YBX1 Indirectly Targets Heterochromatin-Repressed Inflammatory Response-Related Apoptosis Genes through Regulating CBX5 mRNA
Abstract
1. Introduction
2. Results
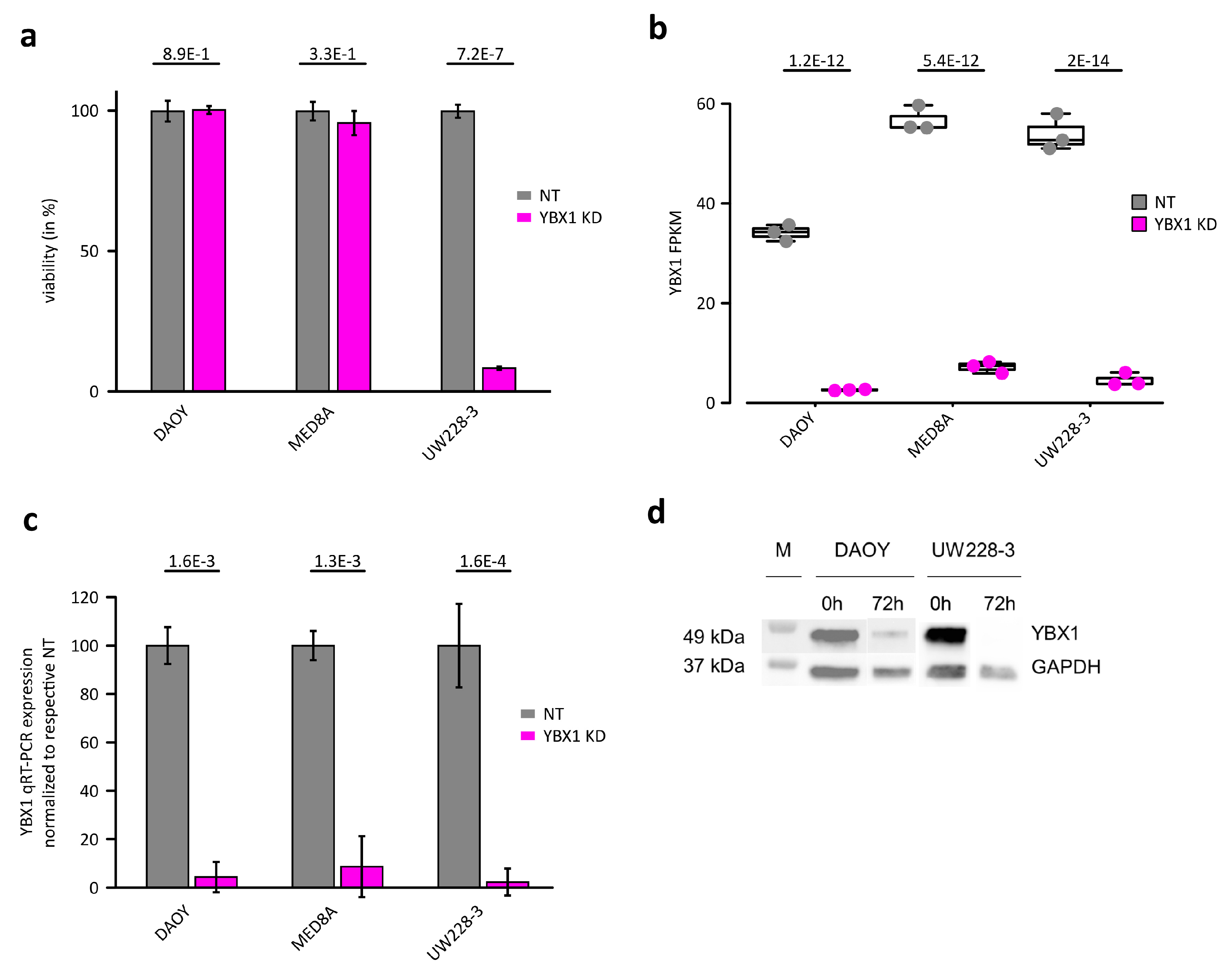
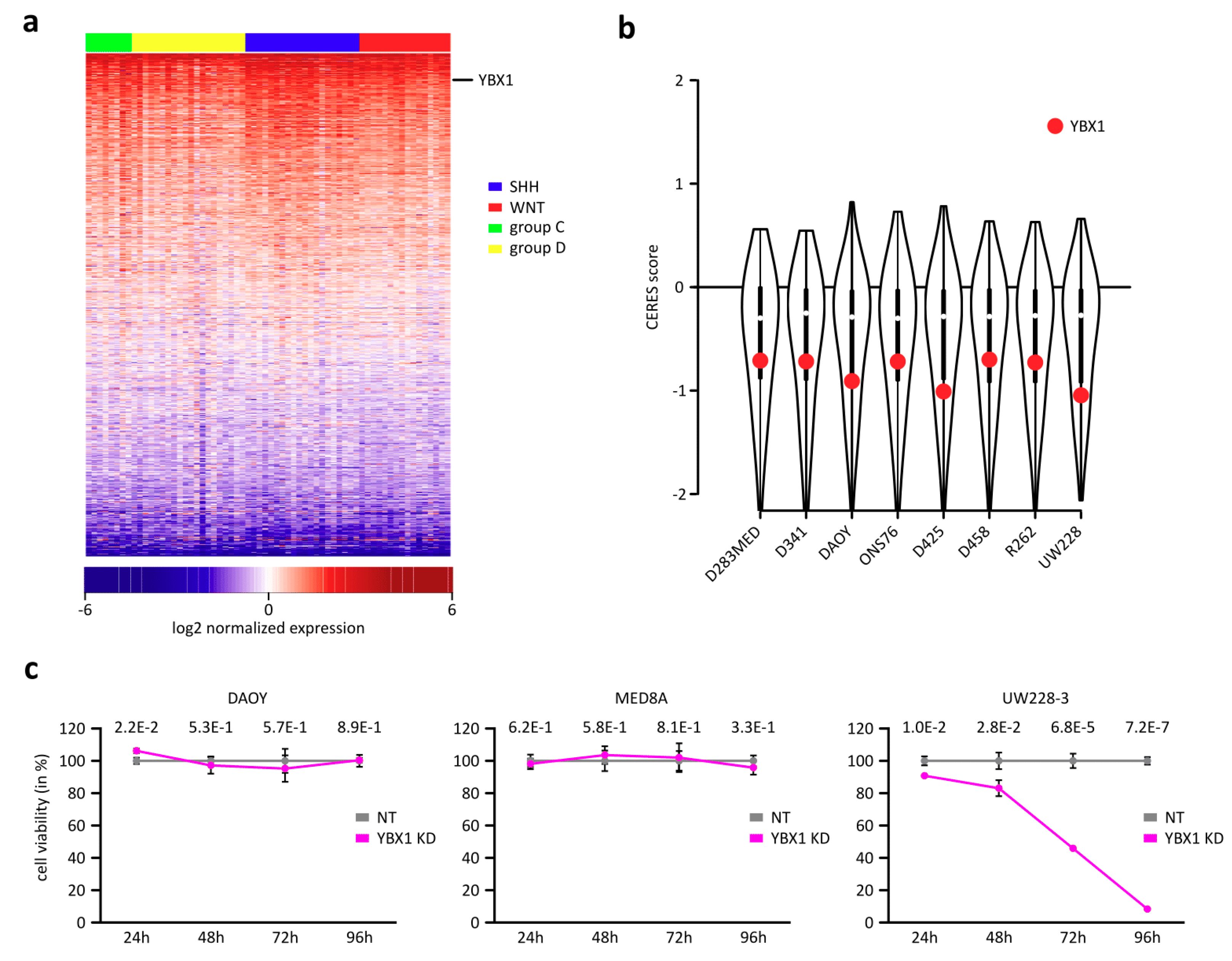
2.1. YBX1 Is an Essential RBP for Cell Survival in Medulloblastomas
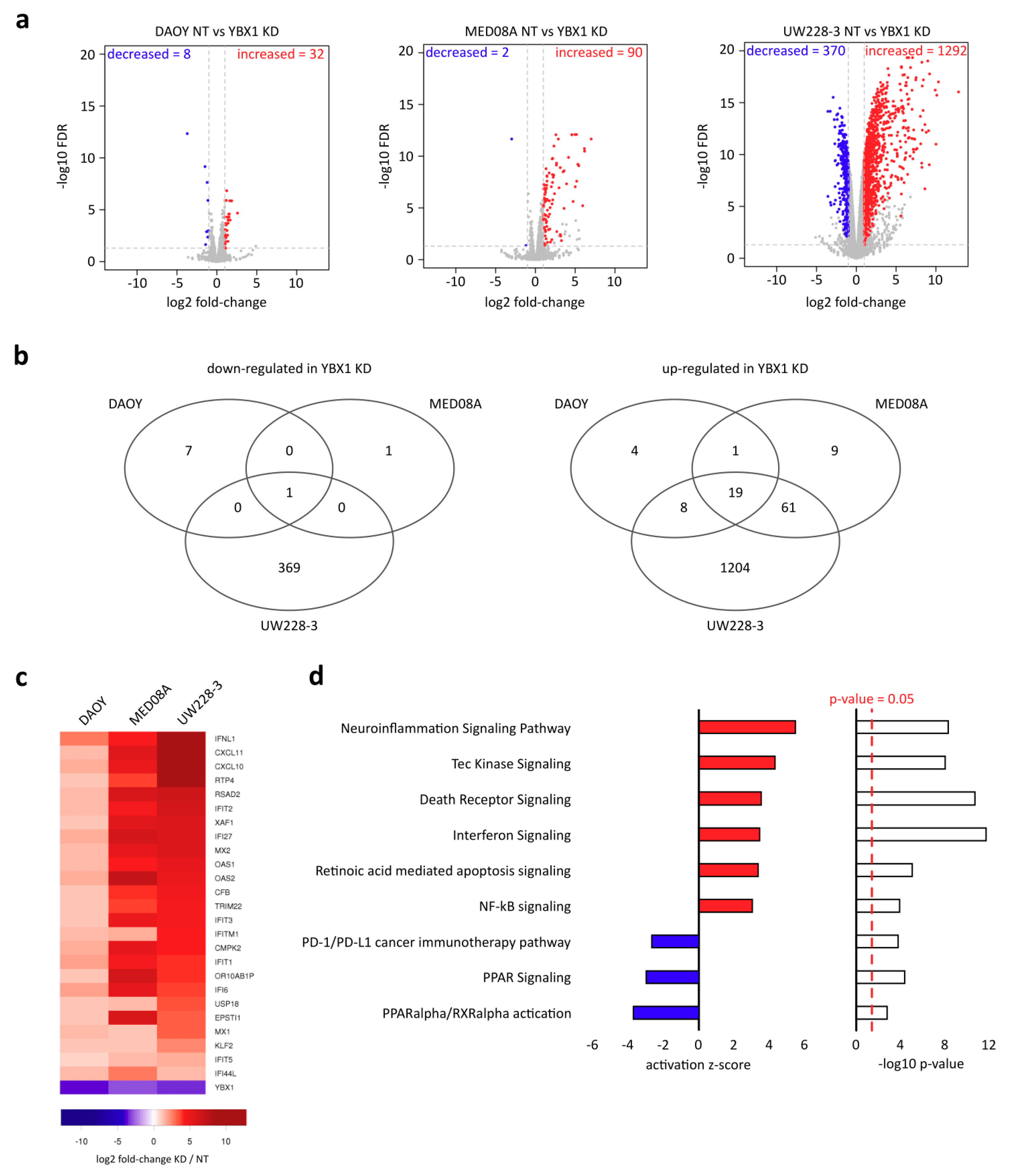
2.2. YBX1 Controls Inflammatory Response and Apoptosis Genes
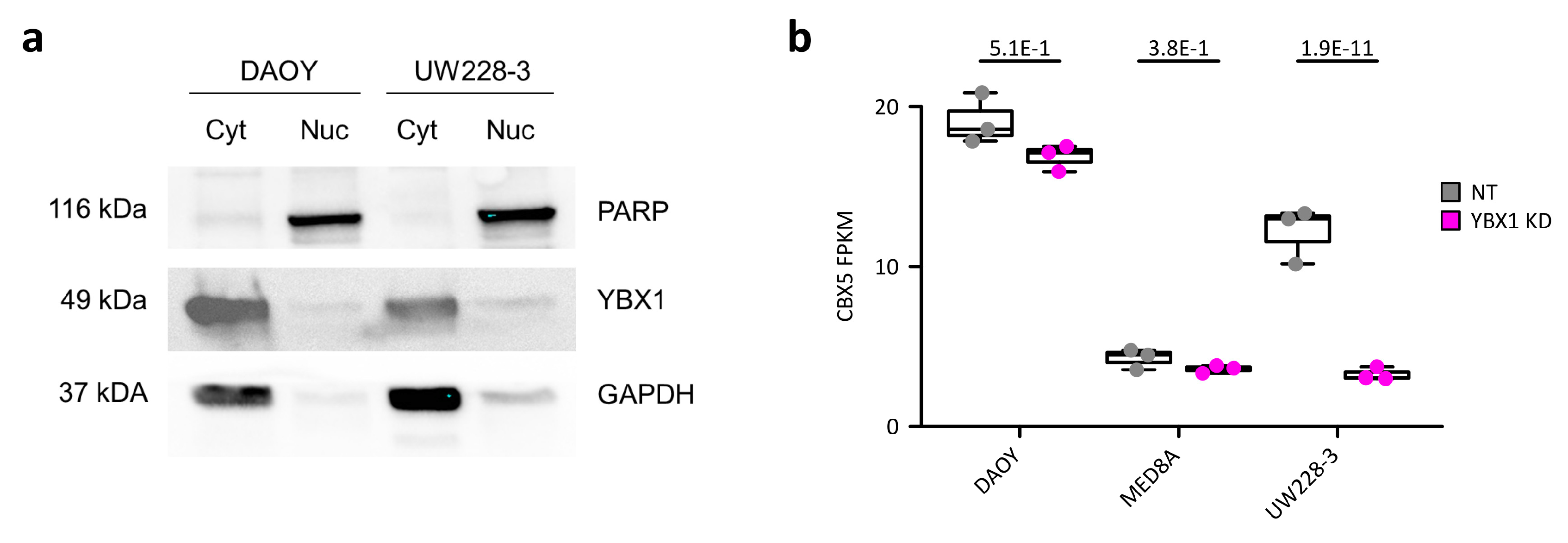
2.3. YBX1 Targets 3′UTRs and Indirectly Regulates Heterochromatin-Repressed Apoptosis-Related Inflammatory Response Genes
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. qRT-PCR
4.3. Western Blot and Protein Localization
4.4. siRNA-Mediated Knockdown
4.5. Cell Viability Assay
4.6. RNA-Seq
4.7. PAR-CLIP
4.8. Bioinformatics
4.8.1. RNA-Seq
4.8.2. Pathway Analysis
4.8.3. PAR-CLIP
4.8.4. ChIP-Seq Data Integration
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CBX5 | Chromobox protein homolog 5 |
| FDR | False discovery rate |
| H3K9me3 | Histone 3 K9 tri-methylation |
| IPA | Ingenuity pathway analysis |
| KD | Knockdown |
| PAR-CLIP | Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation |
| PBS | Phosphate buffered saline |
| RBP | RNA binding protein |
| RNA-seq | RNA sequencing |
| ChIP-seq | Chromatin-immunoprecipitation and sequencing |
| Ssh | Sonic hedgehog signaling |
| Wnt | Wnt signaling |
| YBX1 | Y-box binding protein 1 |
Appendix A

References
- Azzarelli, R.; Simons, B.D.; Philpott, A. The developmental origin of brain tumours: A cellular and molecular framework. Development 2018, 145. [Google Scholar] [CrossRef] [PubMed]
- Bautista, F.; Fioravantti, V.; de Rojas, T.; Carceller, F.; Madero, L.; Lassaletta, A.; Moreno, L. Medulloblastoma in children and adolescents: A systematic review of contemporary phase I and II clinical trials and biology update. Cancer Med. 2017, 6, 2606–2624. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Liao, P.; Rouse, C.; Chen, Y.; Dowling, J.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro-Oncology 2014, 16 (Suppl. 4), iv1–iv63. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro-Oncology 2019, 21, v1–v100. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e736. [Google Scholar] [CrossRef]
