Tooth Enamel and Its Dynamic Protein Matrix
Abstract
1. Introduction
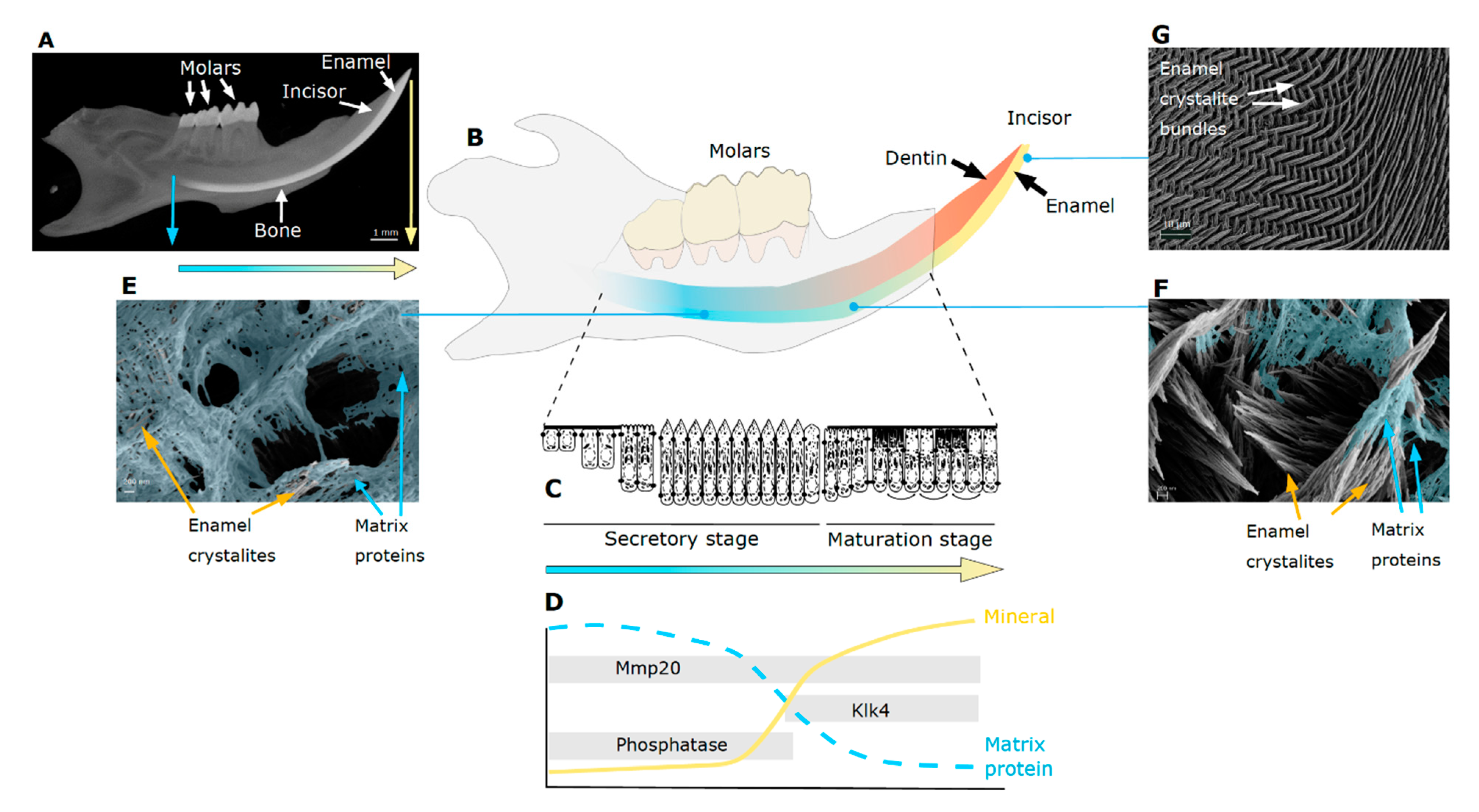
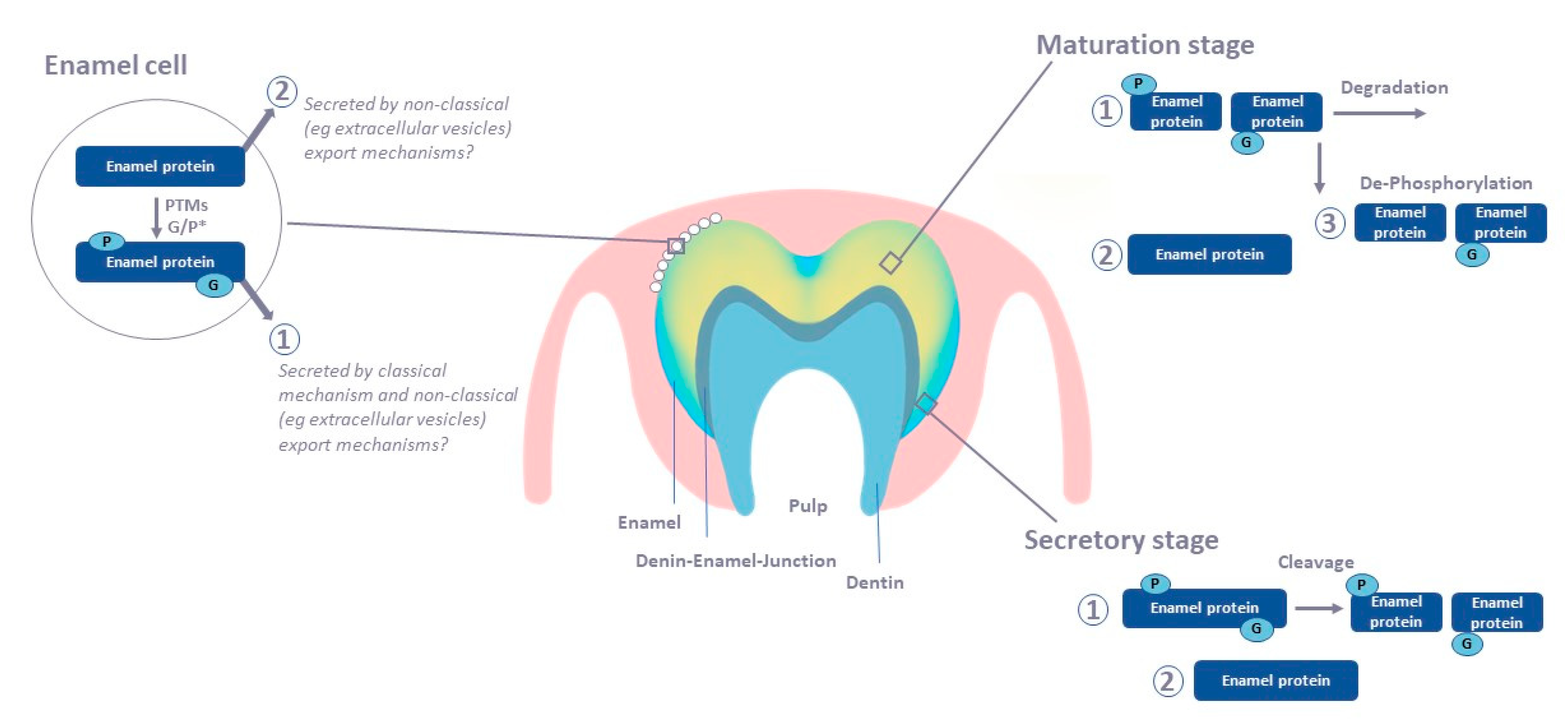
2. Tooth Enamel Formation
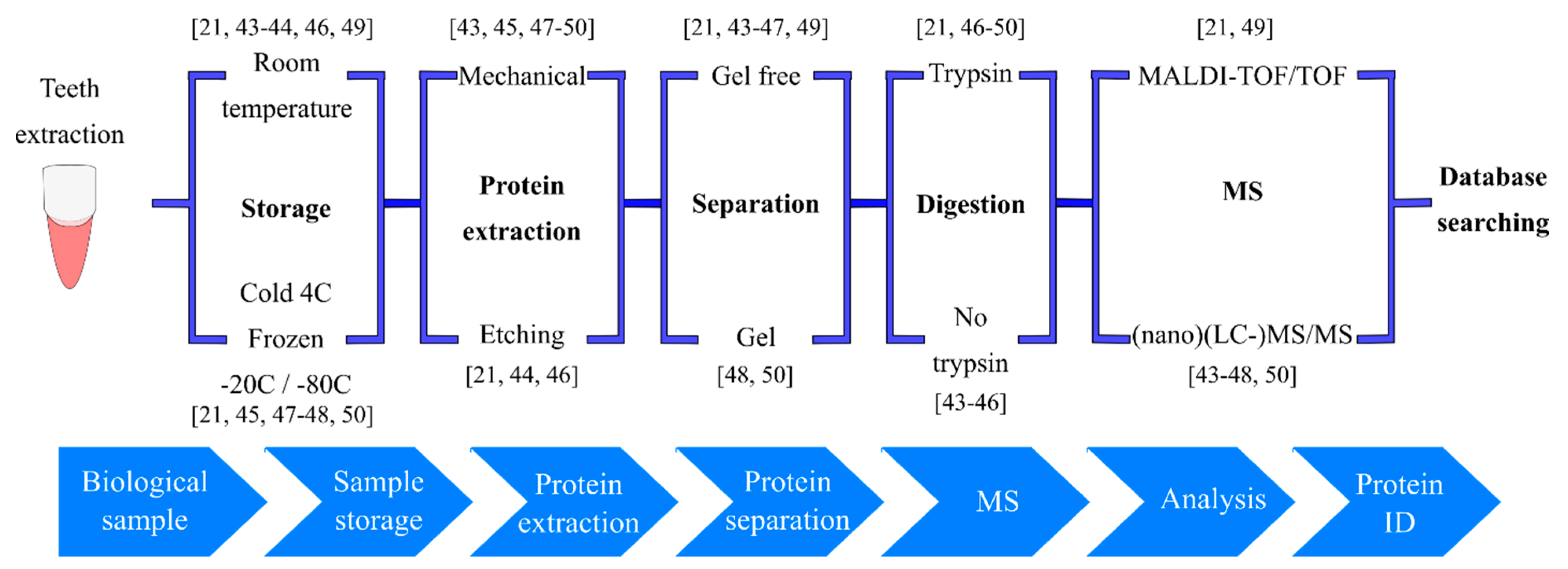
3. Teeth as Records: Enamel Proteomics in Archeology, Forensics and Odontology
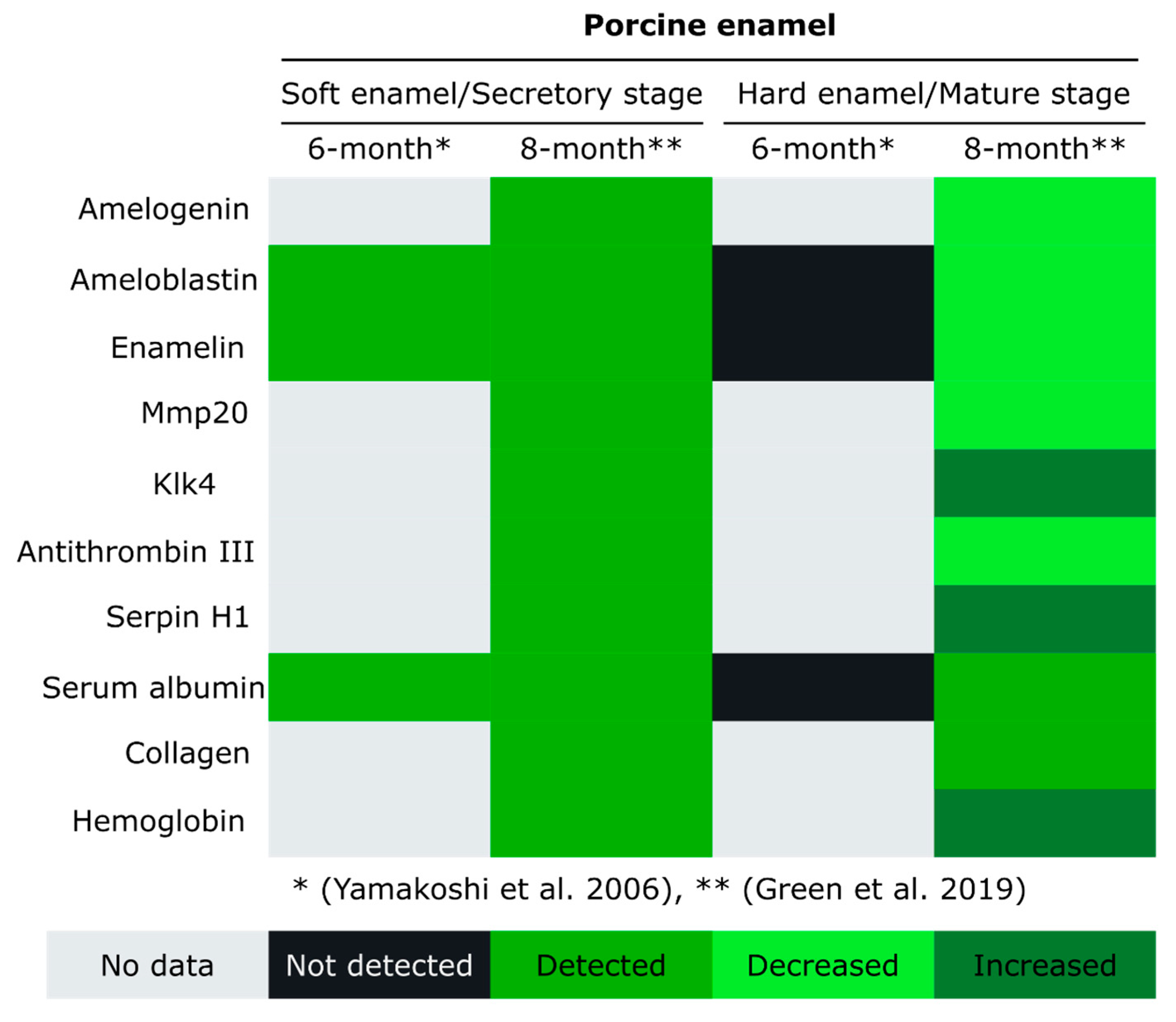
4. Enamel Protein Composition at Different Developmental Stages
5. Structural Enamel Matrix Proteins
6. Enamel Matrix Removal and Proteins Remaining in Erupted Tooth Enamel
7. Fluoride Affects Ameloblast Function and Matrix Composition
8. Animal Models of Enamel Development
9. Chalky Teeth and Serum Albumin in Enamel
10. Proteomic Analysis in Enamel Cells
11. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D-PAGE | Two-dimensional gel electrophoresis |
| AI | Amelogenesis imperfecta |
| AMELX | Amelogenin, isoform X |
| AMELY | Amelogenin, isoform Y |
| DEJ | Dentin-enamel junction |
| EOM | Enamel organic matrix |
| Klk4 | Kallikrein-4 |
| LC-MS/MS | Liquid chromatography coupled to tandem mass spectrometry |
| LRAP | Leucine rich amelogenin peptide |
| MALDI-TOF/TOF | Matrix-Laser Desorption/Ionization Time-Of-Flight/Time-Of-Flight Mass Spectrometry |
| MH | Molar hypomineralization |
| MIH | Molar-incisor hypomineralization |
| Mmp20 | Proteinase matrix metalloproteinase-20 |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| TRAP | Tyrosine-rich amelogenin peptide |
References
- Bartlett, J.D. Dental enamel development: Proteinases and their enamel matrix substrates. ISRN Dent. 2013, 2013, 684607. [Google Scholar] [CrossRef] [PubMed]
- Gunier, R.B.; Mora, A.M.; Smith, D.; Arora, M.; Austin, C.; Eskenazi, B.; Bradman, A. Biomarkers of manganese exposure in pregnant women and children living in an agricultural community in California. Environ. Sci. Technol. 2014, 48, 14695–14702. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.A.; Mountain, R.V.; Pickett, O.R.; Den Besten, P.K.; Bidlack, F.B.; Dunn, E.C. Teeth as Potential New Tools to Measure Early-Life Adversity and Subsequent Mental Health Risk: An Interdisciplinary Review and Conceptual Model. Biol. Psychiatr. 2020, 87, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Queen Mary University of London—Institute of Dentistry—Barts and The London Atlas of Tooth Development and Eruption. Available online: https://www.qmul.ac.uk/dentistry/atlas/ (accessed on 22 June 2020).
