HLA-G: A New Immune Checkpoint in Cancer?
Abstract
1. Introduction
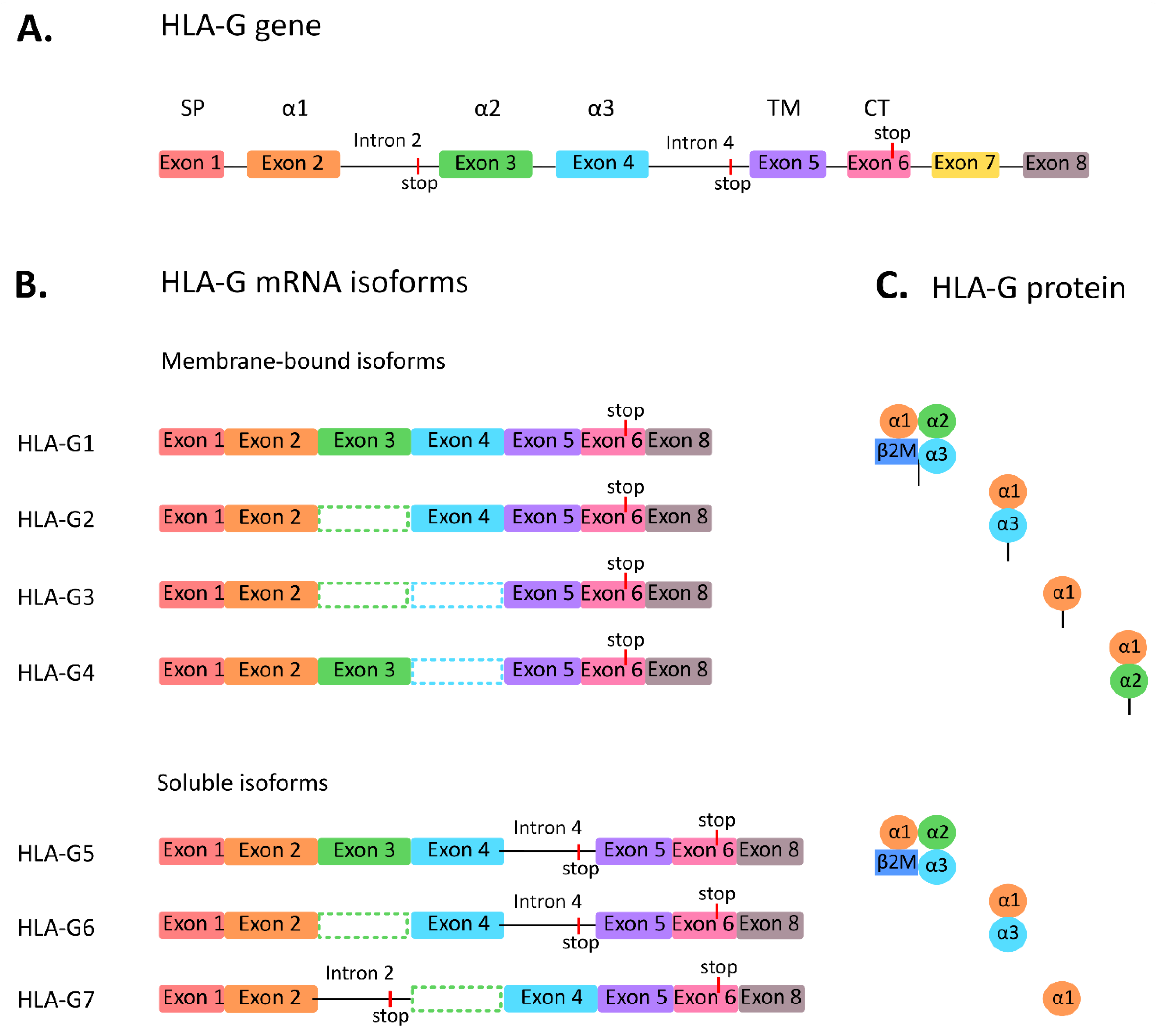
2. HLA-G
3. Receptors of HLA-G
4. Expression of HLA-G in Cancer
5. Function of HLA-G in Cancer
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kovats, S.; Main, E.K.; Librach, C.; Stubblebine, M.; Fisher, S.J.; DeMars, R. A class I antigen, HLA-G, expressed in human trophoblasts. Science 1990, 248, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Rouas-Freiss, N.; Goncalves, R.M.; Menier, C.; Dausset, J.; Carosella, E.D. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc. Natl. Acad. Sci. USA 1997, 94, 11520–11525. [Google Scholar] [CrossRef] [PubMed]
- Shih, K.; Arkenau, H.T.; Infante, J.R. Clinical impact of checkpoint inhibitors as novel cancer therapies. Drugs 2014, 74, 1993–2013. [Google Scholar] [CrossRef] [PubMed]
- Tundo, G.R.; Sbardella, D.; Lacal, P.M.; Graziani, G.; Marini, S. On the Horizon: Targeting Next-Generation Immune Checkpoints for Cancer Treatment. Chemotherapy 2019, 64, 62–80. [Google Scholar] [CrossRef] [PubMed]
- Carosella, E.D.; Rouas-Freiss, N.; Tronik-Le Roux, D.; Moreau, P.; LeMaoult, J. HLA-G: An Immune Checkpoint Molecule. Adv. Immunol. 2015, 127, 33–144. [Google Scholar] [CrossRef]
- Carosella, E.D.; Moreau, P.; Lemaoult, J.; Rouas-Freiss, N. HLA-G: From biology to clinical benefits. Trends Immunol. 2008, 29, 125–132. [Google Scholar] [CrossRef]
- Ishitani, A.; Geraghty, D.E. Alternative splicing of HLA-G transcripts yields proteins with primary structures resembling both class I and class II antigens. Proc. Natl. Acad. Sci. USA 1992, 89, 3947–3951. [Google Scholar] [CrossRef]
- Paul, P.; Cabestre, F.A.; Ibrahim, E.C.; Lefebvre, S.; Khalil-Daher, I.; Vazeux, G.; Quiles, R.M.; Bermond, F.; Dausset, J.; Carosella, E.D. Identification of HLA-G7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6, and -G7 transcripts in human transfected cells. Hum. Immunol. 2000, 61, 1138–1149. [Google Scholar] [CrossRef]
- Swets, M.; Seneby, L.; Boot, A.; van Wezel, T.; Gelderblom, H.; van de Velde, C.J.; van den Elsen, P.J.; Kuppen, P.J. Promoter methylation and mRNA expression of HLA-G in relation to HLA-G protein expression in colorectal cancer. Hum. Immunol. 2016, 77, 764–772. [Google Scholar] [CrossRef]
- Tronik-Le Roux, D.; Renard, J.; Verine, J.; Renault, V.; Tubacher, E.; LeMaoult, J.; Rouas-Freiss, N.; Deleuze, J.F.; Desgrandschamps, F.; Carosella, E.D. Novel landscape of HLA-G isoforms expressed in clear cell renal cell carcinoma patients. Mol. Oncol. 2017, 11, 1561–1578. [Google Scholar] [CrossRef]
- Lin, A.; Yan, W.H. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front. Immunol. 2018, 9, 2164. [Google Scholar] [CrossRef] [PubMed]
