Antibiotics Act with vB_AbaP_AGC01 Phage against Acinetobacter baumannii in Human Heat-Inactivated Plasma Blood and Galleria mellonella Models
Abstract
1. Introduction
2. Results
2.1. Bacteriophage Isolation and Host Range
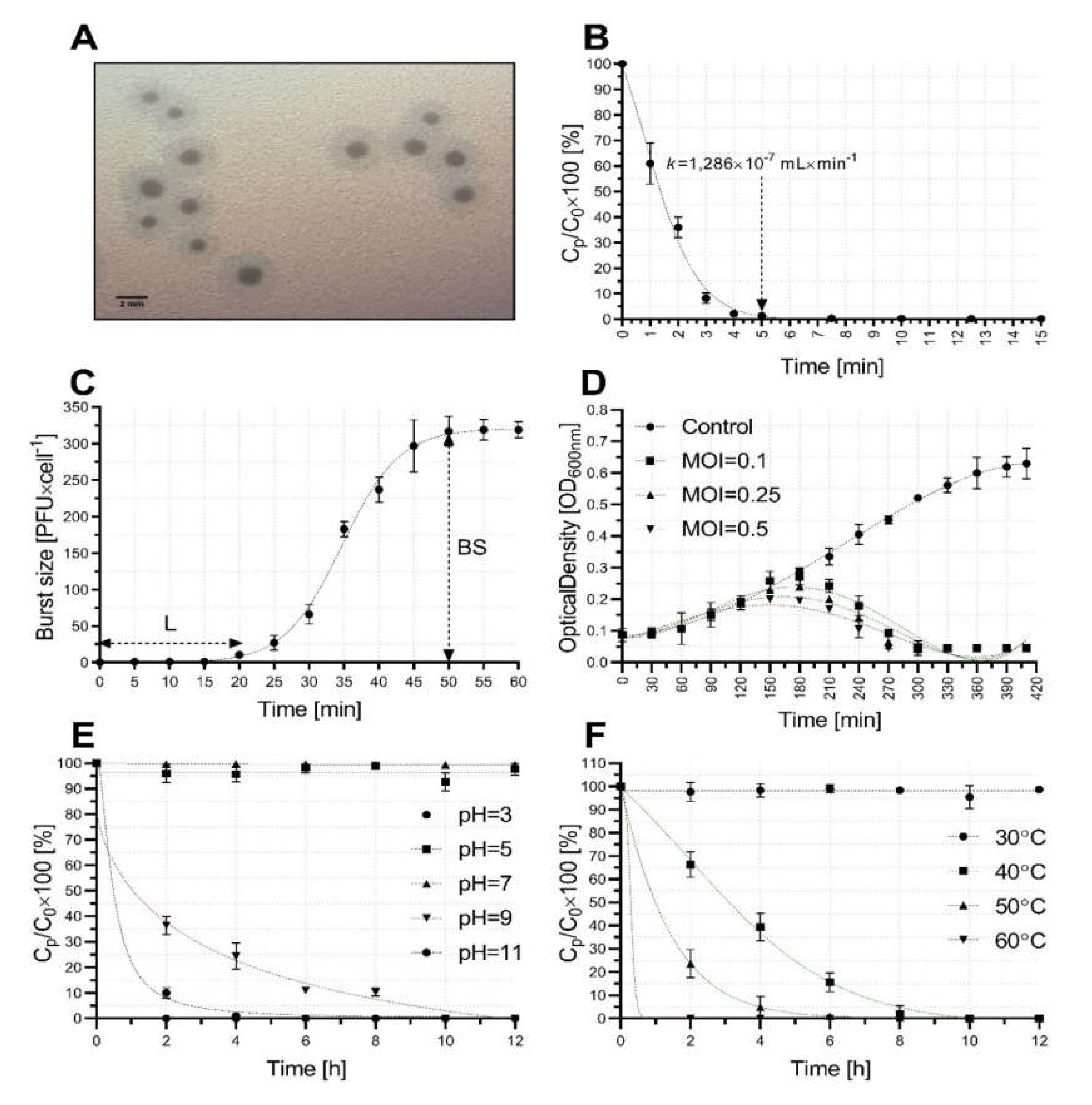
2.2. Bacteriophage Growth and Stability Characterization
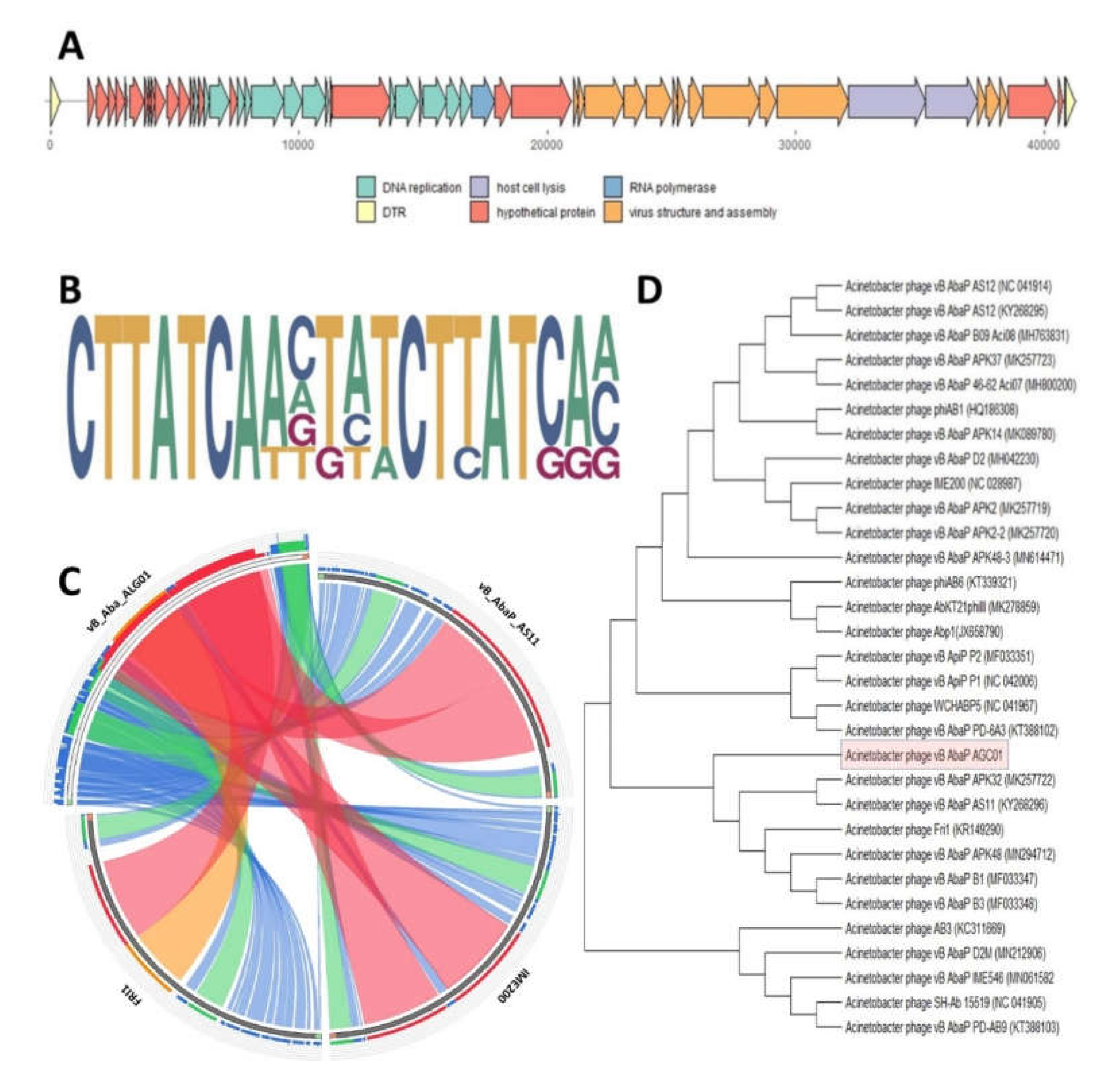
2.3. Bacteriophage Comparative Genomics and Genome Analysis
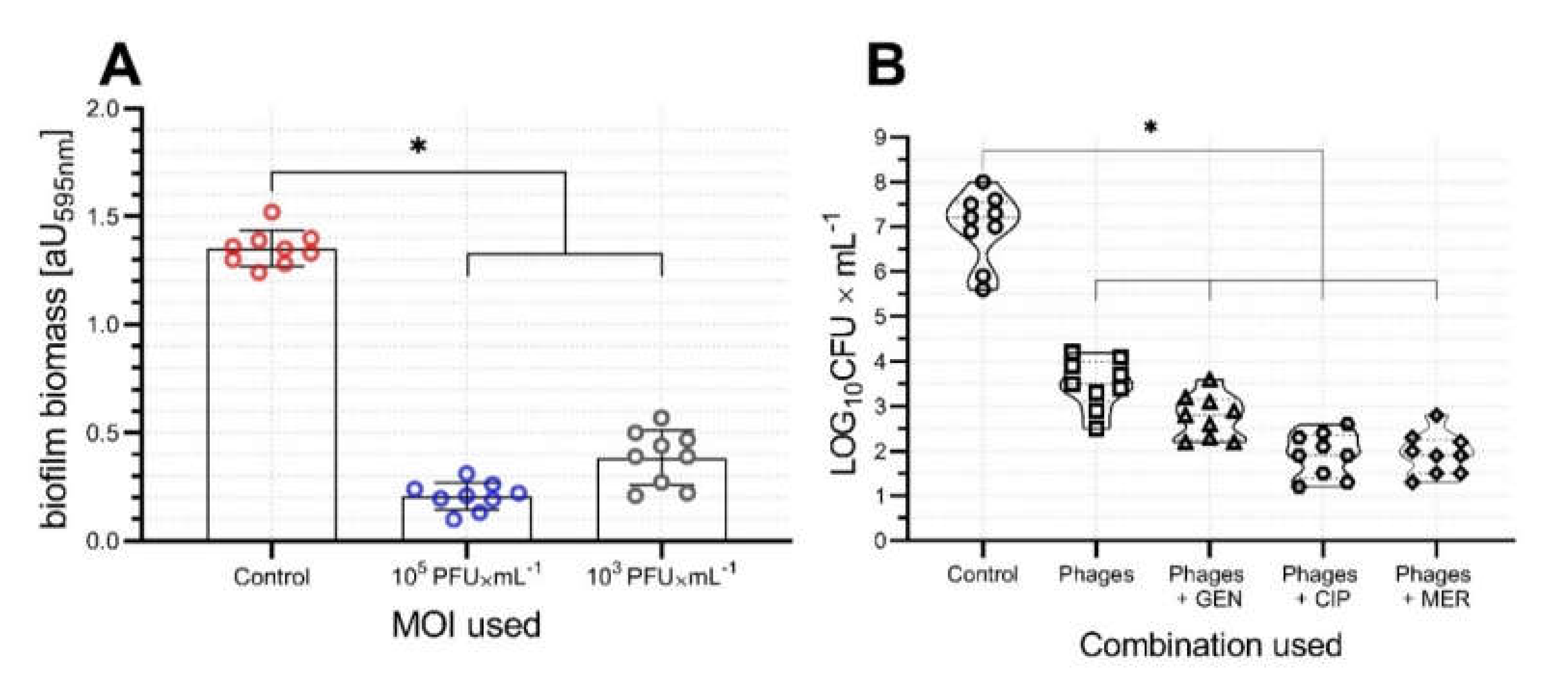
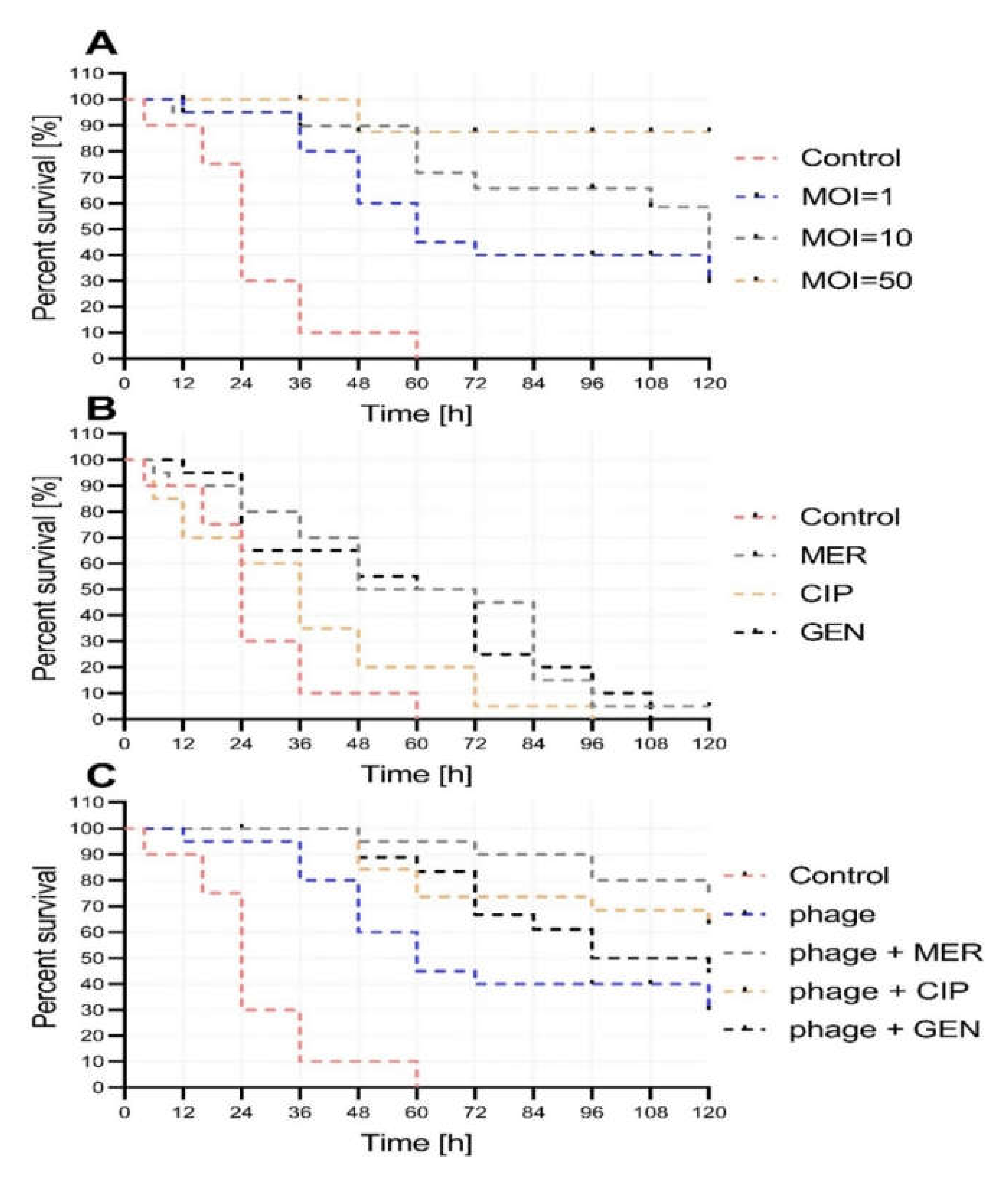
2.4. Activity of vB_AbaP_AGC01 on Biofilm, HIP-B, and G. Mellonella Larva Models
3. Discussion
4. Materials and Methods
4.1. Identification, Storage, and Growth of Bacterial Strains
4.2. Bacteriophage Isolation, Purification, Propagation, and Host Range
4.3. Bacteriophage Characterization
4.4. Analysis of Bacterial Growth and Bacteriophage Activity in Blood Model
4.5. In Vivo Synergy in the Galleria Mellonella Model
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of variance |
| CFU | Colony-forming unit |
| CIP | Ciprofloxacin |
| CLSI | Clinical and Laboratory Standard Institute |
| DLA | Double layer agar |
| GEN | Gentamicin |
| k | Adsorption constant |
| LB | Luria Bertani broth |
| MALDI | Matrix-assisted laser desorption/ionization |
| MDRAB | Multidrug resistant Acinetobacter baumannii |
| MER | Meropenem |
| MOI | Multiplicity of infection |
| NCBI | National Center for Biotechnology Information |
| OD | Optical density |
| ORF | Open reading frame |
| PAS | Phage antibiotic synergy |
| PBS | Phosphate buffered saline |
| PFU | Plaque forming units |
| RNAP | RNA polymerase |
| SD | Standard deviation |
| TOF | Time-of-flight |
| TSA | Trypticase soy agar |
References