- Schuller, U.; Heine, V.M.; Mao, J.; Kho, A.T.; Dillon, A.K.; Han, Y.G.; Huillard, E.; Sun, T.; Ligon, A.H.; Qian, Y.; et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 2008, 14, 123–134. [Google Scholar] [CrossRef]
- Yang, Z.J.; Ellis, T.; Markant, S.L.; Read, T.A.; Kessler, J.D.; Bourboulas, M.; Schuller, U.; Machold, R.; Fishell, G.; Rowitch, D.H.; et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 2008, 14, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Phoenix, T.N.; Patmore, D.M.; Boop, S.; Boulos, N.; Jacus, M.O.; Patel, Y.T.; Roussel, M.F.; Finkelstein, D.; Goumnerova, L.; Perreault, S.; et al. Medulloblastoma Genotype Dictates Blood Brain Barrier Phenotype. Cancer Cell 2016, 29, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, D.; Robinson, G.; Uziel, T.; Gibson, P.; Rehg, J.; Gao, C.; Finkelstein, D.; Qu, C.; Pounds, S.; Ellison, D.W.; et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell 2012, 21, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Moore, C.E.; Wang, J.; Tewari, A.K.; Eroshkin, A.; Cho, Y.J.; Witt, H.; Korshunov, A.; Read, T.A.; Sun, J.L.; et al. An animal model of MYC-driven medulloblastoma. Cancer Cell 2012, 21, 155–167. [Google Scholar] [CrossRef]
- Wang, J.; Wechsler-Reya, R.J. The role of stem cells and progenitors in the genesis of medulloblastoma. Exp. Neurol. 2014, 260, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Erkek, S.; Tong, Y.; Yin, L.; Federation, A.J.; Zapatka, M.; Haldipur, P.; Kawauchi, D.; Risch, T.; Warnatz, H.J.; et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 2016, 530, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Johnston, D.L.; Keene, D.; Strother, D.; Taneva, M.; Lafay-Cousin, L.; Fryer, C.; Scheinemann, K.; Carret, A.S.; Fleming, A.; Afzal, S.; et al. Survival Following Tumor Recurrence in Children With Medulloblastoma. J. Pediatr. Hematol. Oncol. 2018, 40, e159–e163. [Google Scholar] [CrossRef] [PubMed]
