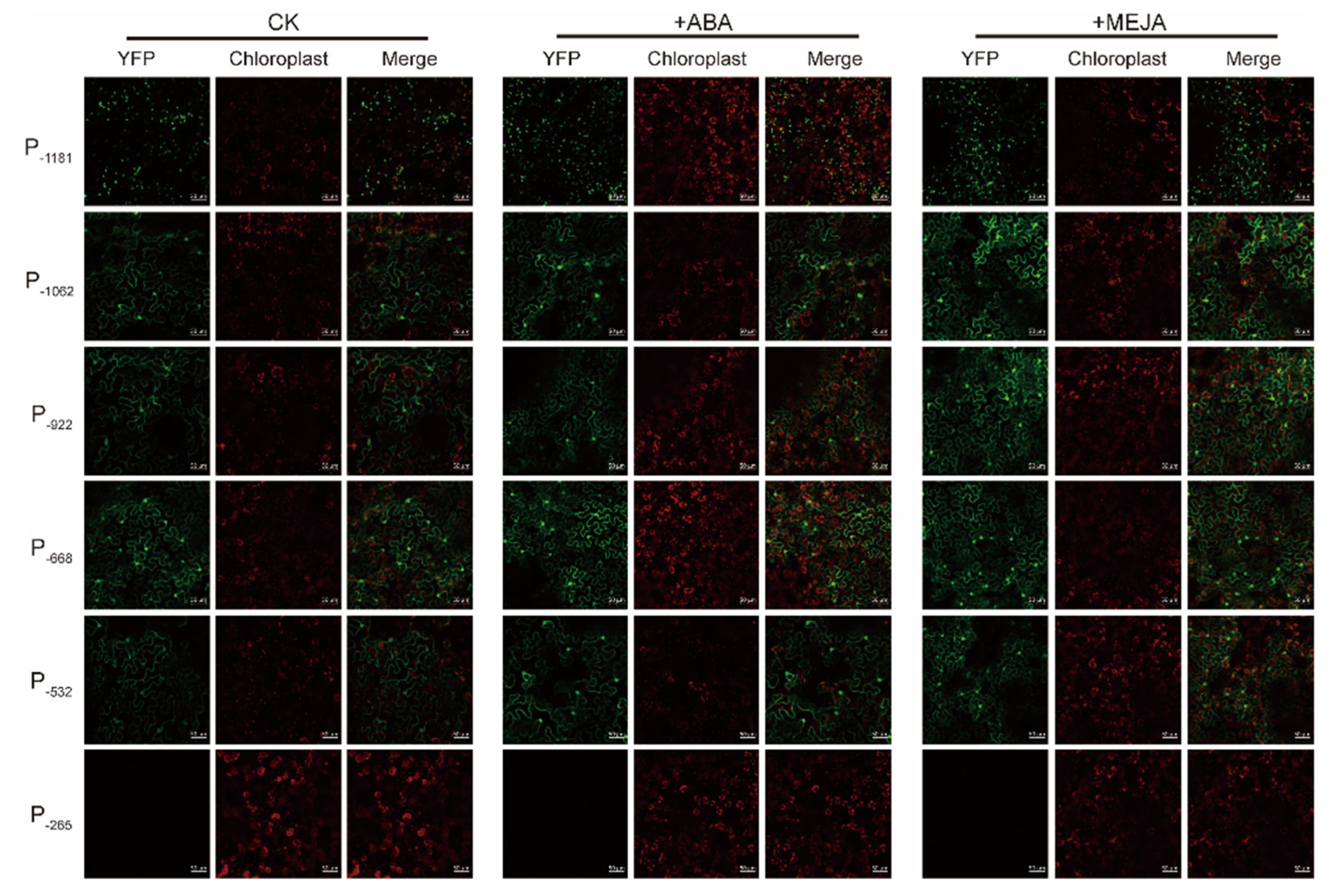
2.2. Characterization of PtAOS1 Promoter
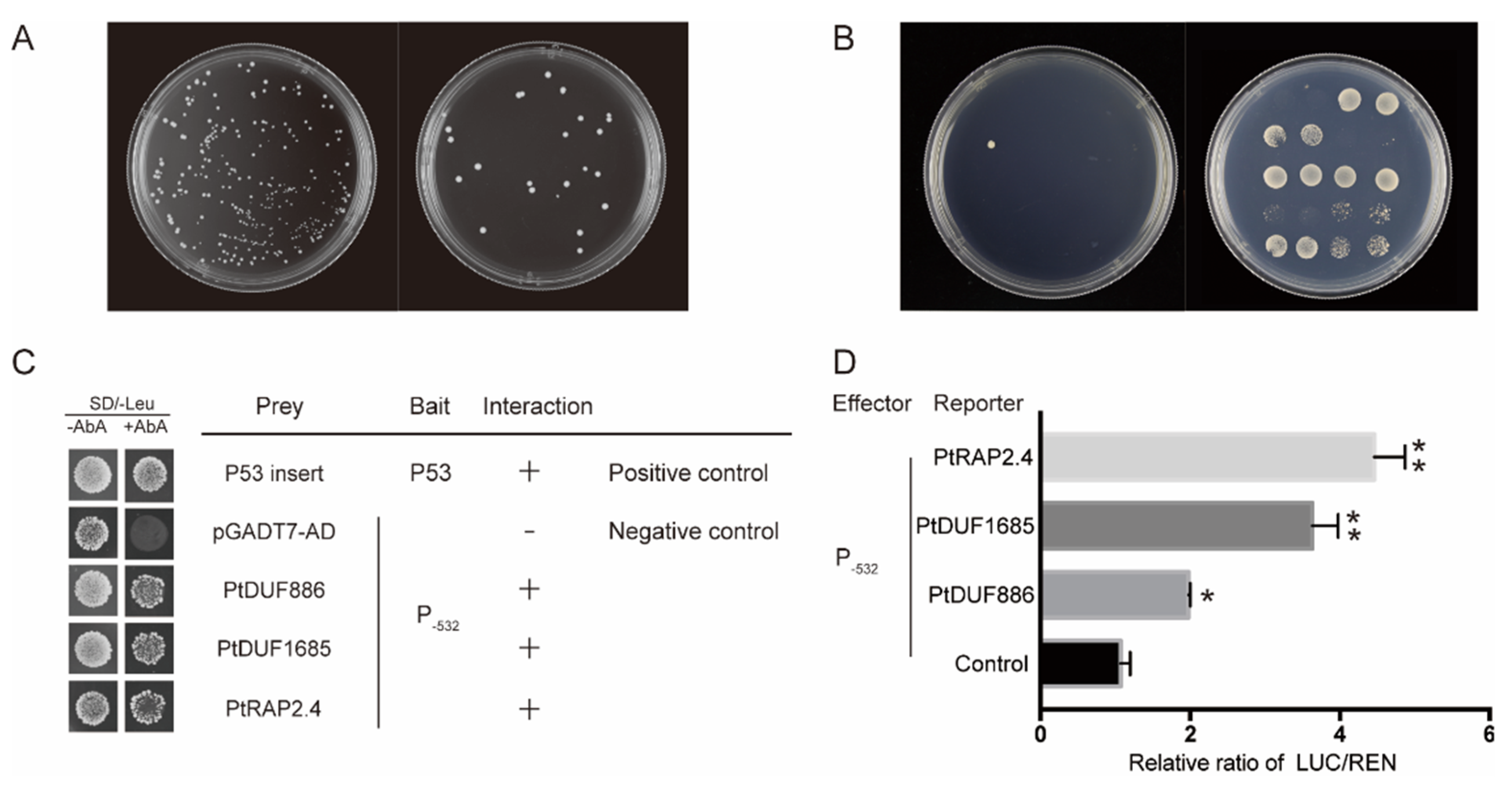
2.3. Proteins Recognizing PtAOS1 Promoter
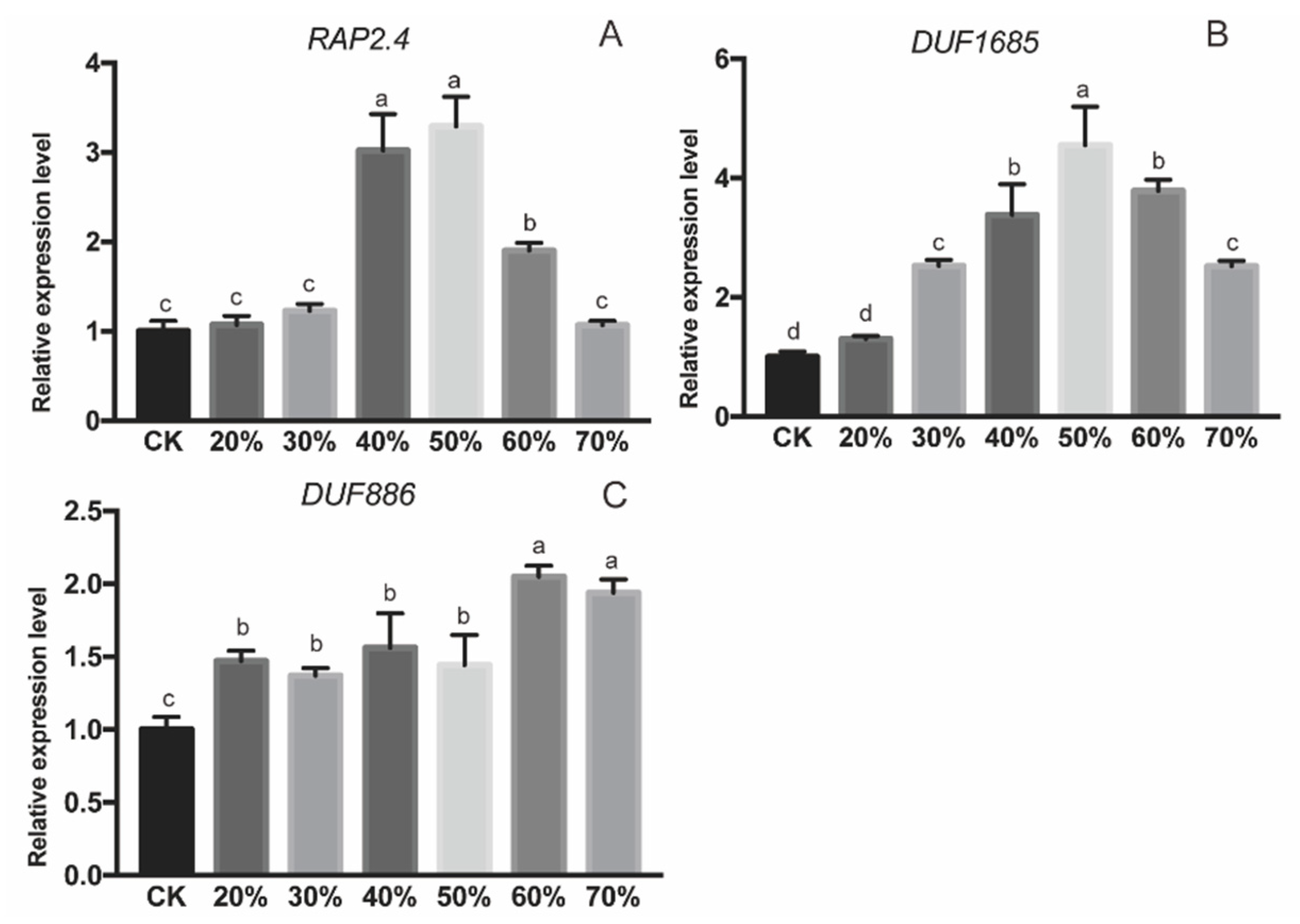
2.4. Expression Profiles of Three Interacting Proteins under Drought Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Cloning and Expression Pattern of PtAOS
4.3. Promoter Isolation and Vector Construction
4.4. Agrobacterium-Mediate Transient Expression in Tobacco
4.5. Construction and Screening of a cDNA Library
4.6. Dual Luciferase Assay
4.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Helder, H.; Miersch, O.; Vreugdenhil, D.; Sembdner, G. Occurrence of hydroxylated jasmonic acids in leaflets of Solanum demissum plants grown under long- and short-day conditions. Physiol. Plant 1993, 88, 647–653. [Google Scholar] [CrossRef]
- Falkenstein, E.; Groth, B.; Mithofer, A.; Weiler, E.W. Methyljasmonate and alpha-linolenic acid are potent inducers of tendril coiling. Planta 1991, 185, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef]
- Toyoda, K.; Kawanishi, Y.; Kawamoto, Y.; Kurihara, C.; Yamagishi, N.; Tamura, A.; Yoshikawa, N.; Inagaki, Y.; Ichinose, Y.; Shiraishi, T. Suppression of mRNAs for lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC) and 12-oxo-phytodienoic acid reductase (OPR) in pea reduces sensitivity to the phytotoxin coronatine and disease development by Mycosphaerella pinodes. J. Gen. Plant Pathol. 2013, 79, 321–334. [Google Scholar] [CrossRef]
- Naor, N.; Gurung, F.B.; Ozalvo, R.; Bucki, P.; Sanadhya, P.; Miyara, S.B. Tight regulation of allene oxide synthase (AOS) and allene oxide cyclase-3 (AOC3) promote Arabidopsis susceptibility to the root-knot nematode Meloidogyne javanica. Eur. J. Plant Pathol. 2018, 150, 149–165. [Google Scholar] [CrossRef]
- Wu, X.; Li, D.; Wang, J.; Shi, J.; Li, R.; Zhao, B.; Ren, S.; Guo, Y.D. Cloning and characterization of a cabbage BoAOS gene with enhanced drought tolerance. N. Z. J. Crop Hort. Sci. 2015, 43, 32–41. [Google Scholar] [CrossRef]
- Alam, M.M.; Nahar, K.; Hasanuzzaman, M.; Fujita, M. Exogenous jasmonic acid modulates the physiology, antioxidant defense and glyoxalase systems in imparting drought stress tolerance in different Brassica species. Plant Biotechnol. Rep. 2014, 8, 279–293. [Google Scholar] [CrossRef]
