BRG1 Is Dispensable for Sertoli Cell Development and Functions in Mice
Abstract
1. Introduction
2. Results
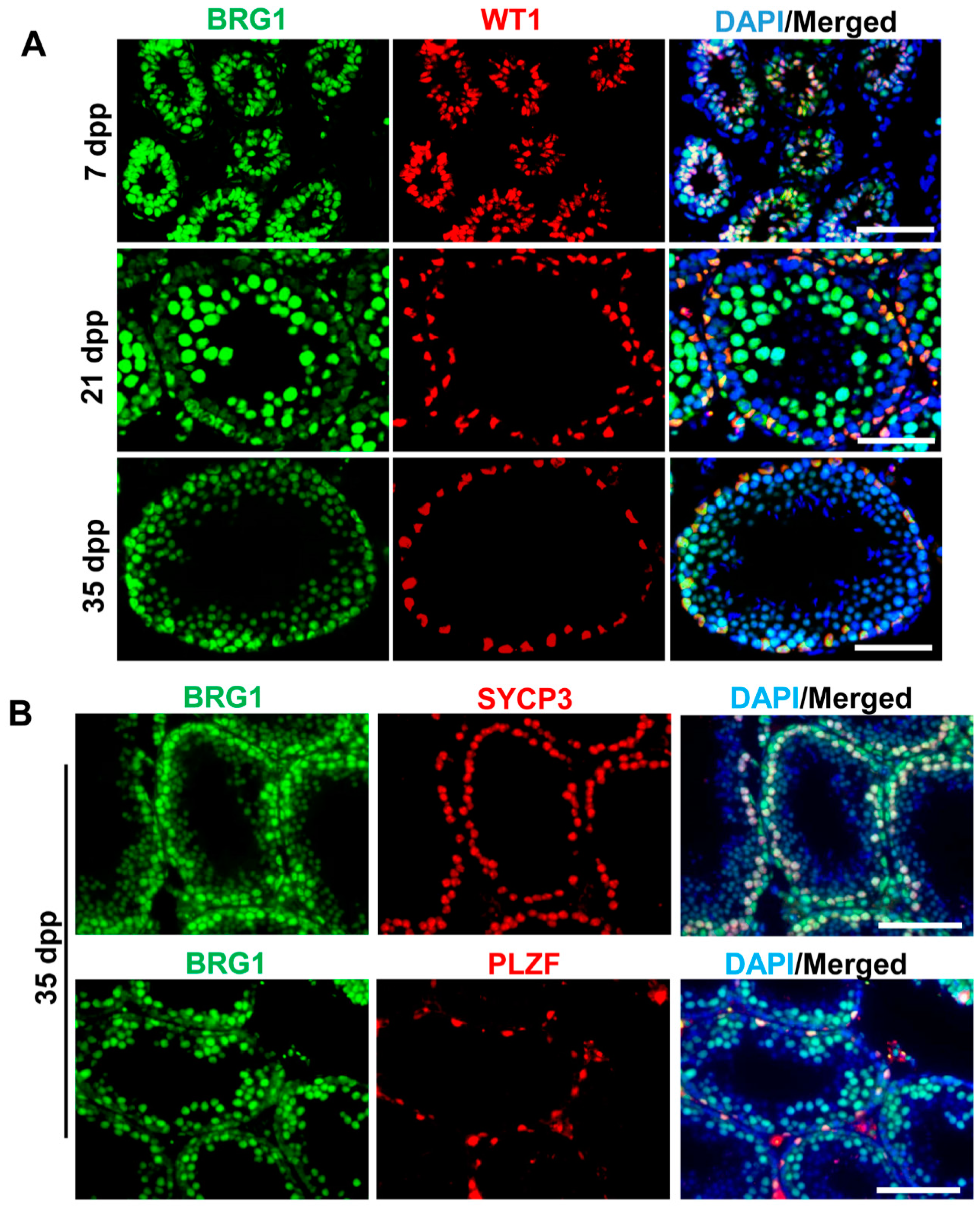
2.1. BRG1 Is Expressed in Both Sertoli Cells and Germ Cells
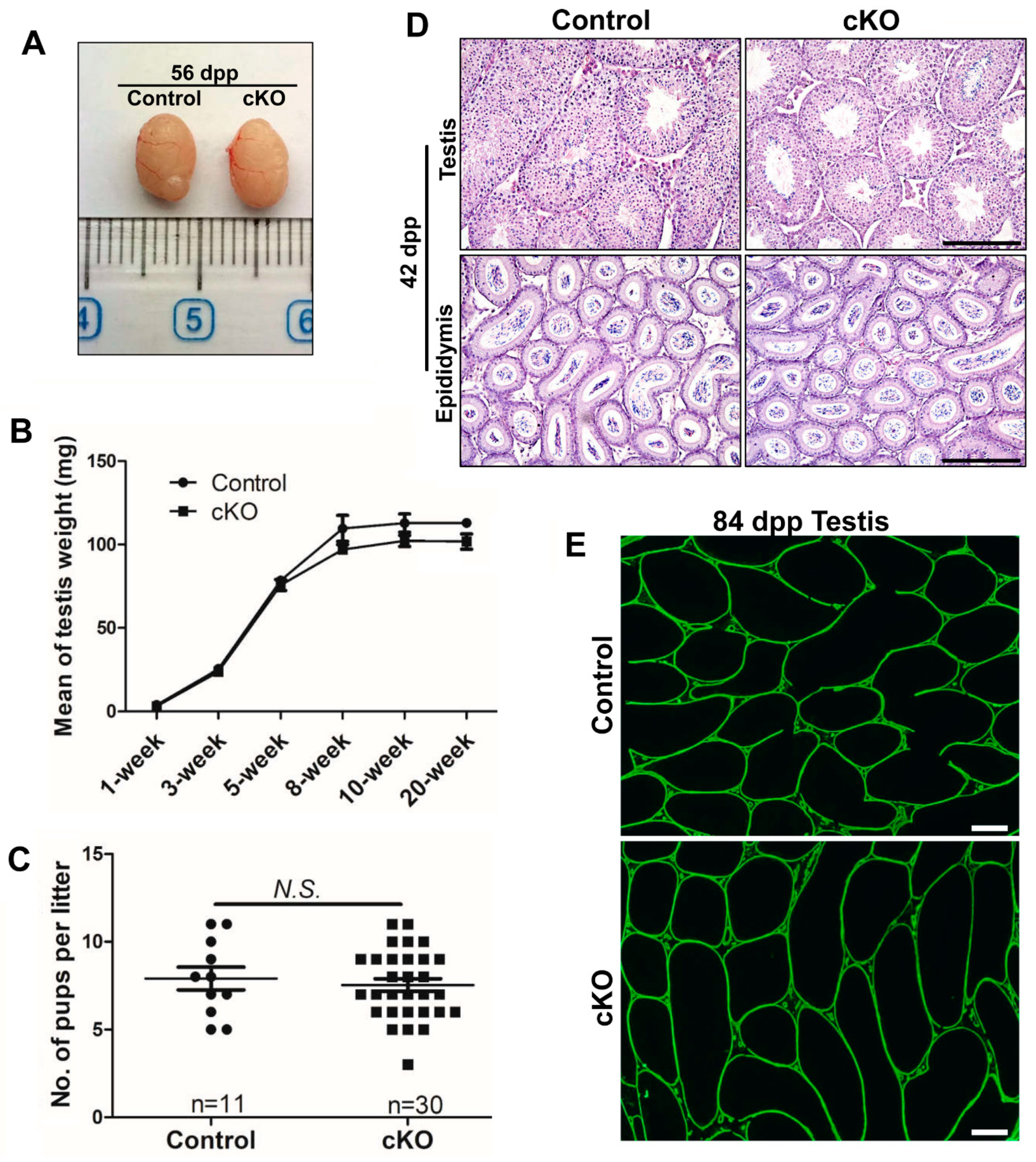
2.2. Partial Brg1 Deletion in Sertoli Cells Does Not Affect Sertoli Cell Development
2.3. Partial Deletion of Brg1 in Sertoli Cells Does Not Affect Spermatogenesis
2.4. Efficient Brg1 Deletion in Sertoli Cells Does Not Affect Spermatogenesis
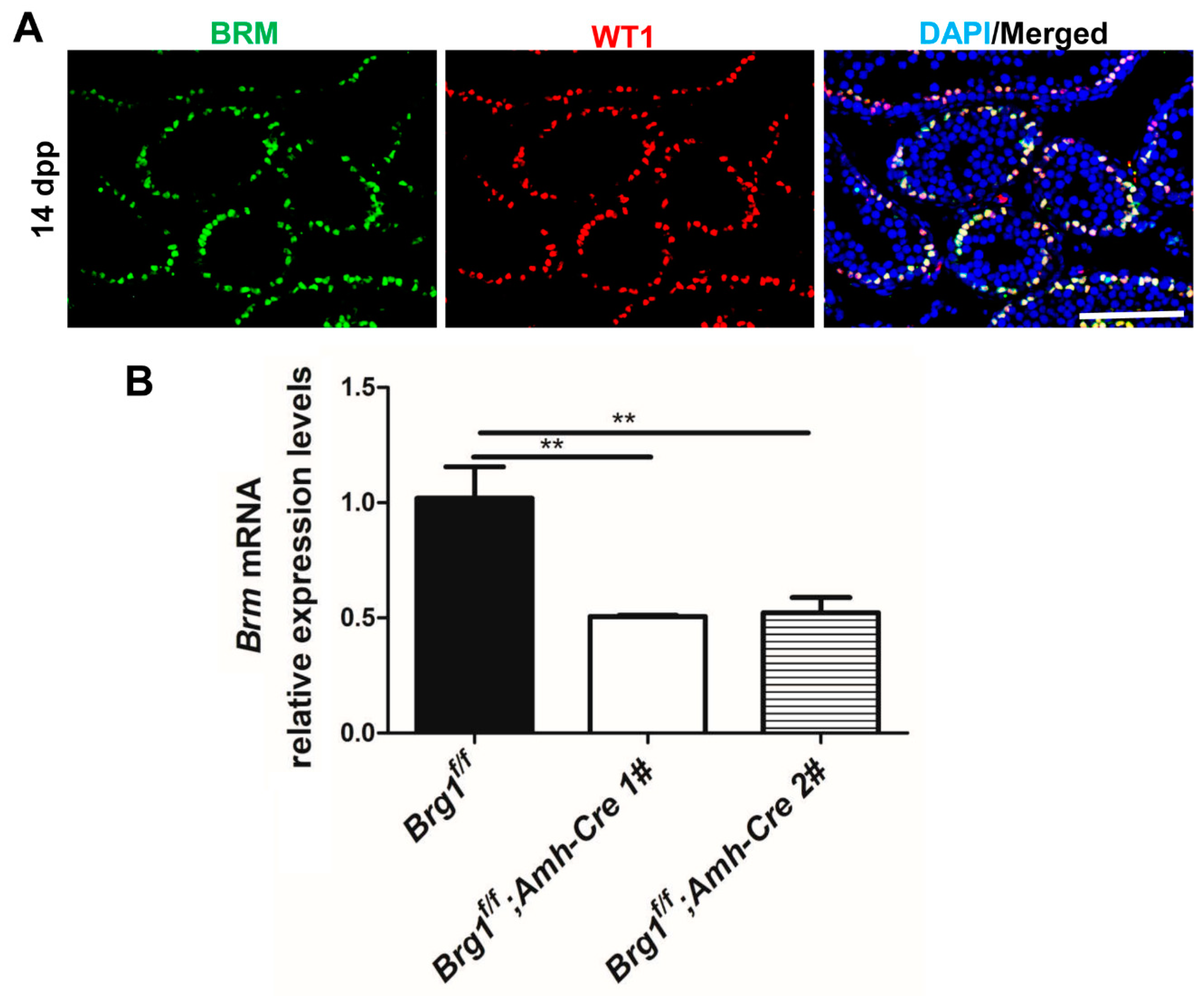
2.5. BRM Is Expressed Exclusively in Sertoli Cells
3. Discussion
4. Materials and Methods
4.1. Mouse Lines, Animal Care and Fertility Test
4.2. Genotyping
4.3. Histology and Immunohistofluorescence (IHF)
4.4. Sertoli Cell Isolation
4.5. Total RNA Extraction, Reverse Transcription, and Real-Time PCR
4.6. Blood-Testis Barrier Integrity Assay
4.7. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- França, L.R.; Hess, R.A.; Dufour, J.M.; Hofmann, M.C.; Griswold, M.D. The Sertoli cell: One hundred fifty years of beauty and plasticity. Andrology 2016, 4, 189–212. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998, 9, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. 50 years of spermatogenesis: Sertoli cells and their interactions with germ cells. Biol. Reprod. 2018, 99, 87–100. [Google Scholar] [CrossRef]
- Nalam, R.L.; Andreu-Vieyra, C.; Braun, R.E.; Akiyama, H.; Matzuk, M.M. Retinoblastoma protein plays multiple essential roles in the terminal differentiation of Sertoli cells. Mol. Endocrinol. 2009, 23, 1900–1913. [Google Scholar] [CrossRef]
- Skinner, M.K.; Griswold, M.D. Sertoli Cell Biology; Elsevier Academic Press: Boston, MA, USA, 2005. [Google Scholar]
- Sharpe, R.M.; McKinnell, C.; Kivlin, C.; Fisher, J.S. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 2003, 125, 769–784. [Google Scholar] [CrossRef]