- Lee, C.-R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.-J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Espinal, P.; Vila, J.; López-Rojas, R.; Rolo, D.; Smani, Y.; Marti, S.; Lavigne, J.-P.; Pantel, A.; Pachón, J. Relationship Between Different Resistance Mechanisms and Virulence in Acinetobacter baumannii. Microb. Drug Resist. 2019, 25, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Moreno, A.; Fernandez, N.; Vallejo, J.A.; Aranda, J.; Adler, B.; Harper, M.; Boyce, J.D.; Bou, G. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014, 58, 518–526. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef]
- Kropinski, A.M. Bacteriophage research—What we have learnt and what still needs to be addressed. Res. Microbiol. 2018, 169, 481–487. [Google Scholar] [CrossRef]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Bruttin, A.; Brüssow, H. Human volunteers receiving Escherichia coli phage T4 orally: A safety test of phage therapy. Antimicrob. Agents Chemother. 2005, 49, 2874–2878. [Google Scholar] [CrossRef]
- Turner, D.; Ackermann, H.W.; Kropinski, A.M.; Lavigne, R.; Sutton, J.M.; Reynolds, D.M. Comparative analysis of 37 Acinetobacter bacteriophages. Viruses 2018, 10, 5. [Google Scholar] [CrossRef]
- Jansen, M.; Wahida, A.; Latz, S.; Krüttgen, A.; Häfner, H.; Buhl, E.M.; Ritter, K.; Horz, H.-P. Enhanced antibacterial effect of the novel T4-like bacteriophage KARL-1 in combination with antibiotics against multi-drug resistant Acinetobacter baumannii. Sci. Rep. 2018, 8, 14140. [Google Scholar] [CrossRef]
- Łoś, J.M.; Golec, P.; Węgrzyn, G.; Węgrzyn, A.; Łoś, M. Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Appl. Environ. Microbiol. 2008, 74, 5113–5120. [Google Scholar] [CrossRef] [PubMed]
