Complex Characterization of Germline Large Genomic Rearrangements of the BRCA1 and BRCA2 Genes in High-Risk Breast Cancer Patients—Novel Variants from a Large National Center
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Patient Selection and Genotyping
4.2. Gene Dosage Analysis
4.3. Breakpoint Resolution of the LGRs
4.4. Haplotyping
4.5. RNA Isolation and RT-PCR
4.6. Allele Imbalance
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| LGR | Large genomic rearrangement |
| MLPA | Multiplex ligation-dependent probe amplification |
| NGS | Next-generation sequencing |
| QMPSF | Quantitative multiplex PCR of short fluorescent fragments |
References
- Karami, F.; Mehdipour, P. A Comprehensive Focus on Global Spectrum ofBRCA1andBRCA2Mutations in Breast Cancer. BioMed Res. Int. 2013, 2013, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Sluiter, M.; Van Rensburg, E.J. Large genomic rearrangements of the BRCA1 and BRCA2 genes: Review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res. Treat. 2010, 125, 325–349. [Google Scholar] [CrossRef]
- Ticha, I.; Kleibl, Z.; Stribrna, J.; Kotlas, J.; Zimovjanova, M.; Mateju, M.; Zikan, M.; Pohlreich, P. Screening for genomic rearrangements in BRCA1 and BRCA2 genes in Czech high-risk breast/ovarian cancer patients: High proportion of population specific alterations in BRCA1 gene. Breast Cancer Res. Treat. 2010, 124, 337–347. [Google Scholar] [CrossRef]
- Concolino, P.; Rizza, R.; Mignone, F.; Costella, A.; Guarino, D.; Carboni, I.; Capoluongo, E.; Santonocito, C.; Urbani, A.; Minucci, A. A comprehensive BRCA1/2 NGS pipeline for an immediate Copy Number Variation (CNV) detection in breast and ovarian cancer molecular diagnosis. Clin. Chim. Acta 2018, 480, 173–179. [Google Scholar] [CrossRef]
- Smith, M.J.; Urquhart, J.; Harkness, E.; Miles, E.K.; Bowers, N.L.; Byers, H.J.; Bulman, M.; Gokhale, C.; Wallace, A.; Newman, W.; et al. The Contribution of Whole Gene Deletions and Large Rearrangements to the Mutation Spectrum in Inherited Tumor Predisposing Syndromes. Hum. Mutat. 2016, 37, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Montagna, M.; Palma, M.D.; Menin, C.; Agata, S.; De Nicolo, A.; Chieco-Bianchi, L.; D’Andrea, E. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum. Mol. Genet. 2003, 12, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Fachal, L.; Blanco, A.; Santamarina, M.; Carracedo, Á.; Vega, A. Large Genomic Rearrangements of BRCA1 and BRCA2 among Patients Referred for Genetic Analysis in Galicia (NW Spain): Delimitation and Mechanism of Three Novel BRCA1 Rearrangements. PLoS ONE 2014, 9, e93306. [Google Scholar] [CrossRef]
- Preisler-Adams, S.; Schönbuchner, I.; Fiebig, B.; Welling, B.; Dworniczak, B.; Weber, B.H.F. Gross rearrangements in BRCA1 but not BRCA2 play a notable role in predisposition to breast and ovarian cancer in high-risk families of German origin. Cancer Genet. Cytogenet. 2006, 168, 44–49. [Google Scholar] [CrossRef]
- Tsaousis, G.N.; Papadopoulou, E.; Apessos, A.; Agiannitopoulos, K.; Pepe, G.; Kampouri, S.; Diamantopoulos, N.; Floros, T.; Iosifidou, R.; Katopodi, O.; et al. Analysis of hereditary cancer syndromes by using a panel of genes: Novel and multiple pathogenic mutations. BMC Cancer 2019, 19, 535. [Google Scholar] [CrossRef]
- Hansen, T.V.O.; Jønson, L.; Albrechtsen, A.; Andersen, M.K.; Ejlertsen, B.; Nielsen, F.C. Large BRCA1 and BRCA2 genomic rearrangements in Danish high risk breast-ovarian cancer families. Breast Cancer Res. Treat. 2009, 115, 315–323. [Google Scholar] [CrossRef]
- Rudnicka, H.; Dębniak, T.; Cybulski, C.; Huzarski, T.; Gronwald, J.; Lubinski, J.; Górski, B. Large BRCA1 and BRCA2 genomic rearrangements in Polish high-risk breast and ovarian cancer families. Mol. Biol. Rep. 2013, 40, 6619–6623. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, A.; Pinheiro, M.; Massena, L.; Santos, C.; Pinto, P.; Rocha, P.; Pinto, C.; Teixeira, M.R. Genomic characterization of two large Alu-mediated rearrangements of the BRCA1 gene. J. Hum. Genet. 2013, 58, 78–83. [Google Scholar] [CrossRef][Green Version]
- Del Valle, J.; Feliubadalo, L.; Nadal, M.; Teulé, A.; Miró, R.; Cuesta, R.; Tornero, E.; Menéndez, M.; Darder, E.; Brunet, J.; et al. Identification and comprehensive characterization of large genomic rearrangements in the BRCA1 and BRCA2 genes. Breast Cancer Res. Treat. 2009, 122, 733–743. [Google Scholar] [CrossRef]