- Moxon-Emre, I.; Taylor, M.D.; Bouffet, E.; Hardy, K.; Campen, C.J.; Malkin, D.; Hawkins, C.; Laperriere, N.; Ramaswamy, V.; Bartels, U.; et al. Intellectual Outcome in Molecular Subgroups of Medulloblastoma. J. Clin. Oncol. 2016, 34, 4161–4170. [Google Scholar] [CrossRef]
- Thomas, A.; Noel, G. Medulloblastoma: Optimizing care with a multidisciplinary approach. J. Multidiscip. Healthc. 2019, 12, 335–347. [Google Scholar] [CrossRef]
- Moxon-Emre, I.; Bouffet, E.; Taylor, M.D.; Laperriere, N.; Scantlebury, N.; Law, N.; Spiegler, B.J.; Malkin, D.; Janzen, L.; Mabbott, D. Impact of craniospinal dose, boost volume, and neurologic complications on intellectual outcome in patients with medulloblastoma. J. Clin. Oncol. 2014, 32, 1760–1768. [Google Scholar] [CrossRef]
- Ladomery, M.; Sommerville, J. A role for Y-box proteins in cell proliferation. Bioessays 1995, 17, 9–11. [Google Scholar] [CrossRef]
- Zou, F.; Tu, R.; Duan, B.; Yang, Z.; Ping, Z.; Song, X.; Chen, S.; Price, A.; Li, H.; Scott, A.; et al. Drosophila YBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs. Proc. Natl. Acad. Sci. USA 2020, 117, 3603–3609. [Google Scholar] [CrossRef]
- Ray, D.; Kazan, H.; Chan, E.T.; Castillo, L.P.; Chaudhry, S.; Talukder, S.; Blencowe, B.J.; Morris, Q.; Hughes, T.R. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009, 27, 667–670. [Google Scholar] [CrossRef]
- Wei, W.-J.; Mu, S.-R.; Heiner, M.; Fu, X.; Cao, L.-J.; Gong, X.-F.; Bindereif, A.; Hui, J. YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res. 2012, 40, 8622–8636. [Google Scholar] [CrossRef]
- Wu, S.-L.; Fu, X.; Huang, J.; Jia, T.-T.; Zong, F.-Y.; Mu, S.-R.; Zhu, H.; Yan, Y.; Qiu, S.; Wu, Q. Genome-wide analysis of YB-1-RNA interactions reveals a novel role of YB-1 in miRNA processing in glioblastoma multiforme. Nucleic Acids Res. 2015, 43, 8516–8528. [Google Scholar] [CrossRef]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.H.; Books, J.T.; Ley, T.J. YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol. Cell. Biol. 2005, 25, 4625–4637. [Google Scholar] [CrossRef] [PubMed]