- Thesleff, I. Developmental biology and building a tooth. Quintessence Int. 2003, 34, 613–620. [Google Scholar]
- Lacruz, R.S.; Habelitz, S.; Wright, J.T.; Paine, M.L. Dental Enamel Formation and Implications for Oral Health and Disease. Physiol. Rev. 2017, 97, 939–993. [Google Scholar] [CrossRef]
- Robinson, C.; Kirkham, J.; Hallsworth, A.S. Volume distribution and concentration of protein, mineral and water in developing bovine enamel. Arch. Oral Biol. 1988, 33, 159–162. [Google Scholar] [CrossRef]
- Smith, C.E. Cellular and chemical events during enamel maturation. Crit. Rev. Oral Biol. Med. 1998, 9, 128–161. [Google Scholar] [CrossRef]
- Bronckers, A.L. Ion Transport by Ameloblasts during Amelogenesis. J. Dent. Res. 2017, 96, 243–253. [Google Scholar] [CrossRef]
- Lacruz, R.S.; Smith, C.E.; Moffatt, P.; Chang, E.H.; Bromage, T.G.; Bringas, P., Jr.; Nanci, A.; Baniwal, S.K.; Zabner, J.; Welsh, M.J.; et al. Requirements for ion and solute transport and pH regulation during enamel maturation. J. Cell. Physiol. 2012, 227, 1776–1785. [Google Scholar] [CrossRef]
- Hu, J.C.; Chun, Y.H.; Al Hazzazzi, T.; Simmer, J.P. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs 2007, 186, 78–85. [Google Scholar] [CrossRef]
- Robinson, C.; Brookes, S.J.; Shore, R.C.; Kirkham, J. The developing enamel matrix: Nature and function. Eur. J. Oral Sci. 1998, 106, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Yates, J.R., 3rd. Recent technical advances in proteomics. F1000Research 2019, 8, F1000. [Google Scholar] [PubMed]
- Graves, P.R.; Haystead, T.A. Molecular biologist’s guide to proteomics. Microbiol. Mol. Biol. Rev. MMBR 2002, 66, 39–63. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Schulte, F.; Lee, K.-H.; Pugach, M.K.; Hardt, M.; Bidlack, F.B. Mapping the Tooth Enamel Proteome and Amelogenin Phosphorylation Onto Mineralizing Porcine Tooth Crowns. Front. Physiol. 2019, 10, 925. [Google Scholar] [CrossRef]
- Kirkham, J.; Brookes, S.J.; Diekwisch, T.G.H.; Margolis, H.C.; Berdal, A.; Hubbard, M.J. Enamel Research: Priorities and Future Directions. Front. Physiol. 2017, 8, 513. [Google Scholar] [CrossRef]
- Members, S.A.B. Enamel biology and future research directions. Eur. J. Oral Sci. 2011, 119, 377–379. [Google Scholar]
- Pandya, M.; Diekwisch, T.G.H. Enamel biomimetics-fiction or future of dentistry. Int. J. Oral Sci. 2019, 11, 8. [Google Scholar] [CrossRef]
- Hillson, S. Archaeology and the study of teeth. Endeavour 1986, 10, 145–149. [Google Scholar] [CrossRef]
- Risnes, S. Growth tracks in dental enamel. J. Hum. Evol. 1998, 35, 331–350. [Google Scholar] [CrossRef]
- Porto, I.M.; Laure, H.J.; Tykot, R.H.; De Sousa, F.B.; Rosa, J.C.; Gerlach, R.F. Recovery and identification of mature enamel proteins in ancient teeth. Eur. J. Oral Sci. 2011, 119 (Suppl. 1), 83–87. [Google Scholar] [CrossRef]
- Welker, F.; Collins, M.J.; Thomas, J.A.; Wadsley, M.; Brace, S.; Cappellini, E.; Turvey, S.T.; Reguero, M.; Gelfo, J.N.; Kramarz, A.; et al. Ancient proteins resolve the evolutionary history of Darwin’s South American ungulates. Nature 2015, 522, 81–84. [Google Scholar] [CrossRef]
- Welker, F.; Ramos-Madrigal, J.; Kuhlwilm, M.; Liao, W.; Gutenbrunner, P.; De Manuel, M.; Samodova, D.; Mackie, M.; Allentoft, M.E.; Bacon, A.M.; et al. Enamel proteome shows that Gigantopithecus was an early diverging pongine. Nature 2019, 576, 262–265. [Google Scholar] [CrossRef]
- Chen, F.; Welker, F.; Shen, C.C.; Bailey, S.E.; Bergmann, I.; Davis, S.; Xia, H.; Wang, H.; Fischer, R.; Freidline, S.E.; et al. A late Middle Pleistocene Denisovan mandible from the Tibetan Plateau. Nature 2019, 569, 409–412. [Google Scholar] [CrossRef]
- Cappellini, E.; Welker, F.; Pandolfi, L.; Ramos-Madrigal, J.; Samodova, D.; Ruther, P.L.; Fotakis, A.K.; Lyon, D.; Moreno-Mayar, J.V.; Bukhsianidze, M.; et al. Early Pleistocene enamel proteome from Dmanisi resolves Stephanorhinus phylogeny. Nature 2019, 574, 103–107. [Google Scholar] [CrossRef]
- Demarchi, B.; Hall, S.; Roncal-Herrero, T.; Freeman, C.L.; Woolley, J.; Crisp, M.K.; Wilson, J.; Fotakis, A.; Fischer, R.; Kessler, B.M.; et al. Protein sequences bound to mineral surfaces persist into deep time. Elife 2016, 5, e17092. [Google Scholar] [CrossRef] [PubMed]
- Tsutaya, T.; Mackie, M.; Koenig, C.; Sato, T.; Weber, A.W.; Kato, H.; Olsen, J.V.; Cappellini, E. Palaeoproteomic identification of breast milk protein residues from the archaeological skeletal remains of a neonatal dog. Sci. Rep. 2019, 9, 12841. [Google Scholar] [CrossRef]
- Dean, M.C.; Humphrey, L.; Groom, A.; Hassett, B. Variation in the timing of enamel formation in modern human deciduous canines. Arch. Oral Biol. 2020, 114, 104719. [Google Scholar] [CrossRef] [PubMed]
- Cleland, T.P.; Schroeter, E.R. A Comparison of Common Mass Spectrometry Approaches for Paleoproteomics. J. Proteome Res. 2018, 17, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Hedenstierna-Jonson, C.; Kjellstrom, A.; Zachrisson, T.; Krzewinska, M.; Sobrado, V.; Price, N.; Gunther, T.; Jakobsson, M.; Gotherstrom, A.; Stora, J. A female Viking warrior confirmed by genomics. Am. J. Phys. Anthropol. 2017, 164, 853–860. [Google Scholar] [CrossRef]
- Loreille, O.; Ratnayake, S.; Bazinet, A.L.; Stockwell, T.B.; Sommer, D.D.; Rohland, N.; Mallick, S.; Johnson, P.L.F.; Skoglund, P.; Onorato, A.J.; et al. Biological Sexing of a 4000-Year-Old Egyptian Mummy Head to Assess the Potential of Nuclear DNA Recovery from the Most Damaged and Limited Forensic Specimens. Genes (Basel) 2018, 9, 135. [Google Scholar] [CrossRef]