- Kamal, F.; Dennis, J.J. Burkholderia cepacia complex phage-antibiotic synergy (PAS): Antibiotics stimulate lytic phage activity. Appl. Environ. Microbiol. 2015, 81, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Wand, M.E.; Briers, Y.; Lavigne, R.; Sutton, J.M.; Reynolds, D.M. Characterisation and genome sequence of the lytic Acinetobacter baumannii bacteriophage vB_AbaS_Loki. PLoS ONE 2017, 12, e0172303. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, D.N.; Elbourne, L.D.H.; Hassan, K.A.; Eijkelkamp, B.A.; Tetu, S.G.; Brown, M.H.; Shah, B.S.; Peleg, A.Y.; Mabbutt, B.C.; Paulsen, I.T. The Complete Genome and Phenome of a Community-Acquired Acinetobacter baumannii. PLoS ONE 2013, 8, e58628. [Google Scholar] [CrossRef]
- Hernandez-Morales, A.C.; Lessor, L.L.; Wood, T.L.; Migl, D.; Mijalis, E.M.; Cahill, J.; Russell, W.K.; Young, R.F.; Gill, J.J. Genomic and Biochemical Characterization of Acinetobacter Podophage Petty Reveals a Novel Lysis Mechanism and Tail-Associated Depolymerase Activity. J. Virol. 2018, 92, e01064-17. [Google Scholar] [CrossRef]
- Łubowska, N.; Grygorcewicz, B.; Kosznik-Kwaśnicka, K.; Zauszkiewicz-Pawlak, A.; Węgrzyn, A.; Dołęgowska, B.; Piechowicz, L. Characterization of the Three New Kayviruses and Their Lytic Activity Against Multidrug-Resistant Staphylococcus aureus. Microorganisms 2019, 7, 471. [Google Scholar] [CrossRef]
- Melo, L.D.R.; Brandão, A.; Akturk, E.; Santos, S.B.; Azeredo, J. Characterization of a new Staphylococcus aureus Kayvirus harboring a lysin active against biofilms. Viruses 2018, 10, 182. [Google Scholar] [CrossRef]
- Szymczak, M.; Grygorcewicz, B.; Karczewska-Golec, J.; Decewicz, P.; Pankowski, J.A.; Országh-Szturo, H.; Bącal, P.; Dołęgowska, B.; Golec, P. Characterization of a Unique Bordetella bronchiseptica vB_BbrP_BB8 Bacteriophage and Its Application as an Antibacterial Agent. Int. J. Mol. Sci. 2020, 21, 1403. [Google Scholar] [CrossRef]