- Vergouwen, R.P.; Jacobs, S.G.; Huiskamp, R.; Davids, J.A.; de Rooij, D.G. Proliferative activity of gonocytes, Sertoli cells and interstitial cells during testicular development in mice. J. Reprod. Fertil. 1991, 93, 233–243. [Google Scholar] [CrossRef]
- Jan, S.Z.; Hamer, G.; Repping, S.; de Rooij, D.G.; van Pelt, A.M.; Vormer, T.L. Molecular control of rodent spermatogenesis. Biochim. Biophys. Acta 2012, 1822, 1838–1850. [Google Scholar] [CrossRef]
- Li, M.W.; Mruk, D.D.; Lee, W.M.; Cheng, C.Y. Connexin 43 is critical to maintain the homeostasis of the blood-testis barrier via its effects on tight junction reassembly. Proc. Natl. Acad. Sci. USA 2010, 107, 17998–18003. [Google Scholar] [CrossRef]
- Feng, L.X.; Ravindranath, N.; Dym, M. Stem cell factor/c-kit up-regulates cyclin D3 and promotes cell cycle progression via the phosphoinositide 3-kinase/p70 S6 kinase pathway in spermatogonia. J. Biol. Chem. 2000, 275, 25572–25576. [Google Scholar] [CrossRef]
- Flanagan, J.G.; Chan, D.C.; Leder, P. Transmembrane form of the kit ligand growth factor is determined by alternative splicing and is missing in the Sld mutant. Cell 1991, 64, 1025–1035. [Google Scholar] [CrossRef]
- Johnston, D.S.; Olivas, E.; DiCandeloro, P.; Wright, W.W. Stage-specific changes in GDNF expression by rat Sertoli cells: A possible regulator of the replication and differentiation of stem spermatogonia. Biol. Reprod. 2011, 85, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Lindahl, M.; Hyvönen, M.E.; Parvinen, M.; de Rooij, D.G.; Hess, M.W.; Raatikainen-Ahokas, A.; Sainio, K.; Rauvala, H.; Lakso, M.; et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000, 287, 1489–1493. [Google Scholar] [CrossRef]