- Kuroki, K.; Matsubara, H.; Kanda, R.; Miyashita, N.; Shiroishi, M.; Fukunaga, Y.; Kamishikiryo, J.; Fukunaga, A.; Fukuhara, H.; Hirose, K.; et al. Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms. J. Immunol. 2019, 203, 3386–3394. [Google Scholar] [CrossRef] [PubMed]
- Clements, C.S.; Kjer-Nielsen, L.; Kostenko, L.; Hoare, H.L.; Dunstone, M.A.; Moses, E.; Freed, K.; Brooks, A.G.; Rossjohn, J.; McCluskey, J. Crystal structure of HLA-G: A nonclassical MHC class I molecule expressed at the fetal-maternal interface. Proc. Natl. Acad. Sci. USA 2005, 102, 3360–3365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zheng, J.; Kang, X.; Deng, M.; Lu, Z.; Kim, J.; Zhang, C. Inhibitory leukocyte immunoglobulin-like receptors in cancer development. Sci. China Life Sci. 2015, 58, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Rouas-Freiss, N.; Moreau, P.; LeMaoult, J.; Carosella, E.D. The dual role of HLA-G in cancer. J. Immunol. Res. 2014, 2014, 359748. [Google Scholar] [CrossRef] [PubMed]
- Baudhuin, J.; Migraine, J.; Faivre, V.; Loumagne, L.; Lukaszewicz, A.C.; Payen, D.; Favier, B. Exocytosis acts as a modulator of the ILT4-mediated inhibition of neutrophil functions. Proc. Natl. Acad. Sci. USA 2013, 110, 17957–17962. [Google Scholar] [CrossRef]
- Gregori, S.; Tomasoni, D.; Pacciani, V.; Scirpoli, M.; Battaglia, M.; Magnani, C.F.; Hauben, E.; Roncarolo, M.G. Differentiation of type 1 T regulatory cells (Tr1) by tolerogenic DC-10 requires the IL-10-dependent ILT4/HLA-G pathway. Blood 2010, 116, 935–944. [Google Scholar] [CrossRef]
- Kostlin, N.; Ostermeir, A.L.; Spring, B.; Schwarz, J.; Marme, A.; Walter, C.B.; Poets, C.F.; Gille, C. HLA-G promotes myeloid-derived suppressor cell accumulation and suppressive activity during human pregnancy through engagement of the receptor ILT4. Eur. J. Immunol. 2017, 47, 374–384. [Google Scholar] [CrossRef]
- Morandi, F.; Rouas-Freiss, N.; Pistoia, V. The emerging role of soluble HLA-G in the control of chemotaxis. Cytokine Growth Factor Rev. 2014, 25, 327–335. [Google Scholar] [CrossRef]
- Agaugue, S.; Carosella, E.D.; Rouas-Freiss, N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood 2011, 117, 7021–7031. [Google Scholar] [CrossRef]