- Bauer, C.M.; Niederstatter, H.; McGlynn, G.; Stadler, H.; Parson, W. Comparison of morphological and molecular genetic sex-typing on mediaeval human skeletal remains. Forensic Sci. Int. Genet. 2013, 7, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Krishan, K.; Chatterjee, P.M.; Kanchan, T.; Kaur, S.; Baryah, N.; Singh, R.K. A review of sex estimation techniques during examination of skeletal remains in forensic anthropology casework. Forensic Sci. Int. 2016, 261, 165-e1. [Google Scholar] [CrossRef] [PubMed]
- Shamim, T. Oral Pathology in Forensic Investigation. J. Int. Soc. Prev. Community Dent. 2018, 8, 1–5. [Google Scholar]
- Ikawa, T.; Kakegawa, A.; Nagano, T.; Ando, H.; Yamakoshi, Y.; Tanabe, T.; Simmer, J.P.; Hu, C.C.; Fukae, M.; Oida, S. Porcine amelogenin is expressed from the X and Y chromosomes. J. Dent. Res. 2005, 84, 144–148. [Google Scholar] [CrossRef]
- Yamakoshi, Y. Porcine Amelogenin: Alternative Splicing, Proteolytic Processing, Protein—Protein Interactions and Possible Functions. J. Oral Biosci. 2011, 53, 275–283. [Google Scholar] [CrossRef]
- Lau, E.C.; Mohandas, T.K.; Shapiro, L.J.; Slavkin, H.C.; Snead, M.L. Human and mouse amelogenin gene loci are on the sex chromosomes. Genomics 1989, 4, 162–168. [Google Scholar] [CrossRef]
- Weikard, R.; Pitra, C.; Kuhn, C. Amelogenin cross-amplification in the family Bovidae and its application for sex determination. Mol. Reprod. Dev. 2006, 73, 1333–1337. [Google Scholar] [CrossRef]
- Tsai, T.C.; Wu, S.H.; Chen, H.L.; Tung, Y.T.; Cheng, W.T.; Huang, J.C.; Chen, C.M. Identification of sex-specific polymorphic sequences in the goat amelogenin gene for embryo sexing. J. Anim. Sci. 2011, 89, 2407–2414. [Google Scholar] [CrossRef][Green Version]
- Salido, E.C.; Yen, P.H.; Koprivnikar, K.; Yu, L.C.; Shapiro, L.J. The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am. J. Hum. Genet. 1992, 50, 303–316. [Google Scholar]
- Nagare, S.P.; Chaudhari, R.S.; Birangane, R.S.; Parkarwar, P.C. Sex determination in forensic identification, a review. J. Forensic Dent. Sci. 2018, 10, 61–66. [Google Scholar]
- Butler, E.; Li, R. Genetic markers for sex identification in forensic DNA analysis. J. Forensic Investig. 2014, 2, 3. [Google Scholar]
- Lugli, F.; Di Rocco, G.; Vazzana, A.; Genovese, F.; Pinetti, D.; Cilli, E.; Carile, M.C.; Silvestrini, S.; Gabanini, G.; Arrighi, S.; et al. Enamel peptides reveal the sex of the Late Antique ‘Lovers of Modena’. Sci. Rep. 2019, 9, 13130. [Google Scholar] [CrossRef] [PubMed]
- Stewart, N.A.; Gerlach, R.F.; Gowland, R.L.; Gron, K.J.; Montgomery, J. Sex determination of human remains from peptides in tooth enamel. Proc. Natl. Acad. Sci. USA 2017, 114, 13649–13654. [Google Scholar] [CrossRef] [PubMed]
- Castiblanco, G.A.; Rutishauser, D.; Ilag, L.L.; Martignon, S.; Castellanos, J.E.; Mejia, W. Identification of proteins from human permanent erupted enamel. Eur. J. Oral Sci. 2015, 123, 390–395. [Google Scholar] [CrossRef]
- Stewart, N.A.; Molina, G.F.; Mardegan Issa, J.P.; Yates, N.A.; Sosovicka, M.; Vieira, A.R.; Line, S.R.P.; Montgomery, J.; Gerlach, R.F. The identification of peptides by nanoLC-MS/MS from human surface tooth enamel following a simple acid etch extraction. RSC Adv. 2016, 6, 61673–61679. [Google Scholar] [CrossRef]
- Jagr, M.; Ergang, P.; Pataridis, S.; Kolrosova, M.; Bartos, M.; Miksik, I. Proteomic analysis of dentin-enamel junction and adjacent protein-containing enamel matrix layer of healthy human molar teeth. Eur. J. Oral Sci. 2019, 127, 112. [Google Scholar] [CrossRef] [PubMed]
- Farah, R.A.; Monk, B.C.; Swain, M.V.; Drummond, B.K. Protein content of molar-incisor hypomineralisation enamel. J. Dent. 2010, 38, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Nielsen-Marsh, C.M.; Stegemann, C.; Hoffmann, R.; Smith, T.; Feeney, R.; Toussaint, M.; Harvati, K.; Panagopoulou, E.; Hublin, J.-J.; Richards, M.P. Extraction and sequencing of human and Neanderthal mature enamel proteins using MALDI-TOF/TOF MS. J. Archaeol. Sci. 2009, 36, 1758–1763. [Google Scholar] [CrossRef]
- Mangum, J.E.; Crombie, F.A.; Kilpatrick, N.; Manton, D.J.; Hubbard, M.J. Surface integrity governs the proteome of hypomineralized enamel. J. Dent. Res. 2010, 89, 1160–1165. [Google Scholar] [CrossRef]
- Parker, G.J.; Yip, J.M.; Eerkens, J.W.; Salemi, M.; Durbin-Johnson, B.; Kiesow, C.; Haas, R.; Buikstra, J.E.; Klaus, H.; Regan, L.A.; et al. Sex estimation using sexually dimorphic amelogenin protein fragments in human enamel. J. Archaeol. Sci. 2019, 101, 169–180. [Google Scholar] [CrossRef]
- Froment, C.; Hourset, M.; Saenz-Oyhereguy, N.; Mouton-Barbosa, E.; Willmann, C.; Zanolli, C.; Esclassan, R.; Donat, R.; Theves, C.; Burlet-Schiltz, O.; et al. Analysis of 5000year-old human teeth using optimized large-scale and targeted proteomics approaches for detection of sex-specific peptides. J. Proteom. 2020, 211, 103548. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Piehowski, P.D.; Shi, T.; Smith, R.D.; Qian, W.J. Advances in microscale separations towards nanoproteomics applications. J. Chromatogr. A 2017, 1523, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Welker, F.; Ramos-Madrigal, J.; Gutenbrunner, P.; Mackie, M.; Tiwary, S.; Rakownikow Jersie-Christensen, R.; Chiva, C.; Dickinson, M.R.; Kuhlwilm, M.; De Manuel, M.; et al. The dental proteome of Homo antecessor. Nature 2020, 580, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Papagerakis, P.; Yamakoshi, Y.; Hu, J.C.; Bartlett, J.D.; Simmer, J.P. Functions of KLK4 and MMP-20 in dental enamel formation. Biol. Chem. 2008, 389, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, H.; Tran, B.; Beniash, E.; Kwak, S.Y.; Margolis, H.C. Proteolysis by MMP20 Prevents Aberrant Mineralization in Secretory Enamel. J. Dent. Res. 2019, 98, 468–475. [Google Scholar] [CrossRef]
- Yamakoshi, Y.; Simmer, J.P.; Bartlett, J.D.; Karakida, T.; Oida, S. MMP20 and KLK4 activation and inactivation interactions in vitro. Arch. Oral Biol. 2013, 58, 1569–1577. [Google Scholar] [CrossRef]