- Pellegrini, M.; Grimaldi, P.; Rossi, P.; Geremia, R.; Dolci, S. Developmental expression of BMP4/ALK3/SMAD5 signaling pathway in the mouse testis: A potential role of BMP4 in spermatogonia differentiation. J. Cell Sci. 2003, 116, 3363–3372. [Google Scholar] [CrossRef] [PubMed]
- Runyan, C.; Schaible, K.; Molyneaux, K.; Wang, Z.; Levin, L.; Wylie, C. Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development 2006, 133, 4861–4869. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Tang, Z.; Li, Y.; Liu, W.; Zhang, S.; Wang, B.; Tian, Y.; Zhao, Y.; Ran, H.; Liu, W.; et al. Deletion of the tyrosine phosphatase Shp2 in Sertoli cells causes infertility in mice. Sci. Rep. 2015, 5, 12982. [Google Scholar] [CrossRef] [PubMed]
- Tarulli, G.A.; Stanton, P.G.; Meachem, S.J. Is the adult Sertoli cell terminally differentiated? Biol. Reprod. 2012, 87, 1–11. [Google Scholar] [CrossRef]
- Trotter, K.W.; Archer, T.K. The BRG1 transcriptional coregulator. Nucl. Recept. Signal 2008, 6, e004. [Google Scholar] [CrossRef]
- Hendricks, K.B.; Shanahan, F.; Lees, E. Role for BRG1 in cell cycle control and tumor suppression. Mol. Cell Biol. 2004, 24, 362–376. [Google Scholar] [CrossRef]
- Vieira, J.M.; Howard, S.; Villa Del Campo, C.; Bollini, S.; Dubé, K.N.; Masters, M.; Barnette, D.N.; Rohling, M.; Sun, X.; Hankins, L.E.; et al. BRG1-SWI/SNF-dependent Regulation of the Wt1 Transcriptional Landscape Mediates Epicardial Activity During Heart Development and Disease. Nat. Commun. 2017, 8, 16034. [Google Scholar] [CrossRef]
- Tsuda, M.; Fukuda, A.; Roy, N.; Hiramatsu, Y.; Leonhardt, L.; Kakiuchi, N.; Hoyer, K.; Ogawa, S.; Goto, N.; Ikuta, K.; et al. The BRG1/SOX9 Axis Is Critical for Acinar Cell-Derived Pancreatic Tumorigenesis. J. Clin. Investig. 2018, 128, 5676. [Google Scholar] [CrossRef]