- Rodríguez, M.; Torres, A.; Borràs, J.; Salvat, M.; Gumà, J.; Rodríguez-Balada, M. Large genomic rearrangements in mutation-negative BRCA families: A population-based study. Clin. Genet. 2010, 78, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Petrij-Bosch, A.; Peelen, T.; Van Vliet, M.; Van Eijk, R.; Olmer, R.; Drüsedau, M.; Hogervorst, F.B.; Hageman, S.; Arts, P.J.; Ligtenberg, M.J.; et al. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat. Genet. 1997, 17, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Agata, S.; Viel, A.; Della Puppa, L.; Cortesi, L.; Fersini, G.; Callegaro, M.; Palma, M.D.; Dolcetti, R.; Federico, M.; Venuta, S.; et al. Prevalence ofBRCA1 genomic rearrangements in a large cohort of Italian breast and breast/ovarian cancer families without detectableBRCA1 andBRCA2 point mutations. Genes Chromosom. Cancer 2006, 45, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Rebbeck, T.R.; Friebel, T.M.; Friedman, E.; Hamann, U.; Huo, D.; Kwong, A.; Olah, E.; Olopade, O.I.; Solano, A.R.; Teo, S.-H.; et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018, 39, 593–620. [Google Scholar] [CrossRef]
- Hömig-Hölzel, C.; Savola, S. Multiplex Ligation-dependent Probe Amplification (MLPA) in Tumor Diagnostics and Prognostics. Diagn. Mol. Pathol. 2012, 21, 189–206. [Google Scholar] [CrossRef]
- Ceulemans, S.; Van Der Ven, K.; Del Favero, J. Targeted Screening and Validation of Copy Number Variations. Adv. Struct. Saf. Stud. 2011, 838, 311–328. [Google Scholar] [CrossRef]
- Povysil, G.; Tzika, A.; Vogt, J.; Haunschmid, V.; Messiaen, L.; Zschocke, J.; Klambauer, G.; Hochreiter, S.; Wimmer, K. panelcn.MOPS: Copy-number detection in targeted NGS panel data for clinical diagnostics. Hum. Mutat. 2017, 38, 889–897. [Google Scholar] [CrossRef]
- Sen, S.K.; Han, K.; Wang, J.; Lee, J.; Wang, H.; Callinan, P.A.; Dyer, M.; Cordaux, R.; Liang, P.; Batzer, M.A. Human Genomic Deletions Mediated by Recombination between Alu Elements. Am. J. Hum. Genet. 2006, 79, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.M.; Lee, M.K.; Szabo, C.I.; Jerome, N.; McEuen, M.; Taylor, M.P.; Hood, L.; King, M.-C. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996, 6, 1029–1049. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Biswas, K.; Habib, L.A.; Kuznetsov, S.; Hamel, N.; Kirchhoff, T.; Wong, N.; Armel, S.; Chong, G.; Narod, S.A.; et al. Functional redundancy of exon 12 ofBRCA2revealed by a comprehensive analysis of the c.6853A>G (p.I2285V) variant. Hum. Mutat. 2009, 30, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Hogervorst, F.B.; Nederlof, P.M.; Gille, J.J.; McElgunn, C.J.; Grippeling, M.; Pruntel, R.; Regnerus, R.; van Welsem, T.; van Spaendonk, R.; Menko, F.H.; et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003, 3, 1449–1453. [Google Scholar]
- Vasickova, P.; Machackova, E.; Lukesova, M.; Damborsky, J.; Horky, O.; Pavlu, H.; Kuklova, J.; Kosinová, V.; Navrátilová, M.; Foretova, L. High occurrence of BRCA1 intragenic rearrangements in hereditary breast and ovarian cancer syndrome in the Czech Republic. BMC Med Genet. 2007, 8, 32. [Google Scholar] [CrossRef]
- Puget, N.; Sinilnikova, O.M.; Stoppa-Lyonnet, M.; Audoynaud, C.; Pages, S.; Lynch, H.T.; Goldgar, D.; Lenoir, G.M.; Mazoyer, S. An Alu-mediated 6-kb duplication in the BRCA1 gene: A new founder mutation? Am. J. Hum. Genet. 1999, 64, 300–302. [Google Scholar] [CrossRef]
- Engert, S.; Wappenschmidt, B.; Betz, B.; Kast, K.; Kutsche, M.; Hellebrand, H.; Goecke, T.O.; Kiechle, M.; Niederacher, D.; Schmutzler, R.K.; et al. MLPA screening in theBRCA1gene from 1,506 German hereditary breast cancer cases: Novel deletions, frequent involvement of exon 17, and occurrence in single early-onset cases. Hum. Mutat. 2008, 29, 948–958. [Google Scholar] [CrossRef]
- Walsh, T.; Casadei, S.; Coats, K.H.; Swisher, E.; Stray, S.M.; Higgins, J.; Roach, K.C.; Mandell, J.; Lee, M.K.; Ciernikova, S.; et al. Spectrum of Mutations in BRCA1, BRCA2, CHEK2, and TP53 in Families at High Risk of Breast Cancer. JAMA 2006, 295, 1379. [Google Scholar] [CrossRef]
- Ratajska, M.; Brozek, I.; Senkus-Konefka, E.; Jassem, J.; Stepnowska, M.; Palomba, G.; Pisano, M.; Casula, M.; Palmieri, G.; Borg, A.; et al. BRCA1 and BRCA2 point mutations and large rearrangements in breast and ovarian cancer families in Northern Poland. Oncol. Rep. 2008, 19, 263–268. [Google Scholar] [CrossRef]