- Smith, C.E.; Richardson, A.S.; Hu, Y.; Bartlett, J.D.; Hu, J.C.; Simmer, J.P. Effect of kallikrein 4 loss on enamel mineralization: Comparison with mice lacking matrix metalloproteinase 20. J. Biol. Chem. 2011, 286, 18149–18160. [Google Scholar] [CrossRef]
- Uchida, T.; Tanabe, T.; Fukae, M.; Shimizu, M.; Yamada, M.; Miake, K.; Kobayashi, S. Immunochemical and immunohistochemical studies, using antisera against porcine 25 kDa amelogenin, 89 kDa enamelin and the 13–17 kDa nonamelogenins, on immature enamel of the pig and rat. Histochemistry 1991, 96, 129–138. [Google Scholar] [CrossRef]
- Gibson, C.W.; Golub, E.; Ding, W.D.; Shimokawa, H.; Young, M.; Termine, J.; Rosenbloom, J. Identification of the leucine-rich amelogenin peptide (LRAP) as the translation product of an alternatively spliced transcript. Biochem. Biophys. Res. Commun. 1991, 174, 1306–1312. [Google Scholar] [CrossRef]
- Bartlett, J.D.; Ball, R.L.; Kawai, T.; Tye, C.E.; Tsuchiya, M.; Simmer, J.P. Origin, splicing and expression of rodent amelogenin exon 8. J. Dent. Res. 2006, 85, 894–899. [Google Scholar] [CrossRef]
- Hu, C.C.; Bartlett, J.D.; Zhang, C.H.; Qian, Q.; Ryu, O.H.; Simmer, J.P. Cloning, cDNA sequence and alternative splicing of porcine amelogenin mRNAs. J. Dent. Res. 1996, 75, 1735–1741. [Google Scholar] [CrossRef]
- Yamakoshi, Y.; Tanabe, T.; Fukae, M.; Shimizu, M. Porcine amelogenins. Calcif. Tissue Int. 1994, 54, 69–75. [Google Scholar] [CrossRef]
- Delgado, S.; Vidal, N.; Veron, G.; Sire, J.Y. Amelogenin, the major protein of tooth enamel: A new phylogenetic marker for ordinal mammal relationships. Mol. Phylogenet. Evol. 2008, 47, 865–869. [Google Scholar] [CrossRef]
- Simmer, J.P. Alternative splicing of amelogenins. Connect. Tissue Res. 1995, 32, 131–136. [Google Scholar] [CrossRef]
- Cho, E.S.; Kim, K.J.; Lee, K.E.; Lee, E.J.; Yun, C.Y.; Lee, M.J.; Shin, T.J.; Hyun, H.K.; Kim, Y.J.; Lee, S.H.; et al. Alteration of conserved alternative splicing in AMELX causes enamel defects. J. Dent. Res. 2014, 93, 980–987. [Google Scholar] [CrossRef]
- Veis, A. Amelogenin gene splice products: Potential signaling molecules. Cell. Mol. Life Sci. 2003, 60, 38–55. [Google Scholar] [CrossRef]
- Lau, E.C.; Simmer, J.P.; Bringas, P., Jr.; Hsu, D.D.; Hu, C.C.; Zeichner-David, M.; Thiemann, F.; Snead, M.L.; Slavkin, H.C.; Fincham, A.G. Alternative splicing of the mouse amelogenin primary RNA transcript contributes to amelogenin heterogeneity. Biochem. Biophys. Res. Commun. 1992, 188, 1253–1260. [Google Scholar] [CrossRef]
- Yamazaki, H.; Beniash, E.; Yamakoshi, Y.; Simmer, J.P.; Margolis, H.C. Protein Phosphorylation and Mineral Binding Affect the Secondary Structure of the Leucine-Rich Amelogenin Peptide. Front. Physiol. 2017, 8, 450. [Google Scholar] [CrossRef]
- Kwak, S.Y.; Kim, S.; Yamakoshi, Y.; Simmer, J.P.; Beniash, E.; Margolis, H.C. Regulation of calcium phosphate formation by native amelogenins in vitro. Connect. Tissue Res. 2014, 55 (Suppl. 1), 21–24. [Google Scholar] [CrossRef]
- Wiedemann-Bidlack, F.B.; Kwak, S.Y.; Beniash, E.; Yamakoshi, Y.; Simmer, J.P.; Margolis, H.C. Effects of phosphorylation on the self-assembly of native full-length porcine amelogenin and its regulation of calcium phosphate formation in vitro. J. Struct. Biol. 2011, 173, 250–260. [Google Scholar] [CrossRef]
- Kwak, S.Y.; Wiedemann-Bidlack, F.B.; Beniash, E.; Yamakoshi, Y.; Simmer, J.P.; Litman, A.; Margolis, H.C. Role of 20-kDa amelogenin (P148) phosphorylation in calcium phosphate formation in vitro. J. Biol. Chem. 2009, 284, 18972–18979. [Google Scholar] [CrossRef]
- Shin, N.Y.; Yamazaki, H.; Beniash, E.; Yang, X.; Margolis, S.S.; Pugach, M.K.; Simmer, J.P.; Margolis, H.C. Amelogenin phosphorylation regulates tooth enamel formation by stabilizing a transient amorphous mineral precursor. J. Biol. Chem. 2020, 295, 1943–1959. [Google Scholar] [CrossRef]
- Gibson, C.W.; Li, Y.; Daly, B.; Suggs, C.; Yuan, Z.A.; Fong, H.; Simmons, D.; Aragon, M.; Kulkarni, A.B.; Wright, J.T. The leucine-rich amelogenin peptide alters the amelogenin null enamel phenotype. Cells Tissues Organs 2009, 189, 169–174. [Google Scholar] [CrossRef]
- Gibson, C.W.; Li, Y.; Suggs, C.; Kuehl, M.A.; Pugach, M.K.; Kulkarni, A.B.; Wright, J.T. Rescue of the murine amelogenin null phenotype with two amelogenin transgenes. Eur. J. Oral Sci. 2011, 119 (Suppl. 1), 70–74. [Google Scholar] [CrossRef][Green Version]
- Stahl, J.; Nakano, Y.; Kim, S.O.; Gibson, C.W.; Le, T.; DenBesten, P. Leucine rich amelogenin peptide alters ameloblast differentiation in vivo. Matrix Biol. 2013, 32, 432–442. [Google Scholar] [CrossRef]
- Veis, A.; Tompkins, K.; Alvares, K.; Wei, K.; Wang, L.; Wang, X.S.; Brownell, A.G.; Jengh, S.M.; Healy, K.E. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J. Biol. Chem. 2000, 275, 41263–41272. [Google Scholar] [CrossRef]
- Boabaid, F.; Gibson, C.W.; Kuehl, M.A.; Berry, J.E.; Snead, M.L.; Nociti, F.H., Jr.; Katchburian, E.; Somerman, M.J. Leucine-rich amelogenin peptide: A candidate signaling molecule during cementogenesis. J. Periodontol. 2004, 75, 1126–1136. [Google Scholar] [CrossRef]
- Xia, Y.; Ren, A.; Pugach, M.K. Truncated amelogenin and LRAP transgenes improve Amelx null mouse enamel. Matrix Biol. 2016, 52, 198–206. [Google Scholar] [CrossRef]