- Shan, C.; Zhou, Y.; Liu, M. Nitric oxide participates in the regulation of the ascorbate-glutathione cycle by exogenous jasmonic acid in the leaves of wheat seedlings under drought stress. Protoplasma 2015, 252, 1397–1405. [Google Scholar] [CrossRef]
- Mohamed, H.I.; Latif, H.H. Improvement of drought tolerance of soybean plants by using methyl jasmonate. Physiol. Mol. Biol. Plants 2017, 23, 545–556. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Pedranzani, H.; Racagni, G.; Alemano, S.; Miersch, O.; Ramírez, I.; Peña-Cortés, H.; Taleisnik, E.; Machado-Domenech, E.; Abdala, G. Salt tolerant tomato plants show increased levels of jasmonic acid. Plant Growth Regul. 2003, 41, 149–158. [Google Scholar] [CrossRef]
- Shahzad, A.N.; Pitann, B.; Ali, H.; Qayyum, M.F.; Fatima, A.; Bakhat, H.F. Maize Genotypes Differing in Salt Resistance Vary in Jasmonic Acid Accumulation During the First Phase of SaltStress. J. Agron. Crop. Sci. 2015, 201, 443–451. [Google Scholar] [CrossRef]
- Qiu, Z.; Guo, J.; Zhu, A.; Zhang, L.; Zhang, M. Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicol. Environ. Saf. 2014, 104, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Abouelsaad, I.; Renault, S. Enhanced oxidative stress in the jasmonic acid-deficient tomato mutant def-1 exposed to NaCl stress. J. Plant Physiol. 2018, 226, 136–144. [Google Scholar] [CrossRef]
- Laudert, D.; Weiler, E.W. Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 1998, 15, 675–684. [Google Scholar] [CrossRef]
- Song, W.C.; Brash, A.R. Purification of an allene oxide synthase and identification of the enzyme as a cytochrome P-450. Science 1991, 253, 781–784. [Google Scholar] [CrossRef]
- Laudert, D.; Schaller, F.; Weiler, E.W. Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta 2000, 211, 163–165. [Google Scholar] [CrossRef]
- Halitschke, R.; Ziegler, J.; Keinanen, M.; Baldwin, I.T. Silencing of hydroperoxide lyase and allene oxide synthase reveals substrate and defense signaling crosstalk in Nicotiana attenuata. Plant J. 2004, 40, 35–46. [Google Scholar] [CrossRef]