- Belogianni, I.; Apessos, A.; Mihalatos, M.; Razis, E.; Labropoulos, S.; Petounis, A.; Gaki, V.; Keramopoulos, A.; Pandis, N.; Kyriacou, K.; et al. Characterization of a novel large deletion and single point mutations in the BRCA1 gene in a Greek cohort of families with suspected hereditary breast cancer. BMC Cancer 2004, 4, 61. [Google Scholar] [CrossRef] [PubMed]
- Neuhausen, S.L.; Mazoyer, S.; Friedman, L.; Stratton, M.; Offit, K.; Caligo, A.; Tomlinson, G.; Cannon-Albright, L.; Bishop, T.; Kelsell, D.; et al. Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61 families: Results of an international study. Am. J. Hum. Genet. 1996, 58, 271–280. [Google Scholar]
- Puget, N.; Gad, S.; Perrin-Vidoz, L.; Sinilnikova, O.M.; Stoppa-Lyonnet, M.; Lenoir, G.M.; Mazoyer, S. Distinct BRCA1 Rearrangements Involving the BRCA1 Pseudogene Suggest the Existence of a Recombination Hot Spot. Am. J. Hum. Genet. 2002, 70, 858–865. [Google Scholar] [CrossRef]
- Van Der Looij, M.; Szabo, C.; Besznyak, I.; Liszka, G.; Csokay, B.; Pulay, T.; Toth, J.; Devilee, P.; King, M.-C.; Olah, E. Prevalence of founderBRCA1 andBRCA2 mutations among breast and ovarian cancer patients in hungary. Int. J. Cancer 2000, 86, 737–740. [Google Scholar] [CrossRef]
- Liu, P.; Carvalho, C.M.; Hastings, P.J.; Lupski, J.R. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 2012, 22, 211–220. [Google Scholar] [CrossRef]
- Kowalczykowski, S.C. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2015, 7, a016410. [Google Scholar] [CrossRef]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef]
- Sinha, S.; Villarreal, D.; Shim, E.Y.; Lee, S.E. Risky business: Microhomology-mediated end joining. Mutat. Res. Mol. Mech. Mutagen. 2016, 788, 17–24. [Google Scholar] [CrossRef]
- Sfeir, A.; Symington, L. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem. Sci. 2015, 40, 701–714. [Google Scholar] [CrossRef]
- Pannunzio, N.R.; Li, S.; Watanabe, G.; Lieber, M.R. Non-homologous end joining often uses microhomology: Implications for alternative end joining. DNA Repair 2014, 17, 74–80. [Google Scholar] [CrossRef]
- Lieber, M.R. The Mechanism of Human Nonhomologous DNA End Joining. J. Biol. Chem. 2007, 283, 1–5. [Google Scholar] [CrossRef]
- Zhang, F.; Khajavi, M.; Connolly, A.; Towne, C.F.; Batish, S.D.; Lupski, J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009, 41, 849–853. [Google Scholar] [CrossRef]
- Mazoyer, S. The Exon 13 Duplication in the BRCA1 Gene Is a Founder Mutation Present in Geographically Diverse Populations. Am. J. Hum. Genet. 2000, 67, 207–212. [Google Scholar] [CrossRef]
- Cerutti, R.; Sahnane, N.; Carnevali, I.; Furlan, D.; Tibiletti, M.G.; Chiaravalli, A.M.; Capella, C. Identification of the first case of germline duplication of BRCA1 exon 13 in an Italian family. Fam. Cancer 2009, 9, 275–282. [Google Scholar] [CrossRef]
- Kremeyer, B.; Soller, M.; Lagerstedt, K.; Maguire, P.; Mazoyer, S.; Nordling, M.; Wahlström, J.; Lindblom, A.; Lagerstedt-Robinson, K. The BRCA1 exon 13 duplication in the Swedish population. Fam. Cancer 2005, 4, 191–194. [Google Scholar] [CrossRef]
- Perrin-Vidoz, L.; Sinilnikova, O.M.; Stoppa-Lyonnet, M.; Lenoir, G.M.; Mazoyer, S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum. Mol. Genet. 2002, 11, 2805–2814. [Google Scholar] [CrossRef] [PubMed]
- Chehade, R.; Pettapiece-Phillips, R.; Salmena, L.; Kotlyar, M.; Jurisica, I.; Narod, S.A.; Akbari, M.R.; Kotsopoulos, J. Reduced BRCA1 transcript levels in freshly isolated blood leukocytes from BRCA1 mutation carriers is mutation specific. Breast Cancer Res. 2016, 18, 87. [Google Scholar] [CrossRef] [PubMed]
- Gad, S.; Bieche, I.; Barrois, M.; Casilli, F.; Pagès-Berhouet, S.; Dehainault, C.; Gauthier-Villars, M.; Bensimon, A.; Aurias, A.; Lidereau, R.; et al. Characterisation of a 161 kb deletion extending from the NBR1 to the BRCA1 genes in a French breast-ovarian cancer family. Hum. Mutat. 2003, 21, 654. [Google Scholar] [CrossRef]
- Xiao, Z.-D.; Liu, X.; Zhuang, L.; Gan, B. NBR2: A former junk gene emerges as a key player in tumor suppression. Mol. Cell. Oncol. 2016, 3, e1187322. [Google Scholar] [CrossRef] [PubMed]
- Papp, J.; Kovacs, M.; Mátrai, Z.; Orosz, E.; Kásler, M.; Børresen-Dale, A.-L.; Olah, E. Contribution of APC and MUTYH mutations to familial adenomatous polyposis susceptibility in Hungary. Fam. Cancer 2016, 15, 85–97. [Google Scholar] [CrossRef]
- Sequence Variant Nomenclature. Available online: http://varnomen.hgvs.org (accessed on 29 June 2020).