- Le Norcy, E.; Lesieur, J.; Sadoine, J.; Rochefort, G.Y.; Chaussain, C.; Poliard, A. Phosphorylated and Non-phosphorylated Leucine Rich Amelogenin Peptide Differentially Affect Ameloblast Mineralization. Front. Physiol. 2018, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Kakegawa, A.; Yamakoshi, Y.; Tsuchiya, S.; Hu, J.C.; Gomi, K.; Arai, T.; Bartlett, J.D.; Simmer, J.P. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J. Dent. Res. 2009, 88, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Fincham, A.G.; Belcourt, A.B.; Termine, J.D.; Butler, W.T.; Cothran, W.C. Dental enamel matrix: Sequences of two amelogenin polypeptides. Biosci. Rep. 1981, 1, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Fincham, A.G.; Hu, Y.Y.; Pavlova, Z.; Slavkin, H.C.; Snead, M.L. Human amelogenins: Sequences of “TRAP” molecules. Calcif. Tissue Int. 1989, 45, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Brookes, S.J.; Robinson, C.; Kirkham, J.; Bonass, W.A. Biochemistry and molecular biology of amelogenin proteins of developing dental enamel. Arch. Oral Biol. 1995, 40, 1–14. [Google Scholar] [CrossRef]
- Baldassarri, M.; Margolis, H.C.; Beniash, E. Compositional determinants of mechanical properties of enamel. J. Dent. Res. 2008, 87, 645–649. [Google Scholar] [CrossRef]
- Smith, C.E.L.; Poulter, J.A.; Antanaviciute, A.; Kirkham, J.; Brookes, S.J.; Inglehearn, C.F.; Mighell, A.J. Amelogenesis Imperfecta; Genes, Proteins and Pathways. Front. Physiol. 2017, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.T.; Carrion, I.A.; Morris, C. The molecular basis of hereditary enamel defects in humans. J. Dent. Res. 2015, 94, 52–61. [Google Scholar] [CrossRef]
- Wright, J.T.; Robinson, C.; Kirkham, J. Enamel protein in smooth hypoplastic amelogenesis imperfecta. Pediatr. Dent. 1992, 14, 331–337. [Google Scholar]
- Wright, J.T.; Butler, W.T. Alteration of enamel proteins in hypomaturation amelogenesis imperfecta. J. Dent. Res. 1989, 68, 1328–1330. [Google Scholar] [CrossRef]
- Wright, J.T.; Hall, K.I.; Yamauche, M. The enamel proteins in human amelogenesis imperfecta. Arch. Oral Biol. 1997, 42, 149–159. [Google Scholar] [CrossRef]
- Takagi, Y.; Fujita, H.; Katano, H.; Shimokawa, H.; Kuroda, T. Immunochemical and biochemical characteristics of enamel proteins in hypocalcified amelogenesis imperfecta. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 1998, 85, 424–430. [Google Scholar] [CrossRef]
- Elhennawy, K.; Manton, D.J.; Crombie, F.; Zaslansky, P.; Radlanski, R.J.; Jost-Brinkmann, P.G.; Schwendicke, F. Structural, mechanical and chemical evaluation of molar-incisor hypomineralization-affected enamel: A systematic review. Arch. Oral Biol. 2017, 83, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.T.; Chen, S.C.; Hall, K.I.; Yamauchi, M.; Bawden, J.W. Protein characterization of fluorosed human enamel. J. Dent. Res. 1996, 75, 1936–1941. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, P.K. Effects of fluoride on protein secretion and removal during enamel development in the rat. J. Dent. Res. 1986, 65, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- DenBesten, P.; Li, W. Chronic fluoride toxicity: Dental fluorosis. Monogr. Oral Sci. 2011, 22, 81–96. [Google Scholar]
- Zhang, J.; Sardana, D.; Li, K.Y.; Leung, K.C.M.; Lo, E.C.M. Topical Fluoride to Prevent Root Caries: Systematic Review with Network Meta-analysis. J. Dent. Res. 2020, 99, 506–513. [Google Scholar] [CrossRef]
- Carey, C.M. Focus on fluorides: Update on the use of fluoride for the prevention of dental caries. J. Evid. Based Dent. Pract. 2014, 14, 95–102. [Google Scholar] [CrossRef]
- Griffin, S.O.; Regnier, E.; Griffin, P.M.; Huntley, V. Effectiveness of fluoride in preventing caries in adults. J. Dent. Res. 2007, 86, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Aoba, T.; Fejerskov, O. Dental fluorosis: Chemistry and biology. Crit. Rev. Oral Biol. Med. 2002, 13, 155–170. [Google Scholar] [CrossRef]
- Srivastava, S.; Flora, S.J.S. Fluoride in Drinking Water and Skeletal Fluorosis: A Review of the Global Impact. Curr. Environ. Health Rep. 2020, 7, 140–146. [Google Scholar] [CrossRef]
- Suzuki, M.; Ikeda, A.; Bartlett, J.D. Sirt1 overexpression suppresses fluoride-induced p53 acetylation to alleviate fluoride toxicity in ameloblasts responsible for enamel formation. Arch. Toxicol. 2018, 92, 1283–1293. [Google Scholar] [CrossRef]
- Suzuki, M.; Bartlett, J.D. Rodent Dental Fluorosis Model: Extraction of Enamel Organ from Rat Incisors. Methods Mol. Biol. 2019, 1922, 335–340. [Google Scholar] [PubMed]
- Bartlett, J.D.; Dwyer, S.E.; Beniash, E.; Skobe, Z.; Payne-Ferreira, T.L. Fluorosis: A new model and new insights. J. Dent. Res. 2005, 84, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Sierant, M.L.; Bartlett, J.D. Stress response pathways in ameloblasts: Implications for amelogenesis and dental fluorosis. Cells 2012, 1, 631–645. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Tye, C.E.; Arun, A.; MacDonald, D.; Chatterjee, A.; Abrazinski, T.; Everett, E.T.; Whitford, G.M.; Bartlett, J.D. Assessment of dental fluorosis in Mmp20 +/− mice. J. Dent. Res. 2011, 90, 788–792. [Google Scholar] [CrossRef]
- Lelis, I.M.P.; Molina, G.F.; Souza, C.; Perez, W.B.; Laure, H.J.; Rosa, J.C.; Gerlach, R.F. Peptide Characterization of Mature Fluorotic and Control Human Enamel. Braz. Dent. J. 2016, 27, 66–71. [Google Scholar] [CrossRef]
- Everett, E.T.; McHenry, M.A.; Reynolds, N.; Eggertsson, H.; Sullivan, J.; Kantmann, C.; Martinez-Mier, E.A.; Warrick, J.M.; Stookey, G.K. Dental fluorosis: Variability among different inbred mouse strains. J. Dent. Res. 2002, 81, 794–798. [Google Scholar] [CrossRef]
- Charone, S.; De Lima Leite, A.; Peres-Buzalaf, C.; Silva Fernandes, M.; Ferreira de Almeida, L.; Zardin Graeff, M.S.; Cardoso de Oliveira, R.; Campanelli, A.P.; Groisman, S.; Whitford, G.M.; et al. Proteomics of Secretory-Stage and Maturation-Stage Enamel of Genetically Distinct Mice. Caries Res. 2016, 50, 24–31. [Google Scholar] [CrossRef]
- Lima Leite, A.; Silva Fernandes, M.; Charone, S.; Whitford, G.M.; Everett, E.T.; Buzalaf, M.A.R. Proteomic Mapping of Dental Enamel Matrix from Inbred Mouse Strains: Unraveling Potential New Players in Enamel. Caries Res. 2018, 52, 78–87. [Google Scholar] [CrossRef]