- Sivasankar, S.; Sheldrick, B.; Rothstein, S.J. Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 2000, 122, 1335–1342. [Google Scholar] [CrossRef]
- Pajerowska-Mukhtar, K.M.; Mukhtar, M.S.; Guex, N.; Halim, V.A.; Rosahl, S.; Somssich, I.E.; Gebhardt, C. Natural variation of potato allene oxide synthase 2 causes differential levels of jasmonates and pathogen resistance in Arabidopsis. Planta 2008, 228, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Guang, Y.L.; Li, J.W.; Wang, F.; Ahammed, G.J.; Yang, Y.X. The CYP74 Gene Family in Watermelon: Genome-Wide Identification and Expression Profiling Under Hormonal Stress and Root-Knot Nematode Infection. Agronomy 2019, 9, 872. [Google Scholar] [CrossRef]
- Mwenda, C.M.; Matsuki, A.; Nishimura, K.; Koeduka, T.; Matsui, K. Spatial expression of the Arabidopsis hydroperoxide lyase gene is controlled differently from that of the allene oxide synthase gene. J. Plant Interact. 2015, 10, 1–10. [Google Scholar] [CrossRef]
- Wang, H.; Gao, Z.J.; Liu, H.L.; Wu, J.J.; Yu, D.Y. Variation in GmAOS1 promoter is associated with soybean defense against insect attack. Euphytica 2014, 196, 365–374. [Google Scholar] [CrossRef]
- Bandara, P.K.; Takahashi, K.; Sato, M.; Matsuura, H.; Nabeta, K. Cloning and functional analysis of an allene oxide synthase in Physcomitrella patens. Biosci. Biotechnol. Biochem. 2009, 73, 2356–2359. [Google Scholar]
- Mei, C.; Qi, M.; Sheng, G.; Yang, Y. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 2006, 19, 1127–1137. [Google Scholar] [CrossRef]
- Zhao, X.; Lu, X.; Nie, Q.; Huang, C.; Xiao, Y.; Xie, S. Effect of Water Stress on Physiological Characteristics, JA Biosynthesis and Correlative Genes Expression in Citrus. Acta Agric. Univ. Jiangxiensis 2013, 35, 530–535. [Google Scholar]
- De Domenico, S.; Bonsegna, S.; Horres, R.; Pastor, V.; Taurino, M.; Poltronieri, P.; Imtiaz, M.; Kahl, G.; Flors, V.; Winter, P.; et al. Transcriptomic analysis of oxylipin biosynthesis genes and chemical profiling reveal an early induction of jasmonates in chickpea roots under drought stress. Plant Physiol. Biochem. 2012, 61, 115–122. [Google Scholar] [CrossRef]