- Lee, C.L.; Guo, Y.; So, K.H.; Vijayan, M.; Guo, Y.; Wong, V.H.; Yao, Y.; Lee, K.F.; Chiu, P.C.; Yeung, W.S. Soluble human leukocyte antigen G5 polarizes differentiation of macrophages toward a decidual macrophage-like phenotype. Hum. Reprod. 2015, 30, 2263–2274. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.H.; Fan, L.A. Residues Met76 and Gln79 in HLA-G alpha1 domain involve in KIR2DL4 recognition. Cell Res. 2005, 15, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Koopman, L.A.; Kopcow, H.D.; Rybalov, B.; Boyson, J.E.; Orange, J.S.; Schatz, F.; Masch, R.; Lockwood, C.J.; Schachter, A.D.; Park, P.J.; et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med. 2003, 198, 1201–1212. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, S.; Long, E.O. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J. Exp. Med. 1999, 189, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Ponte, M.; Cantoni, C.; Biassoni, R.; Tradori-Cappai, A.; Bentivoglio, G.; Vitale, C.; Bertone, S.; Moretta, A.; Moretta, L.; Mingari, M.C. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: Decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc. Natl. Acad. Sci. USA 1999, 96, 5674–5679. [Google Scholar] [CrossRef] [PubMed]
- Faure, M.; Long, E.O. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J. Immunol. 2002, 168, 6208–6214. [Google Scholar] [CrossRef]
- Rajagopalan, S.; Long, E.O. Cellular senescence induced by CD158d reprograms natural killer cells to promote vascular remodeling. Proc. Natl. Acad. Sci. USA 2012, 109, 20596–20601. [Google Scholar] [CrossRef]
- Sarmah, N.; Baruah, M.N.; Baruah, S. Immune Modulation in HLA-G Expressing Head and Neck Squamous Cell Carcinoma in Relation to Human Papilloma Virus Positivity: A Study From Northeast India. Front. Oncol. 2019, 9, 58. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, J.; Qiu, L.; Zhang, P.; Li, J.; Yang, D.; Wei, X.; Han, Y.; Nie, S.; Sun, Y. Co-expression of ILT4/HLA-G in human non-small cell lung cancer correlates with poor prognosis and ILT4-HLA-G interaction activates ERK signaling. Tumour. Biol. 2016, 37, 11187–11198. [Google Scholar] [CrossRef]
- Rouas-Freiss, N.; LeMaoult, J.; Verine, J.; Tronik-Le Roux, D.; Culine, S.; Hennequin, C.; Desgrandchamps, F.; Carosella, E.D. Intratumor heterogeneity of immune checkpoints in primary renal cell cancer: Focus on HLA-G/ILT2/ILT4. Oncoimmunology 2017, 6, e1342023. [Google Scholar] [CrossRef]
- Cai, Z.; Wang, L.; Han, Y.; Gao, W.; Wei, X.; Gong, R.; Zhu, M.; Sun, Y.; Yu, S. Immunoglobulinlike transcript 4 and human leukocyte antigenG interaction promotes the progression of human colorectal cancer. Int. J. Oncol. 2019, 54, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wang, P.; Dai, P.; Jin, B.; Tong, Y.; Lin, H.; Shi, G. Correlation between human leukocyte antigen-G expression and clinical parameters in oral squamous cell carcinoma. Indian J. Cancer 2018, 55, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Zeestraten, E.C.; Reimers, M.S.; Saadatmand, S.; Goossens-Beumer, I.J.; Dekker, J.W.; Liefers, G.J.; van den Elsen, P.J.; van de Velde, C.J.; Kuppen, P.J. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br. J. Cancer 2014, 110, 459–468. [Google Scholar] [CrossRef]