- Evdokimova, V.; Ruzanov, P.; Imataka, H.; Raught, B.; Svitkin, Y.; Ovchinnikov, L.P.; Sonenberg, N. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 2001, 20, 5491–5502. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, M.P.; Ivshina, M.P.; Chernov, K.G.; Kovrigina, E.A.; Evdokimova, V.M.; Thomas, A.A.; Hershey, J.W.; Ovchinnikov, L.P. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J. Biol. Chem. 2003, 278, 13936–13943. [Google Scholar] [CrossRef] [PubMed]
- Coles, L.S.; Bartley, M.A.; Bert, A.; Hunter, J.; Polyak, S.; Diamond, P.; Vadas, M.A.; Goodall, G.J. A multi-protein complex containing cold shock domain (Y-box) and polypyrimidine tract binding proteins forms on the vascular endothelial growth factor mRNA: Potential role in mRNA stabilization. Eur. J. Biochem. 2004, 271, 648–660. [Google Scholar] [CrossRef]
- Dutertre, M.; Sanchez, G.; De Cian, M.-C.; Barbier, J.; Dardenne, E.; Gratadou, L.; Dujardin, G.; Le Jossic-Corcos, C.; Corcos, L.; Auboeuf, D. Cotranscriptional exon skipping in the genotoxic stress response. Nat. Struct. Mol. Biol. 2010, 17, 1358. [Google Scholar] [CrossRef] [PubMed]
- Skalweit, A.; Doller, A.; Huth, A.; Kähne, T.; Persson, P.B.; Thiele, B.-J. Posttranscriptional control of renin synthesis: Identification of proteins interacting with renin mRNA 3′-untranslated region. Circ. Res. 2003, 92, 419–427. [Google Scholar] [CrossRef]
- Lyabin, D.N.; Eliseeva, I.A.; Ovchinnikov, L.P. YB-1 protein: Functions and regulation. Wiley Interdiscip. Rev. RNA 2014, 5, 95–110. [Google Scholar] [CrossRef]
- Faury, D.; Nantel, A.; Dunn, S.E.; Guiot, M.-C.; Haque, T.; Hauser, P.; Garami, M.; Bognár, L.; Hanzély, Z.; Liberski, P.P. Molecular profiling identifies prognostic subgroups of pediatric glioblastoma and shows increased YB-1 expression in tumors. J. Clin. Oncol. 2007, 25, 1196–1208. [Google Scholar] [CrossRef]