- Bultman, S.; Gebuhr, T.; Yee, D.; La Mantia, C.; Nicholson, J.; Gilliam, A.; Randazzo, F.; Metzger, D.; Chambon, P.; Crabtree, G.; et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 2000, 6, 1287–1295. [Google Scholar] [CrossRef]
- Bultman, S.J.; Herschkowitz, J.I.; Godfrey, V.; Gebuhr, T.C.; Yaniv, M.; Perou, C.M.; Magnuson, T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene 2008, 27, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Fedoriw, A.M.; Magnuson, T. An essential role for a mammalian SWI/SNF chromatin-remodeling complex during male meiosis. Development 2012, 139, 1133–1140. [Google Scholar] [CrossRef]
- Wang, J.; Gu, H.; Lin, H.; Chi, T. Essential roles of the chromatin remodeling factor BRG1 in spermatogenesis in mice. Biol. Reprod. 2012, 86, 186. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Barra, J.; Muchardt, C.; Camus, A.; Babinet, C.; Yaniv, M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 1998, 17, 6979–6991. [Google Scholar] [CrossRef]
- Holdcraft, R.W.; Braun, R.E. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development 2004, 131, 459–467. [Google Scholar] [CrossRef]
- Srinivas, S.; Watanabe, T.; Lin, C.S.; William, C.M.; Tanabe, Y.; Jessell, T.M.; Costantini, F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001, 1, 4. [Google Scholar] [CrossRef]
- Lécureuil, C.; Fontaine, I.; Crepieux, P.; Guillou, F. Sertoli and granulosa cell-specific Cre recombinase activity in transgenic mice. Genesis 2002, 33, 114–118. [Google Scholar] [CrossRef]
- Liu, J.G.; Yuan, L.; Brundell, E.; Björkroth, B.; Daneholt, B.; Höög, C. Localization of the N-terminus of SCP1 to the Central Element of the Synaptonemal Complex and Evidence for Direct Interactions Between the N-termini of SCP1 Molecules Organized Head-To-Head. Exp. Cell Res. 1996, 226, 11–19. [Google Scholar] [CrossRef]