- de Kruijf, E.M.; Sajet, A.; van Nes, J.G.; Natanov, R.; Putter, H.; Smit, V.T.; Liefers, G.J.; van den Elsen, P.J.; van de Velde, C.J.; Kuppen, P.J. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J. Immunol. 2010, 185, 7452–7459. [Google Scholar] [CrossRef]
- Yie, S.M.; Yang, H.; Ye, S.R.; Li, K.; Dong, D.D.; Lin, X.M. Expression of human leucocyte antigen G (HLA-G) is associated with prognosis in non-small cell lung cancer. Lung Cancer 2007, 58, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.L.; Zhang, X.; Dong, S.S.; Hu, B.; Han, Q.Y.; Zhang, J.G.; Zhou, W.J.; Lin, A.; Yan, W.H. Predictive value of different proportion of lesion HLA-G expression in colorectal cancer. Oncotarget 2017, 8, 107441–107451. [Google Scholar] [CrossRef]
- Guo, Z.Y.; Lv, Y.G.; Wang, L.; Shi, S.J.; Yang, F.; Zheng, G.X.; Wen, W.H.; Yang, A.G. Predictive value of HLA-G and HLA-E in the prognosis of colorectal cancer patients. Cell Immunol. 2015, 293, 10–16. [Google Scholar] [CrossRef]
- Apps, R.; Gardner, L.; Moffett, A. A critical look at HLA-G. Trends Immunol. 2008, 29, 313–321. [Google Scholar] [CrossRef]
- Polakova, K.; Kuba, D.; Russ, G. The 4H84 monoclonal antibody detecting beta2m free nonclassical HLA-G molecules also binds to free heavy chains of classical HLA class I antigens present on activated lymphocytes. Hum. Immunol. 2004, 65, 157–162. [Google Scholar] [CrossRef]
- Swets, M.; Wouters, A.; Krijgsman, D.; van Vlierberghe, R.L.P.; Boot, A.; van Eendenburg, J.D.; van Wezel, T.; Gelderblom, H.; van de Velde, C.J.H.; van den Elsen, P.J.; et al. HLA-G protein expression in colorectal cancer evaluated by immunohistochemistry and western blot analysis: Its expression characteristics remain enigmatic. Clin. Immunol. 2018, 194, 80–86. [Google Scholar] [CrossRef]
- Lin, A.; Chen, H.X.; Zhu, C.C.; Zhang, X.; Xu, H.H.; Zhang, J.G.; Wang, Q.; Zhou, W.J.; Yan, W.H. Aberrant human leucocyte antigen-G expression and its clinical relevance in hepatocellular carcinoma. J. Cell Mol. Med. 2010, 14, 2162–2171. [Google Scholar] [CrossRef] [PubMed]
- Roelands, J.; Hendrickx, W.; Kuppen, P.J.K.; Mall, R.; Zoppoli, G.; Saad, M.; Halliwill, K. Genomic Landscape of Tumor-Host Interactions with Differential Prognostic and Predictive Connotations. BioRxiv 2019. [Google Scholar] [CrossRef]
- Paul, P.; Cabestre, F.A.; Le Gal, F.A.; Khalil-Daher, I.; Le Danff, C.; Schmid, M.; Mercier, S.; Avril, M.F.; Dausset, J.; Guillet, J.G.; et al. Heterogeneity of HLA-G gene transcription and protein expression in malignant melanoma biopsies. Cancer Res. 1999, 59, 1954–1960. [Google Scholar] [PubMed]