- Global Variome Shared LOVD, BRCA1. Available online: https://databases.lovd.nl/shared/genes/BRCA1 (accessed on 29 June 2020).
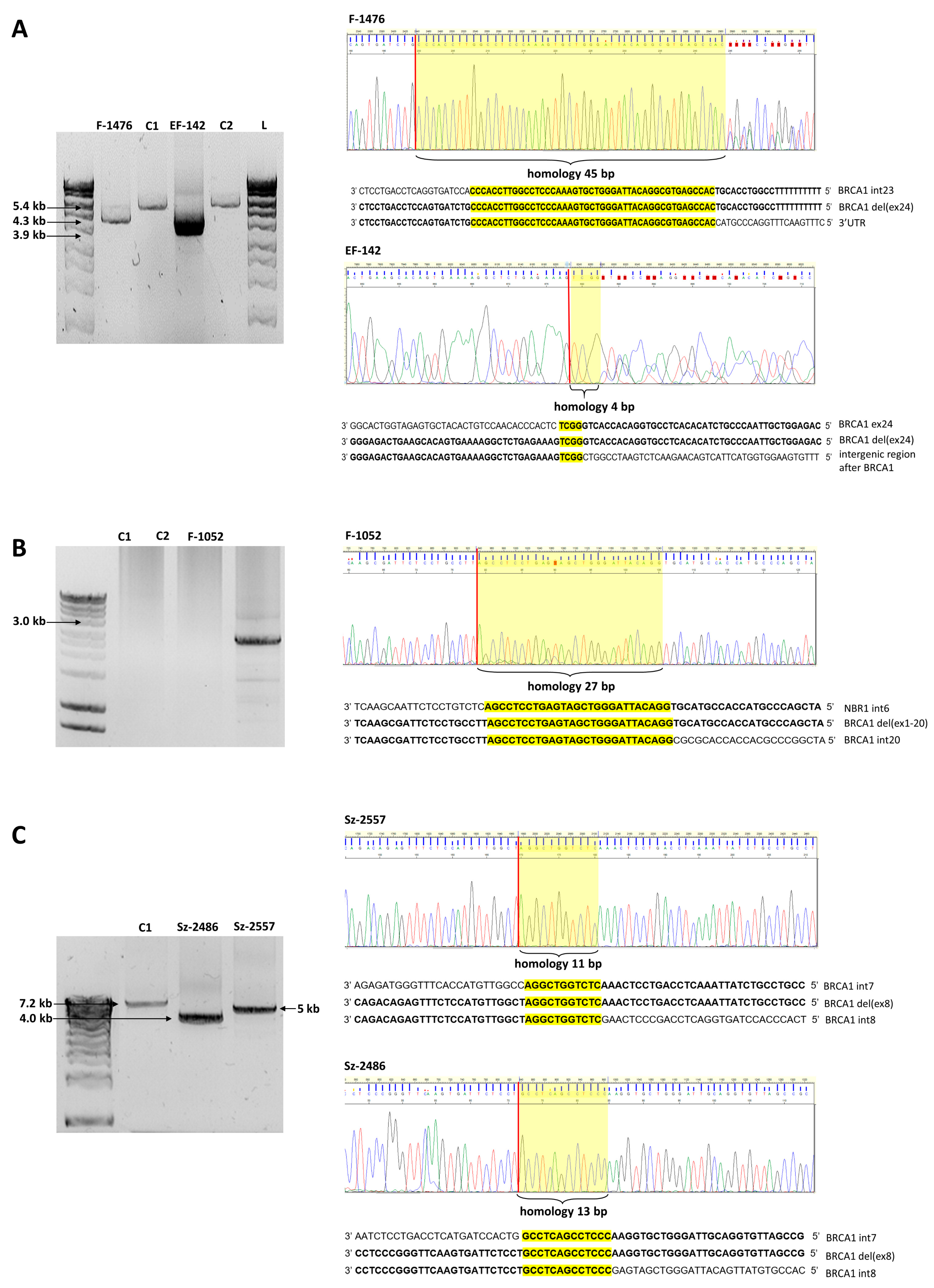
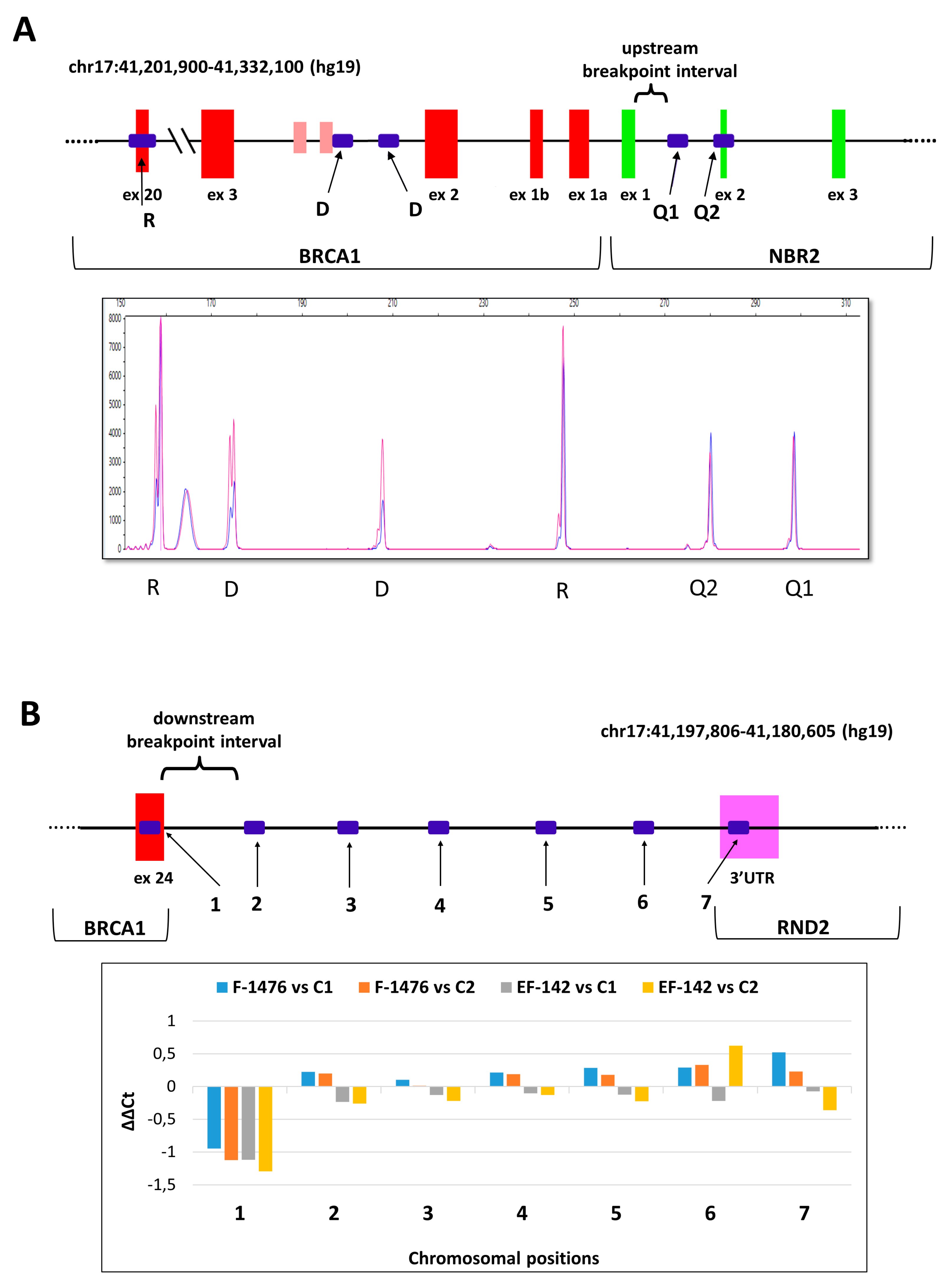
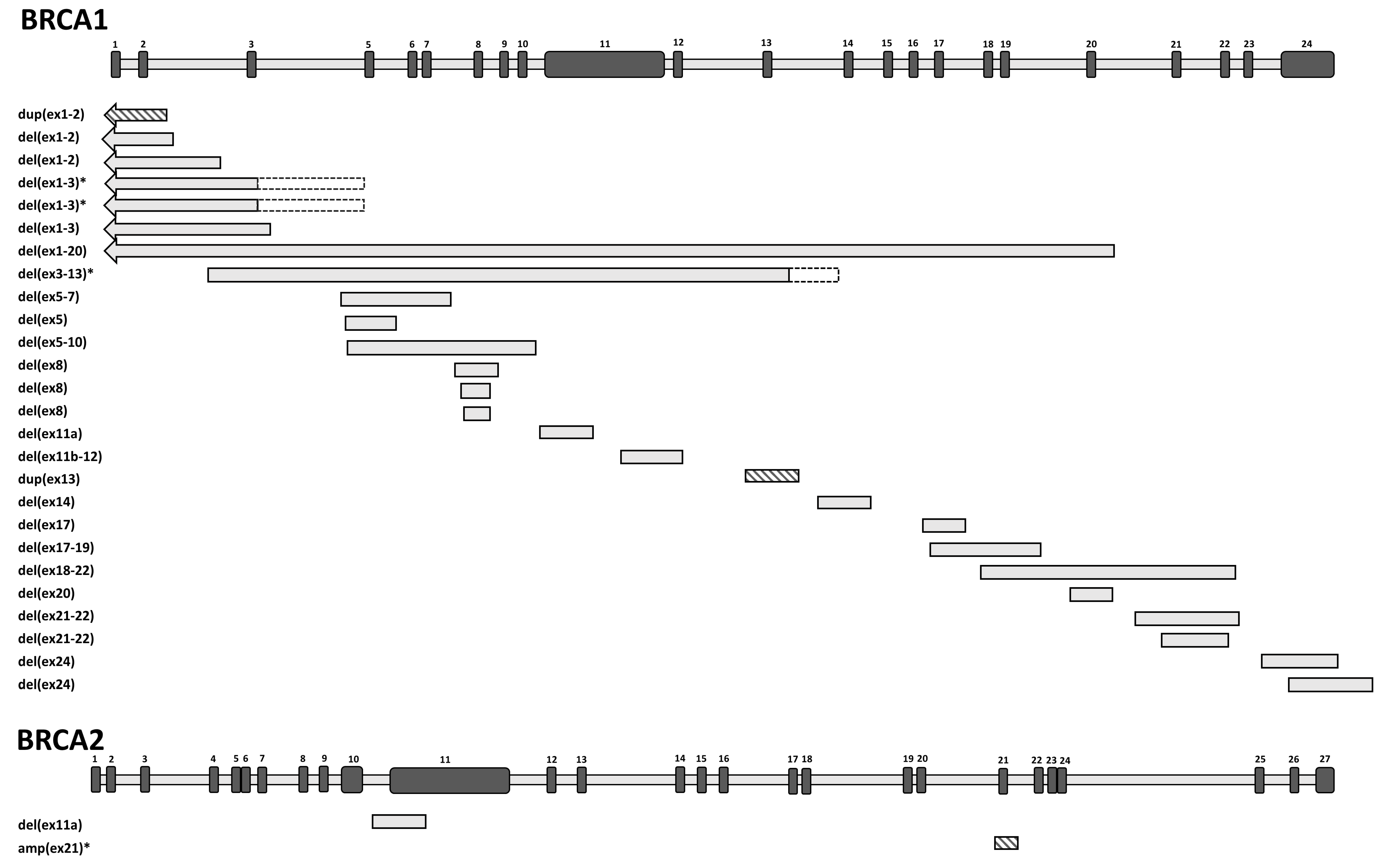
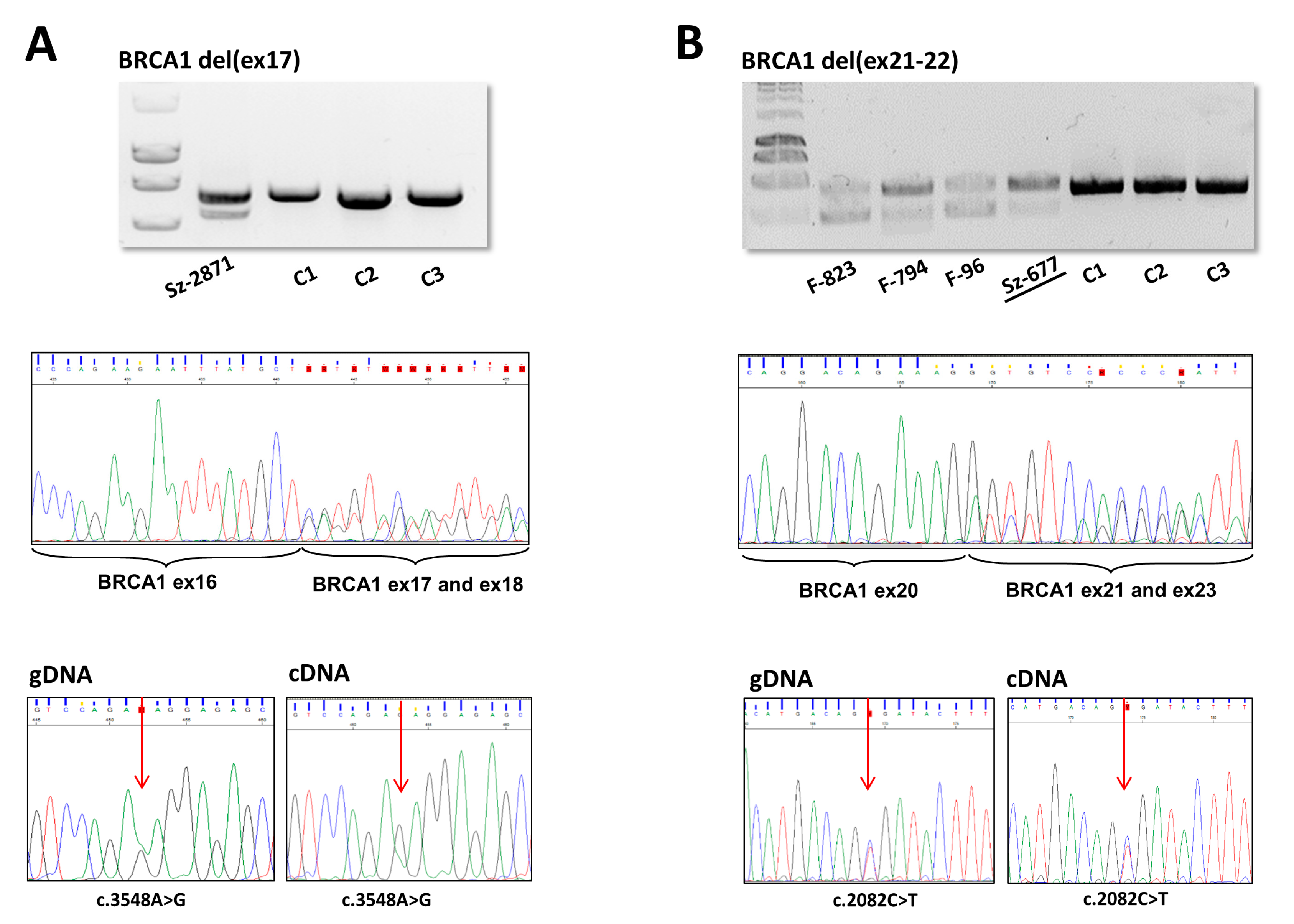
| Number of Probands | Gene | Running Name | CNV | HGVS Name | RNA | Protein | Reference | Upstream Breakpoint | Downstream Breakpoint | Homology | Inferred Rearrangement Mechanism |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | BRCA1 | BRCA1 dup(ex1-2) | dup | NG_005905.2:g.90060_97318dup | NA | NA | [7] | AluY | AluYk4 | 48 bp | NAHR |
| 1 | BRCA1 | BRCA1 del(ex1-2) | del | NG_005905.2:g.59988_96300del | NA | NA | NA | PsiBRCA1 (AluSp) | BRCA1 intron 2 (AluSp) | 15 bp | NAHR |
| 1 | BRCA1 | BRCA1 del(ex1-2) | del | NG_005905.2:g.59885_96193del | NA | NA | NA | PsiBRCA1 | BRCA1 intron 2 | 62 bp | NAHR |
| 1 | BRCA1 | BRCA1 del(ex1-3) | del | NG_005905.2:g.(24943_26402)_(102259_111450)del | NA | NA | NA | NA | NA | NA | NA |
| 1 | BRCA1 | BRCA1 del(ex1-3) | del | NG_005905.2:g.(88971_92304)_(102259_111450)del | NA | NA | NA | NA | NA | NA | NA |