- Ito, S.; Nagata, K. Biology of Hsp47 (Serpin H1), a collagen-specific molecular chaperone. Semin. Cell Dev. Biol. 2017, 62, 142–151. [Google Scholar] [CrossRef]
- Charone, S.; Kuchler, E.C.; Leite, A.L.; Silva Fernandes, M.; Taioqui Pela, V.; Martini, T.; Brondino, B.M.; Magalhaes, A.C.; Dionisio, T.J.; Carlos, F.S.; et al. Analysis of Polymorphisms in Genes Differentially Expressed in the Enamel of Mice with Different Genetic Susceptibilities to Dental Fluorosis. Caries Res. 2019, 53, 228–233. [Google Scholar] [CrossRef]
- Yamakoshi, Y.; Hu, J.C.; Zhang, H.; Iwata, T.; Yamakoshi, F.; Simmer, J.P. Proteomic analysis of enamel matrix using a two-dimensional protein fractionation system. Eur. J. Oral Sci. 2016, 114 (Suppl. 1), 266–271, discussion 285-6, 382. [Google Scholar] [CrossRef] [PubMed]
- Barre-Sinoussi, F.; Montagutelli, X. Animal models are essential to biological research: Issues and perspectives. Future Sci. OA 2015, 1, FSO63. [Google Scholar] [CrossRef] [PubMed]
- De Teruel, J.D.; Alcolea, A.; Hernandez, A.; Ruiz, A.J. Comparison of chemical composition of enamel and dentine in human, bovine, porcine and ovine teeth. Arch. Oral Biol. 2015, 60, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Pugach, M.K.; Gibson, C.W. Analysis of enamel development using murine model systems: Approaches and limitations. Front. Physiol. 2014, 5, 313. [Google Scholar] [CrossRef]
- Ortiz-Ruiz, A.J.; Teruel-Fernandez, J.D.; Alcolea-Rubio, L.A.; Hernandez-Fernandez, A.; Martinez-Beneyto, Y.; Gispert-Guirado, F. Structural differences in enamel and dentin in human, bovine, porcine and ovine teeth. Ann. Anat. 2018, 218, 7–17. [Google Scholar] [CrossRef]
- Bartlett, J.D.; Skobe, Z.; Nanci, A.; Smith, C.E. Matrix metalloproteinase 20 promotes a smooth enamel surface, a strong dentino-enamel junction and a decussating enamel rod pattern. Eur. J. Oral Sci. 2011, 119, 199–205. [Google Scholar] [CrossRef]
- Nunez, S.M.; Chun, Y.P.; Ganss, B.; Hu, Y.; Richardson, A.S.; Schmitz, J.E.; Fajardo, R.; Yang, J.; Hu, J.C.; Simmer, J.P. Maturation stage enamel malformations in Amtn and Klk4 null mice. Matrix Biol. 2016, 52, 219–233. [Google Scholar] [CrossRef]
- Hu, Y.; Smith, C.E.; Richardson, A.S.; Bartlett, J.D.; Hu, J.C.; Simmer, J.P. MMP20, KLK4 and MMP20/KLK4 double null mice define roles for matrix proteases during dental enamel formation. Mol. Genet. Genomic Med. 2016, 4, 178–196. [Google Scholar] [CrossRef]
- Pandya, M.; Liu, H.; Dangaria, S.J.; Zhu, W.; Li, L.L.; Pan, S.; Abufarwa, M.; Davis, R.G.; Guggenheim, S.; Keiderling, T.; et al. Integrative Temporo-Spatial, Mineralogic, Spectroscopic and Proteomic Analysis of Postnatal Enamel Development in Teeth with Limited Growth. Front. Physiol. 2017, 8, 793. [Google Scholar] [CrossRef]
- Robinson, C.; Kirkham, J.; Weatherell, J.A.; Richards, A.; Josephsen, K.; Fejerskov, O. Developmental stages in permanent porcine enamel. Acta Anat. (Basel) 1987, 128, 1–10. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Fang, D.; Shi, S. The miniature pig: A useful large animal model for dental and orofacial research. Oral Dis. 2007, 13, 530–537. [Google Scholar] [CrossRef]
- Fincham, A.G.; Moradian-Oldak, J.; Simmer, J.P. The structural biology of the developing dental enamel matrix. J. Struct. Biol. 1999, 126, 270–299. [Google Scholar] [CrossRef] [PubMed]
- Acil, Y.; Mobasseri, A.E.; Warnke, P.H.; Terheyden, H.; Wiltfang, J.; Springer, I. Detection of mature collagen in human dental enamel. Calcif. Tissue Int. 2005, 76, 121–126. [Google Scholar] [CrossRef] [PubMed]
- McGuire, J.D.; Walker, M.P.; Mousa, A.; Wang, Y.; Gorski, J.P. Type VII collagen is enriched in the enamel organic matrix associated with the dentin-enamel junction of mature human teeth. Bone 2014, 63, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Felszeghy, S.; Hollo, K.; Modis, L.; Lammi, M.J. Type X collagen in human enamel development: A possible role in mineralization. Acta Odontol. Scand. 2000, 58, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tannukit, S.; Zhu, D.; Snead, M.L.; Paine, M.L. Enamel matrix protein interactions. J. Bone Miner. Res. 2005, 20, 1032–1040. [Google Scholar] [CrossRef]
- Hubbard, M.J. Molar hypomineralization: What is the US experience? J. Am. Dent. Assoc. 2018, 149, 329–330. [Google Scholar] [CrossRef]
- Schwendicke, F.; Elhennawy, K.; Reda, S.; Bekes, K.; Manton, D.J.; Krois, J. Global burden of molar incisor hypomineralization. J. Dent. 2018, 68, 10–18. [Google Scholar] [CrossRef]
- Weerheijm, K.L.; Duggal, M.; Mejare, I.; Papagiannoulis, L.; Koch, G.; Martens, L.C.; Hallonsten, A.L. Judgement criteria for molar incisor hypomineralisation (MIH) in epidemiologic studies: A summary of the European meeting on MIH held in Athens, 2003. Eur. J. Paediatr. Dent. 2003, 4, 110–113. [Google Scholar]
- Almuallem, Z.; Busuttil-Naudi, A. Molar incisor hypomineralisation (MIH)—An overview. Br. Dent. J. 2018, 225, 601–609. [Google Scholar] [CrossRef]
- Rao, M.H.; Aluru, S.C.; Jayam, C.; Bandlapalli, A.; Patel, N. Molar Incisor Hypomineralization. J. Contemp. Dent. Pract. 2016, 17, 609–613. [Google Scholar]
- Hubbard, M.J.; Mangum, J.E.; Perez, V.A.; Nervo, G.J.; Hall, R.K. Molar Hypomineralisation: A Call to Arms for Enamel Researchers. Front. Physiol. 2017, 8, 546. [Google Scholar] [CrossRef]
- Fatturi, A.L.; Wambier, L.M.; Chibinski, A.C.; Assuncao, L.; Brancher, J.A.; Reis, A.; Souza, J.F. A systematic review and meta-analysis of systemic exposure associated with molar incisor hypomineralization. Community Dent. Oral Epidemiol. 2019, 47, 407–415. [Google Scholar] [CrossRef] [PubMed]
- The D3 Group (D3G) Whar Are Chalky Teeth? Available online: https://www.thed3group.org/what-are-chalky-teeth.html (accessed on 22 June 2020).