- Wang, P.; Ma, L.L.; Li, Y.; Wang, S.A.; Li, L.F.; Yang, R.T. Transcriptome analysis reveals sunflower cytochrome P450 CYP93A1 responses to high salinity treatment at the seedling stage. Genes Genom. 2017, 39, 581–591. [Google Scholar] [CrossRef]
- Laudert, D.; Pfannschmidt, U.; Lottspeich, F.; Hollander-Czytko, H.; Weiler, E.W. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol. Biol. 1996, 31, 323–335. [Google Scholar] [CrossRef]
- Howe, G.A.; Lee, G.I.; Itoh, A.; Li, L.; DeRocher, A.E. Cytochrome P450-dependent metabolism of oxylipins in tomato. Cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol. 2000, 123, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Dumin, W.; Rostas, M.; Winefield, C. Identification and functional characterisation of an allene oxide synthase from grapevine (Vitis vinifera L. Sauvignon blanc). Mol. Biol. Rep. 2018, 45, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Harms, K.; Atzorn, R.; Brash, A.; Kühn, H.; Wasternack, C.; Willmitzer, L.; Pena-Cortés, H. Expression of a Flax Allene Oxide Synthase cDNA Leads to Increased Endogenous Jasmonic Acid (JA) Levels in Transgenic Potato Plants but Not to a Corresponding Activation of JA-Responding Genes. Plant Cell 1995, 7, 1645–1654. [Google Scholar] [CrossRef]
- Peng, Q.; Zhou, Y.; Liao, Y.; Zeng, L.; Xu, X.; Jia, Y.; Dong, F.; Li, J.; Tang, J.; Yang, Z. Functional Characterization of An Allene Oxide Synthase Involved in Biosynthesis of Jasmonic Acid and Its Influence on Metabolite Profiles and Ethylene Formation in Tea (Camellia sinensis) Flowers. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Liu, H.H.; Wang, Y.G.; Wang, S.P.; Li, H.J.; Xin, Q.G. Improved zinc tolerance of tobacco by transgenic expression of an allene oxide synthase gene from hexaploid wheat. Acta Physiol. Plant. 2014, 36, 2433–2440. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Novina, C.D.; Roy, A.L. Core promoters and transcriptional control. Trends Genet. 1996, 12, 351–355. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Freitas, E.O.; Melo, B.P.; Lourenco-Tessutti, I.T.; Arraes, F.B.M.; Amorim, R.M.; Lisei-de-Sa, M.E.; Costa, J.A.; Leite, A.G.B.; Faheem, M.; Ferreira, M.A.; et al. Identification and characterization of the GmRD26 soybean promoter in response to abiotic stresses: Potential tool for biotechnological application. BMC Biotechnol. 2019, 19, 79. [Google Scholar] [CrossRef]