| 1 | BRCA1 | BRCA1 del(ex1-3) | del | NG_005905.2:g.84958_106171del | NA | NA | NA | AluY | AluY | 4 bp | MMEJ/SSA |
| 1 | BRCA1 | BRCA1 del(ex1-20) | del | NG_005905.2:g.33502_166230del | NA | NA | NA | AluSx | AluSg | 27 bp | MMEJ/SSA |
| 1 | BRCA1 | BRCA1 del(ex3-13) | del | LRG_292t1:c.(81-2037_81-1)_(4357+1_4358-1)del | NA | NA | NA | NA | NA | NA | NA |
| 1 | BRCA1 | BRCA1 del(ex5) | del | LRG_292t1:c.135-30_212+136del | NA | NA | [8] | Non-Alu | Non-Alu | 9 bp | NHEJ |
| 1 | BRCA1 | BRCA1 del(ex5-7) | del | LRG_292t1:c.135-1004_441+1608del | NA | NA | [8] | AluSz6 | AluSc5 | 15 bp | MMEJ/SSA |
| 1 | BRCA1 | BRCA1 del(ex5-10) | del | LRG_292t1:c.135-4505_670+361delins35 | LRG_292t1:r.135_670del | p.(Lys45AsnfsTer3) | [3] | AluSx/AluY | AluY/AluJb | 10 bp/29 bp | FoSTeS |
| 2 | BRCA1 | BRCA1 del(ex8) | del | LRG_292t1:c.441+1521_547+392del | LRG_292t1:r.442_547del | p.(Gln148AspfsTer50) | NA | AluSc5 | AluSp | 13 bp | MMEJ/SSA |
| 2 | BRCA1 | BRCA1 del(ex8) | del | LRG_292t1:c.442-1102_547+252del | NA | NA | [24] | AluSx | AluSp | 26 bp | MMEJ/SSA |
| 1 | BRCA1 | BRCA1 del(ex8) | del | LRG_292t1:c.442-1830_547+295del | NA | NA | NA | AluSz | AluSp | 11 bp | MMEJ/SSA |