- Mohazab, L.; Koivisto, L.; Jiang, G.; Kytomaki, L.; Haapasalo, M.; Owen, G.R.; Wiebe, C.; Xie, Y.; Heikinheimo, K.; Yoshida, T.; et al. Critical role for alphavbeta6 integrin in enamel biomineralization. J. Cell Sci. 2013, 126 Pt 3, 732–744. [Google Scholar] [CrossRef]
- Masurier, N.; Arama, D.P.; El Amri, C.; Lisowski, V. Inhibitors of kallikrein-related peptidases: An overview. Med. Res. Rev. 2018, 38, 655–683. [Google Scholar] [CrossRef]
- Robinson, C.; Brookes, S.J.; Kirkham, J.; Shore, R.C.; Bonass, W.A. Uptake and metabolism of albumin by rodent incisor enamel in vivo and postmortem: Implications for control of mineralization by albumin. Calcif. Tissue Int. 1994, 55, 467–472. [Google Scholar] [CrossRef]
- Robinson, C.; Kirkham, J.; Fincham, A. The enamelin/non-amelogenin problem. A brief review. Connect. Tissue Res. 1989, 22, 93–100. [Google Scholar] [CrossRef]
- Nanci, A.; Fortin, M.; Ghitescu, L. Endocytotic functions of ameloblasts and odontoblasts: Immunocytochemical and tracer studies on the uptake of plasma proteins. Anat. Rec. 1996, 245, 219–234. [Google Scholar] [CrossRef]
- Robinson, C.; Shore, R.C.; Bonass, W.A.; Brookes, S.J.; Boteva, E.; Kirkham, J. Identification of human serum albumin in human caries lesions of enamel: The role of putative inhibitors of remineralisation. Caries Res. 1998, 32, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Menanteau, J.; Gregoire, M.; Daculsi, G.; Jans, I. In vitro albumin binding on apatite crystals from developing enamel. Bone Miner. 1987, 3, 137–141. [Google Scholar]
- Garnett, J.; Dieppe, P. The effects of serum and human albumin on calcium hydroxyapatite crystal growth. Biochem. J. 1990, 266, 863–868. [Google Scholar] [PubMed]
- Robinson, C.; Kirkham, J.; Brookes, S.J.; Shore, R.C. The role of albumin in developing rodent dental enamel: A possible explanation for white spot hypoplasia. J. Dent. Res. 1992, 71, 1270–1274. [Google Scholar] [CrossRef]
- Robinson, C.; Brookes, S.J.; Kirkham, J.; Bonass, W.A.; Shore, R.C. Crystal growth in dental enamel: The role of amelogenins and albumin. Adv. Dent. Res. 1996, 10, 173–179, discussion 179-80. [Google Scholar] [CrossRef]
- Hubbard, M.J.; Faught, M.J.; Carlisle, B.H.; Stockwell, P.A. ToothPrint, a proteomic database for dental tissues. Proteomics 2001, 1, 132–135. [Google Scholar] [CrossRef]
- Hubbard, M.J. Proteomic analysis of enamel cells from developing rat teeth: Big returns from a small tissue. Electrophoresis 1998, 19, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, M.J. Calbindin28kDa and calmodulin are hyperabundant in rat dental enamel cells. Identification of the protein phosphatase calcineurin as a principal calmodulin target and of a secretion-related role for calbindin28kDa. Eur. J. Biochem. 1995, 230, 68–79. [Google Scholar] [CrossRef]
- Hubbard, M.J. Abundant calcium homeostasis machinery in rat dental enamel cells. Up-regulation of calcium store proteins during enamel mineralization implicates the endoplasmic reticulum in calcium transcytosis. Eur. J. Biochem. 1996, 239, 611–623. [Google Scholar] [CrossRef]
- Hubbard, M.J.; McHugh, N.J. Mitochondrial ATP synthase F1-beta-subunit is a calcium-binding protein. FEBS Lett. 1996, 391, 323–329. [Google Scholar] [CrossRef]
- Hubbard, M.J.; Kon, J.C. Proteomic analysis of dental tissues. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002, 771, 211–220. [Google Scholar] [CrossRef]
- Mangum, J.E.; Veith, P.D.; Reynolds, E.C.; Hubbard, M.J. Towards second-generation proteome analysis of murine enamel-forming cells. Eur. J. Oral Sci. 2006, 114 (Suppl. 1), 259–265, discussion 285-286, 382. [Google Scholar] [CrossRef]
- Haag, A.M. Mass Analyzers and Mass Spectrometers. Adv. Exp. Med. Biol. 2016, 919, 157–169. [Google Scholar]

| Peptide(s) | Contemporary Molar (Trypsin) | Contemporary Molar (No Trypsin) | Mummy (Trypsin) | Mummy (No Trypsin) |
|---|---|---|---|---|
| IRPPYPSYGYEPMG | [45,46] | |||
| LPPHPGHPGYIN | [21,46] | [21] | ||
| LPPHPGHPGYINF | [46] | [45,46] | ||
| LPPHPGHPGYINFSYEVLTPLK | [46] | [45] | ||
| M(ox)PLPPHPGH (AMELX/AMELY) | [43,46] | [43,46] | ||
| M(ox)PLPPHPGHPGYINF | [46] | [43,46] | [43] | |
| MPLPPHPGHPG | [21,46] | [45,46] | ||
| MPLPPHPGHPGYIN | [46] | [45,46] | ||
| MPLPPHPGHPGYINF | [45,46] | [21] | ||
| MPLPPHPGHPGYINFSYEVLTPLK | [46] | [45] | ||
| PHPGHPGYINF | [46] | [45,46] | [46] | |
| SIRPPYPSY | [46] | [43,46] | [43,44,46] | |
| SIRPPYPSYGYEP | [45,46] | |||
| SIRPPYPSYGYEPM | [45,46] | |||
| SIRPPYPSYGYEPMG | [43,45,46] | [43] | ||
| SM(ox)IRPPY (AMELY) | [43,46] | [43,44] | ||
| SYEVLTPLK (AMELX/AMELY) | [46] | [43,45,46] | [43,46] | |
| SYEVLTPLKWYQSIRPPYP | [46] | [43] | [43] | |
| WYQSIRPPYP | [46] | [21] | ||
| YEVLTPLK | [46] | [45,46] | [46] | |
| YEVLTPLKWY (AMELX/AMELY) | [43,46] | [43,46] |
| Condition | Proteins (Peptides Identified) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amelogenin, X Isoform | Amelogenin, Y Isoform | Ameloblastin | Enamelin | Serum Albumin | Hemoglobin Subunit Alpha | Hemoglobin Subunit Beta | Collagen Alpha-1(I) Chain | Collagen Alpha-1(III) Chain | Collagen Alpha-2(I) Chain | Antithrombin-III | Alpha-1-Antitrypsin | |
| Contemporary (trypsin) | [21,46] | [21,46] | [46,47,48] | [46] | [46,47,48] | [46,47] | [46,47] | [46,47,48] | [47] | [46,47,48] | [47] | [46,47,48] |
| Contemporary (no trypsin) | [43,44,45,46] | [43,44,45,46] | [43,44,45,46] | [43,44,45,46] | [45,46] | [46] | [46] | [43] | [45] | |||
| Mummy (no trypsin) | [43,46] | [43,46] | [43,46] | [43,46] | [43,46] | |||||||
| Mummy (trypsin) | [21] | |||||||||||
| Model | References | Pros | Cons |
|---|---|---|---|
| Human: modern, archeological  | [2,3,21,34,37,40,41,43,44,45,46,47,48,49,50,51,52,54,86,88,89,90,91,92,93,95,99,100,106] |
|
|
Pig/miniature pig | [15,35,36,71,112,121,122] |
|
|
Mouse/rat | [37,56,57,58,75,94,102,105,107,108,109,111,115,117,118,119,120] |
|
|
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil-Bona, A.; Bidlack, F.B. Tooth Enamel and Its Dynamic Protein Matrix. Int. J. Mol. Sci. 2020, 21, 4458. https://doi.org/10.3390/ijms21124458
Gil-Bona A, Bidlack FB. Tooth Enamel and Its Dynamic Protein Matrix. International Journal of Molecular Sciences. 2020; 21(12):4458. https://doi.org/10.3390/ijms21124458
Chicago/Turabian StyleGil-Bona, Ana, and Felicitas B. Bidlack. 2020. "Tooth Enamel and Its Dynamic Protein Matrix" International Journal of Molecular Sciences 21, no. 12: 4458. https://doi.org/10.3390/ijms21124458
APA StyleGil-Bona, A., & Bidlack, F. B. (2020). Tooth Enamel and Its Dynamic Protein Matrix. International Journal of Molecular Sciences, 21(12), 4458. https://doi.org/10.3390/ijms21124458