- Reches, A.; Berhani, O.; Mandelboim, O. A Unique Regulation Region in the 3’ UTR of HLA-G with a Promising Potential. Int. J. Mol. Sci. 2020, 21, 900. [Google Scholar] [CrossRef] [PubMed]
- Castelli, E.C.; Veiga-Castelli, L.C.; Yaghi, L.; Moreau, P.; Donadi, E.A. Transcriptional and posttranscriptional regulations of the HLA-G gene. J. Immunol. Res. 2014, 2014, 734068. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jin, X.; Wang, J. miR148a modulates the viability, migration and invasion of oral squamous cell carcinoma cells by regulating HLAG expression. Mol. Med. Rep. 2019, 20, 795–801. [Google Scholar] [CrossRef]
- Seliger, B. Role of microRNAs on HLA-G expression in human tumors. Hum. Immunol. 2016, 77, 760–763. [Google Scholar] [CrossRef]
- Jasinski-Bergner, S.; Stoehr, C.; Bukur, J.; Massa, C.; Braun, J.; Huttelmaier, S.; Spath, V.; Wartenberg, R.; Legal, W.; Taubert, H.; et al. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology 2015, 4, e1008805. [Google Scholar] [CrossRef]
- Roelands, J.; Hendrickx, W.; Zoppoli, G.; Mall, R.; Saad, M.; Halliwill, K.; Curigliano, G.; Rinchai, D.; Decock, J.; Delogu, L.G.; et al. Oncogenic states dictate the prognostic and predictive connotations of intratumoral immune response. J. Immunother Cancer 2020, 8. [Google Scholar] [CrossRef]
- Wang, E.; Worschech, A.; Marincola, F.M. The immunologic constant of rejection. Trends Immunol. 2008, 29, 256–262. [Google Scholar] [CrossRef]
- Galon, J.; Angell, H.K.; Bedognetti, D.; Marincola, F.M. The continuum of cancer immunosurveillance: Prognostic, predictive, and mechanistic signatures. Immunity 2013, 39, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, W.; Simeone, I.; Anjum, S.; Mokrab, Y.; Bertucci, F.; Finetti, P.; Curigliano, G.; Seliger, B.; Cerulo, L.; Tomei, S.; et al. Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. Oncoimmunology 2017, 6, e1253654. [Google Scholar] [CrossRef]
- Roelands, J.; Decock, J.; Boughorbel, S.; Rinchai, D.; Maccalli, C.; Ceccarelli, M.; Black, M.; Print, C.; Chou, J.; Presnell, S.; et al. A collection of annotated and harmonized human breast cancer transcriptome datasets, including immunologic classification. F1000Res 2017, 6, 296. [Google Scholar] [CrossRef]
- Michielsen, C.; Hangelbroek, R.W.J.; Feskens, E.J.M.; Afman, L.A. Disentangling the Effects of Monounsaturated Fatty Acids from Other Components of a Mediterranean Diet on Serum Metabolite Profiles: A Randomized Fully Controlled Dietary Intervention in Healthy Subjects at Risk of the Metabolic Syndrome. Mol. Nutr. Food Res. 2019, 63, e1801095. [Google Scholar] [CrossRef] [PubMed]