- Tanaka, H.; Pereira, L.A.; Nozaki, M.; Tsuchida, J.; Sawada, K.; Mori, H.; Nishimune, Y. A germ cell-specific nuclear antigen recognized by a monoclonal antibody raised against mouse testicular germ cells. Int. J. Androl. 1997, 20, 361–366. [Google Scholar] [CrossRef]
- Heidaran, M.A.; Showman, R.M.; Kistler, W.S. A Cytochemical Study of the Transcriptional and Translational Regulation of Nuclear Transition Protein 1 (TP1), a Major Chromosomal Protein of Mammalian Spermatids. J. Cell Biol. 1988, 106, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.J.; Gallagher, S.J.; Foreman, O.; Dannenberg, J.H.; Depinho, R.A. Braun RE. Sin3a is required by sertoli cells to establish a niche for undifferentiated spermatogonia, germ cell tumors, and spermatid elongation. Stem Cells 2010, 28, 1424–1434. [Google Scholar] [CrossRef] [PubMed]
- Muchardt, C.; Yaniv, M. When the SWI/SNF complex remodels...the cell cycle. Oncogene 2001, 20, 3067–3075. [Google Scholar] [CrossRef]
- Ho, L.; Ronan, J.L.; Wu, J.; Staahl, B.T.; Chen, L.; Kuo, A.; Lessard, J.; Nesvizhskii, A.I.; Ranish, J.; Crabtree, G.R. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA 2009, 106, 5181–5186. [Google Scholar] [CrossRef]
- Kaeser, M.D.; Aslanian, A.; Dong, M.; Yates, J.R., 3rd; Emerson, B.M. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 2008, 283, 32254–32263. [Google Scholar] [CrossRef] [PubMed]
- Kidder, B.L.; Palmer, S.; Knott, J.G. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 2009, 27, 317–328. [Google Scholar] [CrossRef]
- Zhang, X.; Li, B.; Li, W.; Ma, L.; Zheng, D.; Li, L.; Yang, W.; Chu, M.; Chen, W.; Mailman, R.B.; et al. Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Rep. 2014, 3, 460–474. [Google Scholar] [CrossRef]