| 1 | BRCA1 | BRCA1 del(ex11a) | del | LRG_292t1:c.671-216_901del | NA | NA | NA | non-Alu | non-Alu | 4 bp | NHEJ |
| 1 | BRCA1 | BRCA1 del(ex11b-12) | del | LRG_292t1:c.1644_4185+3618del | NA | NA | [25] | non-Alu | L1 | 3 bp | NHEJ |
| 8 | BRCA1 | BRCA1 dup(ex13) | dup | LRG_292t1:c.4186-1787_4357+4122dup | NA | NA | [26] | AluSx | AluSx1 | 23 bp | unknown |
| 1 | BRCA1 | BRCA1 del(ex14) | del | LRG_292t1:c.4357+1661_4485-338del | NA | NA | NA | AluJo | AluSx1 | 10 bp | MMEJ/SSA |
| 4 | BRCA1 | BRCA1 del(ex17) | del | LRG_292t1:c.4986+726_5074+84del | LRG_292t1:r.4987_5074del | p.(Val1665SerfsTer7) | [27,28] | AluSp | AluSc | 10 bp | MMEJ/SSA |
| 1 | BRCA1 | BRCA1 del(ex17-19) | del | LRG_292t1:c.4987-365_5194-484del | NA | NA | [29] | AluY | AluY | 43 bp | MMEJ/SSA |
| 1 | BRCA1 | BRCA1 del(ex20) | del | LRG_292t1:c.5213_5278-2753delinsA † | NA | NA | [30] | non-Alu | AluSp | no | NHEJ |
| 2 | BRCA1 | BRCA1 del(ex18-22) | del | LRG_292t1:c.5075-1135_5406+346del | LRG_292t1:r.5075_5406del | p.(Asp1692GlyfsTer26) | [3] | AluY | AluSz | 7 bp | MMEJ/SSA |