- Yang, H.; Liang, L.; Lin, S.; Jia, S. Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 2010, 10, 131. [Google Scholar] [CrossRef]
- Lai, M.J.; Chang, K.C.; Huang, S.W.; Luo, C.H.; Chiou, P.Y.; Wu, C.C.; Lin, N.T. The tail associated protein of Acinetobacter baumannii phage qab6 is the host specificity determinant possessing exopolysaccharide depolymerase activity. PLoS ONE 2016, 11, e0153361. [Google Scholar] [CrossRef]
- Liu, Y.; Mi, Z.; Mi, L.; Huang, Y.; Li, P.; Liu, H.; Yuan, X.; Niu, W.; Jiang, N.; Bai, C.; et al. Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ 2019, 7, e6173. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Calap, P.; Beamud, B.; Mora-Quilis, L.; González-Candelas, F.; Sanjuán, R. Isolation and Characterization of Two Klebsiella pneumoniae Phages Encoding Divergent Depolymerases. Int. J. Mol. Sci. 2020, 21, 3160. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Costa, A.R.; Konstantinides, N.; Ferreira, A.; Akturk, E.; Sillankorva, S.; Nemec, A.; Shneider, M.; Dötsch, A.; Azeredo, J. Ability of phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains. Environ. Microbiol. 2017, 19, 5060–5077. [Google Scholar] [CrossRef] [PubMed]
- Bedi, M.S.; Verma, V.; Chhibber, S. Amoxicillin and specific bacteriophage can be used together for eradication of biofilm of Klebsiella pneumoniae B5055. World J. Microbiol. Biotechnol. 2009, 25, 1145–1151. [Google Scholar] [CrossRef]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages—Review. Folia Microbiol. (Praha) 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Grygorcewicz, B.; Grudziński, M.; Wasak, A.; Augustyniak, A.; Pietruszka, A.; Nawrotek, P. Bacteriophage-mediated reduction of Salmonella Enteritidis in swine slurry. Appl. Soil Ecol. 2017, 119, 179–182. [Google Scholar] [CrossRef]