- Chatterjee, M.; Rancso, C.; Stühmer, T.; Eckstein, N.; Andrulis, M.; Gerecke, C.; Lorentz, H.; Royer, H.-D.; Bargou, R.C. The Y-box binding protein YB-1 is associated with progressive disease and mediates survival and drug resistance in multiple myeloma. Blood 2008, 111, 3714–3722. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Kawahara, A.; Basaki, Y.; Hattori, S.; Nakashima, K.; Nakano, K.; Shirouzu, K.; Kohno, K.; Yanagawa, T.; Yamana, H. Expression of HER2 and estrogen receptor α depends upon nuclear localization of Y-box binding protein-1 in human breast cancers. Cancer Res. 2008, 68, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Mantwill, K.; Naumann, U.; Seznec, J.; Girbinger, V.; Lage, H.; Surowiak, P.; Beier, D.; Mittelbronn, M.; Schlegel, J.; Holm, P.S. YB-1 dependent oncolytic adenovirus efficiently inhibits tumor growth of glioma cancer stem like cells. J. Transl. Med. 2013, 11, 216. [Google Scholar] [CrossRef] [PubMed]
- Stratford, A.L.; Habibi, G.; Astanehe, A.; Jiang, H.; Hu, K.; Park, E.; Shadeo, A.; Buys, T.P.; Lam, W.; Pugh, T. Epidermal growth factor receptor (EGFR) is transcriptionally induced by the Y-box binding protein-1 (YB-1) and can be inhibited with Iressa in basal-like breast cancer, providing a potential target for therapy. Breast Cancer Res. 2007, 9, R61. [Google Scholar] [CrossRef] [PubMed]
- Remke, M.; Hielscher, T.; Korshunov, A.; Northcott, P.A.; Bender, S.; Kool, M.; Westermann, F.; Benner, A.; Cin, H.; Ryzhova, M. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J. Clin. Oncol. 2011, 2011, 2036–2798. [Google Scholar] [CrossRef]
- Liao, J.-Y.; Yang, B.; Zhang, Y.-C.; Wang, X.-J.; Ye, Y.; Peng, J.-W.; Yang, Z.-Z.; He, J.-H.; Zhang, Y.; Hu, K. EuRBPDB: A comprehensive resource for annotation, functional and oncological investigation of eukaryotic RNA binding proteins (RBPs). Nucleic Acids Res. 2020, 48, D307–D313. [Google Scholar] [CrossRef]
- Behan, F.M.; Iorio, F.; Picco, G.; Goncalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Jacobsen, P.F.; Jenkyn, D.J.; Papadimitriou, J.M. Establishment of a human medulloblastoma cell line and its heterotransplantation into nude mice. J. Neuropathol. Exp. Neurol. 1985, 44, 472–485. [Google Scholar] [CrossRef]
- Keles, G.E.; Berger, M.S.; Srinivasan, J.; Kolstoe, D.D.; Bobola, M.S.; Silber, J.R. Establishment and characterization of four human medulloblastoma-derived cell lines. Oncol. Res. 1995, 7, 493–503. [Google Scholar]