- Stalberg, K.; Ellerstom, M.; Ezcurra, I.; Ablov, S.; Rask, L. Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 1996, 199, 515–519. [Google Scholar] [CrossRef]
- Baranowskij, N.; Frohberg, C.; Prat, S.; Willmitzer, L. A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 1994, 13, 5383–5392. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, B.; Davydov, O.; Knight, H.; Galon, Y.; Knight, M.R.; Fluhr, R.; Fromm, H. Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 2006, 18, 2733–2748. [Google Scholar] [CrossRef]
- Roy, S.; Choudhury, S.R.; Singh, S.K.; Das, K.P. Functional analysis of light-regulated promoter region of AtPollambda gene. Planta 2012, 235, 411–432. [Google Scholar] [CrossRef]
- Morishima, A. Identification of preferred binding sites of a light-inducible DNA-binding factor (MNF1) within 5′-upstream sequence of C4-type phosphoenolpyruvate carboxylase gene in maize. Plant Mol. Biol. 1998, 38, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Wei, R.; Li, Y.; Wang, R.; Tang, Y.; Wang, L.; Zhu, H.; Wang, Y.; Zhang, C. Molecular cloning and functional characterization of a seed-specific VvbetaVPE gene promoter from Vitis vinifera. Planta 2019, 250, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, J.; Lian, J.; Zhang, Y.; Zhu, L.; Zeng, D.L.; Guo, L.B.; Yu, L.; Xu, G.H.; Qian, Q. Functional characterization of OsHAK1 promoter in response to osmotic/drought stress by deletion analysis in transgenic rice. Plant Growth Regul. 2019, 88, 241–251. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, M.; Hu, J.; Wang, W.; Fu, X.; Liu, J.H. PtrABF of Poncirus trifoliata functions in dehydration tolerance by reducing stomatal density and maintaining reactive oxygen species homeostasis. J. Exp. Bot. 2015, 66, 5911–5927. [Google Scholar] [CrossRef]
- Rouster, J.; Leah, R.; Mundy, J.; Cameron-Mills, V. Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J. 1997, 11, 513–523. [Google Scholar] [CrossRef]