- Taube, J.M.; Klein, A.; Brahmer, J.R.; Xu, H.; Pan, X.; Kim, J.H.; Chen, L.; Pardoll, D.M.; Topalian, S.L.; Anders, R.A. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 2014, 20, 5064–5074. [Google Scholar] [CrossRef] [PubMed]
- Pazmany, L.; Mandelboim, O.; Vales-Gomez, M.; Davis, D.M.; Reyburn, H.T.; Strominger, J.L. Protection from natural killer cell-mediated lysis by HLA-G expression on target cells. Science 1996, 274, 792–795. [Google Scholar] [CrossRef]
- Wiendl, H.; Mitsdoerffer, M.; Hofmeister, V.; Wischhusen, J.; Bornemann, A.; Meyermann, R.; Weiss, E.H.; Melms, A.; Weller, M. A functional role of HLA-G expression in human gliomas: An alternative strategy of immune escape. J. Immunol. 2002, 168, 4772–4780. [Google Scholar] [CrossRef]
- Lin, A.; Yan, W.H.; Xu, H.H.; Gan, M.F.; Cai, J.F.; Zhu, M.; Zhou, M.Y. HLA-G expression in human ovarian carcinoma counteracts NK cell function. Ann. Oncol. 2007, 18, 1804–1809. [Google Scholar] [CrossRef]
- Lin, A.; Zhang, X.; Xu, H.H.; Xu, D.P.; Ruan, Y.Y.; Yan, W.H. HLA-G expression is associated with metastasis and poor survival in the Balb/c nu/nu murine tumor model with ovarian cancer. Int. J. Cancer 2012, 131, 150–157. [Google Scholar] [CrossRef]
- Loumagne, L.; Baudhuin, J.; Favier, B.; Montespan, F.; Carosella, E.D.; Rouas-Freiss, N. In vivo evidence that secretion of HLA-G by immunogenic tumor cells allows their evasion from immunosurveillance. Int. J. Cancer 2014, 135, 2107–2117. [Google Scholar] [CrossRef]
- Rutten, M.J.; Dijk, F.; Savci-Heijink, C.D.; Buist, M.R.; Kenter, G.G.; van de Vijver, M.J.; Jordanova, E.S. HLA-G expression is an independent predictor for improved survival in high grade ovarian carcinomas. J. Immunol. Res. 2014, 2014, 274584. [Google Scholar] [CrossRef] [PubMed]
- Reimers, M.S.; Engels, C.C.; Putter, H.; Morreau, H.; Liefers, G.J.; van de Velde, C.J.; Kuppen, P.J. Prognostic value of HLA class I, HLA-E, HLA-G and Tregs in rectal cancer: A retrospective cohort study. BMC Cancer 2014, 14, 486. [Google Scholar] [CrossRef] [PubMed]
- Rebmann, V.; Konig, L.; Nardi Fda, S.; Wagner, B.; Manvailer, L.F.; Horn, P.A. The Potential of HLA-G-Bearing Extracellular Vesicles as a Future Element in HLA-G Immune Biology. Front. Immunol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Caumartin, J.; Favier, B.; Daouya, M.; Guillard, C.; Moreau, P.; Carosella, E.D.; LeMaoult, J. Trogocytosis-based generation of suppressive NK cells. EMBO J. 2007, 26, 1423–1433. [Google Scholar] [CrossRef]