- Bazan, N.G.; Halabi, A.; Ertel, M.; Petasis, N.A. Neuroinflammation. In Basic Neurochemistry; Elsevier: Amsterdam, The Netherlands, 2012; pp. 610–620. [Google Scholar]
- Bannister, A.J.; Zegerman, P.; Partridge, J.F.; Miska, E.A.; Thomas, J.O.; Allshire, R.C.; Kouzarides, T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001, 410, 120–124. [Google Scholar] [CrossRef]
- Kwon, S.H.; Workman, J.L. The heterochromatin protein 1 (HP1) family: Put away a bias toward HP1. Mol. Cells (Springer Sci. Bus. Media BV) 2008, 26, 217–227. [Google Scholar]
- Dey, A.; Robitaille, M.; Remke, M.; Maier, C.; Malhotra, A.; Gregorieff, A.; Wrana, J.L.; Taylor, M.D.; Angers, S.; Kenney, A.M. YB-1 is elevated in medulloblastoma and drives proliferation in Sonic hedgehog-dependent cerebellar granule neuron progenitor cells and medulloblastoma cells. Oncogene 2016, 35, 4256–4268. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Li, Q.; Bing, Z.; Zhang, Y.; Niu, L.; Yin, H.; Yuan, G.; Pan, Y. Comprehensive analysis of a microRNA expression profile in pediatric medulloblastoma. Mol. Med. Rep. 2017, 15, 4109–4115. [Google Scholar] [CrossRef] [PubMed]
- Kuwano, M.; Shibata, T.; Watari, K.; Ono, M. Oncogenic Y-box binding protein-1 as an effective therapeutic target in drug-resistant cancer. Cancer Sci. 2019, 110, 1536–1543. [Google Scholar] [CrossRef]
- Di, C.; Liao, S.; Adamson, D.C.; Parrett, T.J.; Broderick, D.K.; Shi, Q.; Lengauer, C.; Cummins, J.M.; Velculescu, V.E.; Fults, D.W. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005, 65, 919–924. [Google Scholar] [PubMed]
- Chang, Q.; Chen, Z.; You, J.; McNutt, M.A.; Zhang, T.; Han, Z.; Zhang, X.; Gong, E.; Gu, J. All-trans-retinoic acid induces cell growth arrest in a human medulloblastoma cell line. J. Neuro-Oncol. 2007, 84, 263–267. [Google Scholar] [CrossRef]
- Hallahan, A.R.; Pritchard, J.I.; Chandraratna, R.A.; Ellenbogen, R.G.; Geyer, J.R.; Overland, R.P.; Strand, A.D.; Tapscott, S.J.; Olson, J.M. BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat. Med. 2003, 9, 1033–1038. [Google Scholar] [CrossRef]