| 9 | BRCA1 | BRCA1 del(ex21-22) | del | LRG_292t1:c.5278-492_5407-128delins236 | LRG_292t1:r.5278_5406del | p.(Ile1760_Thr1802) | [25] | non-Alu/AluSx | AluSx/AluJb | 26 bp | FoSTeS |
| 1 | BRCA1 | BRCA1 del(ex21-22) | del | LRG_292t1:c.5277+2114_5407-689del | LRG_292t1:r.5278_5406del | p.(Ile1760_Thr1802) | NA | AluSq2 | AluSc | 8 bp | MMEJ/SSA |
| 1 | BRCA1 | BRCA1 del(ex24) | del | LRG_292t1:c.5506_*1383+36del | NA | NA | NA | non-Alu | non-Alu | 4 bp | NHEJ |
| 1 | BRCA1 | BRCA1 del(ex24) | del | LRG_292t1:c.5468-364_*749del | NA | NA | NA | AluSx | AluSx1 | 45 bp | MMEJ/SSA |
| 1 | BRCA2 | BRCA2 del(ex11a) | del | LRG_293t1:c.1910-92_3888del | NA | NA | NA | non-Alu | non-Alu | no | NHEJ |
| 1 | BRCA2 | BRCA2 amp(ex21) | amp | LRG_293t1:c.(8633-70_8633-1)_ (8754+78_8754+122)amp | NA | NA | NA | NA | NA | NA | unknown |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bozsik, A.; Pócza, T.; Papp, J.; Vaszkó, T.; Butz, H.; Patócs, A.; Oláh, E. Complex Characterization of Germline Large Genomic Rearrangements of the BRCA1 and BRCA2 Genes in High-Risk Breast Cancer Patients—Novel Variants from a Large National Center. Int. J. Mol. Sci. 2020, 21, 4650. https://doi.org/10.3390/ijms21134650