- Ma, L.; Green, S.I.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Metals Enhance the Killing of Bacteria by Bacteriophage in Human Blood. Sci. Rep. 2018, 8, 2326. [Google Scholar] [CrossRef]
- Blasco, L.; Ambroa, A.; Lopez, M.; Fernandez-Garcia, L.; Bleriot, I.; Trastoy, R.; Ramos-Vivas, J.; Coenye, T.; Fernandez-Cuenca, F.; Vila, J.; et al. Combined use of the Ab105-2ϕ∆CI lytic mutant phage and different antibiotics in clinical isolates of multi-resistant Acinetobacter baumannii. Microorganisms 2019, 7, 556. [Google Scholar] [CrossRef]
- Pletzer, D.; Mansour, S.C.; Hancock, R.E.W. Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog. 2018, 14, e1007084. [Google Scholar] [CrossRef]
- Desbois, A.P.; Coote, P.J. Wax moth larva (Galleria mellonella): An in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 2011, 66, 1785–1790. [Google Scholar] [CrossRef]
- Seed, K.D.; Dennis, J.J. Experimental bacteriophage therapy increases survival of Galleria mellonella larvae infected with clinically relevant strains of the Burkholderia cepacia complex. Antimicrob. Agents Chemother. 2009, 53, 2205–2208. [Google Scholar] [CrossRef] [PubMed]
- Abbasifar, R.; Kropinski, A.M.; Sabour, P.M.; Chambers, J.R.; MacKinnon, J.; Malig, T.; Griffiths, M.W. Efficiency of bacteriophage therapy against Cronobacter sakazakii in Galleria mellonella (greater wax moth) larvae. Arch. Virol. 2014, 159, 2253–2261. [Google Scholar] [CrossRef] [PubMed]