- Gumireddy, K.; Sutton, L.N.; Phillips, P.C.; Reddy, C.D. All-trans-Retinoic Acid-induced Apoptosis in Human Medulloblastoma Activation of Caspase-3/Poly (ADP-ribose) Polymerase 1 Pathway. Clin. Cancer Res. 2003, 9, 4052–4059. [Google Scholar]
- Liu, J.; Guo, L.; Jun-Wei, L.; Liu, N.; Li, H. All-trans retinoic acid modulates fas expression and enhances chemosensitivity of human medulloblastoma cells. Int. J. Mol. Med. 2000, 5, 145–154. [Google Scholar] [CrossRef]
- Tao, Z.; Ruan, H.; Sun, L.; Kuang, D.; Song, Y.; Wang, Q.; Wang, T.; Hao, Y.; Chen, K. Targeting the YB-1/PD-L1 Axis to Enhance Chemotherapy and Antitumor Immunity. Cancer Immunol. Res. 2019, 7, 1135–1147. [Google Scholar] [CrossRef]
- Ivanov, D.P.; Coyle, B.; Walker, D.A.; Grabowska, A.M. In vitro models of medulloblastoma: Choosing the right tool for the job. J. Biotechnol. 2016, 236, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Saksouk, N.; Hajdari, S.; Perez, Y.; Pratlong, M.; Barrachina, C.; Graber, C.; Grégoire, D.; Zavoriti, A.; Sarrazin, A.; Pirot, N. The mouse HP1 proteins are essential for preventing liver tumorigenesis. Oncogene 2020, 39, 2676–2691. [Google Scholar] [CrossRef]
- Higashijima, Y.; Matsui, Y.; Shimamura, T.; Nakaki, R.; Nagai, N.; Tsutsumi, S.; Abe, Y.; Link, V.M.; Osaka, M.; Yoshida, M. Coordinated demethylation of H3K9 and H3K27 is required for rapid inflammatory responses of endothelial cells. EMBO J. 2020. [Google Scholar] [CrossRef]
- Khan, M.I.; Adhami, V.M.; Lall, R.K.; Sechi, M.; Joshi, D.C.; Haidar, O.M.; Syed, D.N.; Siddiqui, I.A.; Chiu, S.Y.; Mukhtar, H. YB-1 expression promotes epithelial-to-mesenchymal transition in prostate cancer that is inhibited by a small molecule fisetin. Oncotarget 2014, 5, 2462–2474. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Hannus, M.; Beitzinger, M.; Engelmann, J.C.; Weickert, M.-T.; Spang, R.; Hannus, S.; Meister, G. siPools: Highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014, 42, 8049–8061. [Google Scholar] [CrossRef]