- Aviles-Arnaut, H.; Delano-Frier, J.P. Characterization of the tomato prosystemin promoter: Organ-specific expression, hormone specificity and methyl jasmonate responsiveness by deletion analysis in transgenic tobacco plants. J. Integr. Plant Biol. 2012, 54, 15–32. [Google Scholar] [CrossRef]
- Li, J.; Liang, D.; Li, M.; Ma, F. Light and abiotic stresses regulate the expression of GDP-L-galactose phosphorylase and levels of ascorbic acid in two kiwifruit genotypes via light-responsive and stress-inducible cis-elements in their promoters. Planta 2013, 238, 535–547. [Google Scholar] [CrossRef]
- Mukherjee, K.; Choudhury, A.R.; Gupta, B.; Gupta, S.; Sengupta, D.N. An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice. BMC Plant Biol. 2006, 6, 18. [Google Scholar] [CrossRef]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Fan, M.; Yang, M.; Zhao, J.; Zhang, W.; Su, Y.; Xiao, L.; Deng, H.; Xie, D. Injury Activates Ca2+ /Calmodulin-Dependent Phosphorylation of JAV1-JAZ8-WRKY51 Complex for Jasmonate Biosynthesis. Mol. Cell 2018, 70, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Shim, Y.; Kang, K.; An, G.; Paek, N.C. Rice DNA-Binding One Zinc Finger 24 (OsDOF24) Delays Leaf Senescence in a Jasmonate-Mediated Pathway. Plant Cell Physiol. 2019, 60, 2065–2076. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coggill, P.; Finn, R.D. DUFs: Families in search of function. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 1148–1152. [Google Scholar] [CrossRef]
- Li, L.H.; Lv, M.M.; Li, X.; Ye, T.Z.; He, X.; Rong, S.H.; Dong, Y.L.; Guan, Y.; Gao, X.L.; Zhu, J.Q.; et al. The Rice OsDUF810 Family: OsDUF810.7 May be Involved in the Tolerance to Salt and Drought. Mol. Biol. 2018, 52, 489–496. [Google Scholar] [CrossRef]
- Li, L.H.; Lv, M.M.; Zhao, L.; Ye, T.Z.; Xu, J.H.; Cai, L.J.; Xie, C.; Gao, X.L.; Huang, Z.J.; Zhu, J.Q.; et al. Molecular characterization and function analysis of the rice OsDUF829 family. Biotechnol. Biotechnol. Equip. 2018, 32, 550–557. [Google Scholar] [CrossRef]