- Beeton, M.L.L.; Alves, D.R.R.; Enright, M.C.C.; Jenkins, A.T.A.T.A. Assessing phage therapy against Pseudomonas aeruginosa using a Galleria mellonella infection model. Int. J. Antimicrob. Agents 2015, 46, 196–200. [Google Scholar] [CrossRef]
- Jeon, J.; Park, J.H.; Yong, D. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 2019, 19, 70. [Google Scholar] [CrossRef]
- Tsai, C.J.Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 5594, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Segall, A.M.; Roach, D.R.; Strathdee, S.A. Stronger together? Perspectives on phage-antibiotic synergy in clinical applications of phage therapy. Curr. Opin. Microbiol. 2019, 51, 46–50. [Google Scholar] [CrossRef]
- Twest, R.; Kropinski, A.M.; Van Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. Methods Mol. Biol. 2009, 501, 15–21. [Google Scholar]
- Adriaenssens, E.M.; Rodney Brister, J. How to name and classify your phage: An informal guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by the Direct Plating Plaque Assay. In Bacteriophages; Humana Press: Totowa, NJ, SAD, 2009; Volume 501, ISBN 978-1-58829-682-5. [Google Scholar]
- Mirzaei, M.K.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Kropinski, A.M. Measurement of the rate of attachment of bacteriophage to cells. Methods Mol. Biol. 2009, 501, 151–155. [Google Scholar]
- Grygorcewicz, B.; Chajecka-Wierzchowska, W.; Augustyniak, A.; Wasak, A.; Stachurska, X.; Nawrotek, P.; Dołęgowska, B. In-milk inactivation of Escherichia coli O157: H7 by the environmental lytic bacteriophage ECPS-6. J. Food Saf. 2019, 40, e12747. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Boil. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Sun, W.D.; Volckaert, G. PHIRE, a deterministic approach to reveal regulatory elements in bacteriophage genomes. Bioinformatics 2004, 20, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Darzentas, N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grygorcewicz, B.; Roszak, M.; Golec, P.; Śleboda-Taront, D.; Łubowska, N.; Górska, M.; Jursa-Kulesza, J.; Rakoczy, R.; Wojciuk, B.; Dołęgowska, B. Antibiotics Act with vB_AbaP_AGC01 Phage against Acinetobacter baumannii in Human Heat-Inactivated Plasma Blood and Galleria mellonella Models. Int. J. Mol. Sci. 2020, 21, 4390. https://doi.org/10.3390/ijms21124390
Grygorcewicz B, Roszak M, Golec P, Śleboda-Taront D, Łubowska N, Górska M, Jursa-Kulesza J, Rakoczy R, Wojciuk B, Dołęgowska B. Antibiotics Act with vB_AbaP_AGC01 Phage against Acinetobacter baumannii in Human Heat-Inactivated Plasma Blood and Galleria mellonella Models. International Journal of Molecular Sciences. 2020; 21(12):4390. https://doi.org/10.3390/ijms21124390
Chicago/Turabian StyleGrygorcewicz, Bartłomiej, Marta Roszak, Piotr Golec, Daria Śleboda-Taront, Natalia Łubowska, Martyna Górska, Joanna Jursa-Kulesza, Rafał Rakoczy, Bartosz Wojciuk, and Barbara Dołęgowska. 2020. "Antibiotics Act with vB_AbaP_AGC01 Phage against Acinetobacter baumannii in Human Heat-Inactivated Plasma Blood and Galleria mellonella Models" International Journal of Molecular Sciences 21, no. 12: 4390. https://doi.org/10.3390/ijms21124390
APA StyleGrygorcewicz, B., Roszak, M., Golec, P., Śleboda-Taront, D., Łubowska, N., Górska, M., Jursa-Kulesza, J., Rakoczy, R., Wojciuk, B., & Dołęgowska, B. (2020). Antibiotics Act with vB_AbaP_AGC01 Phage against Acinetobacter baumannii in Human Heat-Inactivated Plasma Blood and Galleria mellonella Models. International Journal of Molecular Sciences, 21(12), 4390. https://doi.org/10.3390/ijms21124390