- Spitzer, J.; Hafner, M.; Landthaler, M.; Ascano, M.; Farazi, T.; Wardle, G.; Nusbaum, J.; Khorshid, M.; Burger, L.; Zavolan, M.; et al. PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation): A step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzym. 2014, 539, 113–161. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kloetgen, A.; Borkhardt, A.; Hoell, J.I.; McHardy, A.C. The PARA-suite: PAR-CLIP specific sequence read simulation and processing. PeerJ 2016, 4, e2619. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Dündar, F.; Diehl, S.; Grüning, B.A.; Manke, T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kloetgen, A.; Duggimpudi, S.; Schuschel, K.; Hezaveh, K.; Picard, D.; Schaal, H.; Remke, M.; Klusmann, J.-H.; Borkhardt, A.; McHardy, A.C.; et al. YBX1 Indirectly Targets Heterochromatin-Repressed Inflammatory Response-Related Apoptosis Genes through Regulating CBX5 mRNA. Int. J. Mol. Sci. 2020, 21, 4453. https://doi.org/10.3390/ijms21124453
Kloetgen A, Duggimpudi S, Schuschel K, Hezaveh K, Picard D, Schaal H, Remke M, Klusmann J-H, Borkhardt A, McHardy AC, et al. YBX1 Indirectly Targets Heterochromatin-Repressed Inflammatory Response-Related Apoptosis Genes through Regulating CBX5 mRNA. International Journal of Molecular Sciences. 2020; 21(12):4453. https://doi.org/10.3390/ijms21124453
Chicago/Turabian StyleKloetgen, Andreas, Sujitha Duggimpudi, Konstantin Schuschel, Kebria Hezaveh, Daniel Picard, Heiner Schaal, Marc Remke, Jan-Henning Klusmann, Arndt Borkhardt, Alice C. McHardy, and et al. 2020. "YBX1 Indirectly Targets Heterochromatin-Repressed Inflammatory Response-Related Apoptosis Genes through Regulating CBX5 mRNA" International Journal of Molecular Sciences 21, no. 12: 4453. https://doi.org/10.3390/ijms21124453
APA StyleKloetgen, A., Duggimpudi, S., Schuschel, K., Hezaveh, K., Picard, D., Schaal, H., Remke, M., Klusmann, J.-H., Borkhardt, A., McHardy, A. C., & Hoell, J. I. (2020). YBX1 Indirectly Targets Heterochromatin-Repressed Inflammatory Response-Related Apoptosis Genes through Regulating CBX5 mRNA. International Journal of Molecular Sciences, 21(12), 4453. https://doi.org/10.3390/ijms21124453