- Wang, L.; Shen, R.; Chen, L.T.; Liu, Y.G. Characterization of a novel DUF1618 gene family in rice. J. Integr. Plant Biol. 2014, 56, 151–158. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Pre, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Vogel, M.O.; Moore, M.; Konig, K.; Pecher, P.; Alsharafa, K.; Lee, J.; Dietz, K.J. Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 2014, 26, 1151–1165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Qi, L.; Wang, A.; Ye, X.; Du, L.; Liang, H.; Xin, Z.; Zhang, Z. The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 2014, 12, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Van der Fits, L.; Memelink, J. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 2001, 25, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Zarei, A.; Korbes, A.P.; Younessi, P.; Montiel, G.; Champion, A.; Memelink, J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 2011, 75, 321–331. [Google Scholar] [CrossRef]
- Yu, Z.X.; Li, J.X.; Yang, C.Q.; Hu, W.L.; Wang, L.J.; Chen, X.Y. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 2012, 5, 353–365. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, J.; Liu, L.; Ma, X.; Li, F.; Tang, C.; Li, Z.; Lü, B.; Zhou, T.; Lian, X.; Chang, Y.; et al. Characterization of PtAOS1 Promoter and Three Novel Interacting Proteins Responding to Drought in Poncirus trifoliata. Int. J. Mol. Sci. 2020, 21, 4705. https://doi.org/10.3390/ijms21134705
Xiong J, Liu L, Ma X, Li F, Tang C, Li Z, Lü B, Zhou T, Lian X, Chang Y, et al. Characterization of PtAOS1 Promoter and Three Novel Interacting Proteins Responding to Drought in Poncirus trifoliata. International Journal of Molecular Sciences. 2020; 21(13):4705. https://doi.org/10.3390/ijms21134705
Chicago/Turabian StyleXiong, Jiang, Lian Liu, Xiaochuan Ma, Feifei Li, Chaolan Tang, Zehang Li, Biwen Lü, Tie Zhou, Xuefei Lian, Yuanyuan Chang, and et al. 2020. "Characterization of PtAOS1 Promoter and Three Novel Interacting Proteins Responding to Drought in Poncirus trifoliata" International Journal of Molecular Sciences 21, no. 13: 4705. https://doi.org/10.3390/ijms21134705
APA StyleXiong, J., Liu, L., Ma, X., Li, F., Tang, C., Li, Z., Lü, B., Zhou, T., Lian, X., Chang, Y., Tang, M., Xie, S., & Lu, X. (2020). Characterization of PtAOS1 Promoter and Three Novel Interacting Proteins Responding to Drought in Poncirus trifoliata. International Journal of Molecular Sciences, 21(13), 4705. https://doi.org/10.3390/ijms21134705