- Brown, R.; Kabani, K.; Favaloro, J.; Yang, S.; Ho, P.J.; Gibson, J.; Fromm, P.; Suen, H.; Woodland, N.; Nassif, N.; et al. CD86+ or HLA-G+ can be transferred via trogocytosis from myeloma cells to T cells and are associated with poor prognosis. Blood 2012, 120, 2055–2063. [Google Scholar] [CrossRef]
- Tilburgs, T.; Evans, J.H.; Crespo, A.C.; Strominger, J.L. The HLA-G cycle provides for both NK tolerance and immunity at the maternal-fetal interface. Proc. Natl. Acad. Sci. USA 2015, 112, 13312–13317. [Google Scholar] [CrossRef]
- Cao, M.; Yie, S.M.; Liu, J.; Ye, S.R.; Xia, D.; Gao, E. Plasma soluble HLA-G is a potential biomarker for diagnosis of colorectal, gastric, esophageal and lung cancer. Tissue Antigens 2011, 78, 120–128. [Google Scholar] [CrossRef]
- Konig, L.; Kasimir-Bauer, S.; Hoffmann, O.; Bittner, A.K.; Wagner, B.; Manvailer, L.F.; Schramm, S.; Bankfalvi, A.; Giebel, B.; Kimmig, R.; et al. The prognostic impact of soluble and vesicular HLA-G and its relationship to circulating tumor cells in neoadjuvant treated breast cancer patients. Hum. Immunol. 2016, 77, 791–799. [Google Scholar] [CrossRef]

| HLA-G mAbs | Specificity | Applications |
|---|---|---|
| 4H84 | An epitope in the HLA-G α1 domain * | IHC(P), IP, WB, ICC, ELISA |
| MEM-G/1 | Denatured HLA-G heavy chain, all isoforms * | IHC(F/P), WB |
| MEM-G/2 | Free heavy chain of all HLA-G isoforms | IHC(F/P), WB |
| MEM-G/9 | Native form of HLA-G1 and HLA-G5 isoform associated with β2M | IHC(F), IP, ELISA, FC |
| MEM-G/11 | HLA-G1 | IHC(F), IP, ELISA, FC, ICC |
| 01G | HLA-G1 | IHC(F), IP, ICC, FC, ELISA |
| 87G | HLA-G1 and HLA-G5 | IHC(F), FC, ELISA |
| 2A12 | HLA-G5 and HLA-G6 | IHC(F/P), WB, FC, ELISA |
| 5A6G7 | HLA-G5 and HLA-G6 | IHC(F/P), WB, FC, ELISA, ICC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krijgsman, D.; Roelands, J.; Hendrickx, W.; Bedognetti, D.; Kuppen, P.J.K. HLA-G: A New Immune Checkpoint in Cancer? Int. J. Mol. Sci. 2020, 21, 4528. https://doi.org/10.3390/ijms21124528
Krijgsman D, Roelands J, Hendrickx W, Bedognetti D, Kuppen PJK. HLA-G: A New Immune Checkpoint in Cancer? International Journal of Molecular Sciences. 2020; 21(12):4528. https://doi.org/10.3390/ijms21124528
Chicago/Turabian StyleKrijgsman, Daniëlle, Jessica Roelands, Wouter Hendrickx, Davide Bedognetti, and Peter J. K. Kuppen. 2020. "HLA-G: A New Immune Checkpoint in Cancer?" International Journal of Molecular Sciences 21, no. 12: 4528. https://doi.org/10.3390/ijms21124528
APA StyleKrijgsman, D., Roelands, J., Hendrickx, W., Bedognetti, D., & Kuppen, P. J. K. (2020). HLA-G: A New Immune Checkpoint in Cancer? International Journal of Molecular Sciences, 21(12), 4528. https://doi.org/10.3390/ijms21124528