Bozsik A, Pócza T, Papp J, Vaszkó T, Butz H, Patócs A, Oláh E. Complex Characterization of Germline Large Genomic Rearrangements of the BRCA1 and BRCA2 Genes in High-Risk Breast Cancer Patients—Novel Variants from a Large National Center. International Journal of Molecular Sciences. 2020; 21(13):4650. https://doi.org/10.3390/ijms21134650
Chicago/Turabian StyleBozsik, Anikó, Tímea Pócza, János Papp, Tibor Vaszkó, Henriett Butz, Attila Patócs, and Edit Oláh. 2020. "Complex Characterization of Germline Large Genomic Rearrangements of the BRCA1 and BRCA2 Genes in High-Risk Breast Cancer Patients—Novel Variants from a Large National Center" International Journal of Molecular Sciences 21, no. 13: 4650. https://doi.org/10.3390/ijms21134650
APA StyleBozsik, A., Pócza, T., Papp, J., Vaszkó, T., Butz, H., Patócs, A., & Oláh, E. (2020). Complex Characterization of Germline Large Genomic Rearrangements of the BRCA1 and BRCA2 Genes in High-Risk Breast Cancer Patients—Novel Variants from a Large National Center. International Journal of Molecular Sciences, 21(13), 4650. https://doi.org/10.3390/ijms21134650