- Lessard, J.; Wu, J.I.; Ranish, J.A.; Wan, M.; Winslow, M.M.; Staahl, B.T.; Wu, H.; Aebersold, R.; Graef, I.A.; Crabtree, G.R. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 2007, 55, 201–215. [Google Scholar] [CrossRef]
- Staahl, B.T.; Tang, J.; Wu, W.; Sun, A.; Gitler, A.D.; Yoo, A.S.; Crabtree, G.R. Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J. Neurosci. 2013, 33, 10348–10361. [Google Scholar] [CrossRef]
- Ninkovic, J.; Steiner-Mezzadri, A.; Jawerka, M.; Akinci, U.; Masserdotti, G.; Petricca, S.; Fischer, J.; von Holst, A.; Beckers, J.; Lie, C.D.; et al. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell 2013, 13, 403–418. [Google Scholar] [CrossRef]
- King, H.A.; Trotter, K.W.; Archer, T.K. Chromatin remodeling during glucocorticoid receptor regulated transactivation. Biochim. Biophys. Acta 2012, 1819, 716–726. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Ru, B.; Zhang, Y.; Chan, W.L.; Chow, S.C.; Zhang, J.; Lo, C.; Lui, W.Y. Sertoli cell-specific coxsackievirus and adenovirus receptor knockout regulates cell adhesion and gene transcription via β-catenin inactivation and Cdc42 activation. FASEB J. 2019, 33, 7588–7602. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.R.; Hao, X.X.; Zhang, Y.; Deng, S.L.; Wang, Z.P.; Wang, Y.Q.; Wang, X.X.; Liu, Y.X. Androgen receptor in Sertoli cells regulates DNA double-strand break repair and chromosomal synapsis of spermatocytes partially through intercellular EGF-EGFR signaling. Oncotarget. 2016, 7, 18722–18735. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Chen, Z.; Wang, C.; Xiong, Z.; Zhao, W.; Jia, C.; Lin, J.; Lin, Y.; Yuan, W.; Zhao, A.Z.; et al. Rictor Regulates Spermatogenesis by Controlling Sertoli Cell Cytoskeletal Organization and Cell Polarity in the Mouse Testis. Endocrinology 2015, 156, 4244–4256. [Google Scholar] [CrossRef]
- Ghaffari, R.; Di Bona, K.R.; Riley, C.L.; Richburg, J.H. Copper transporter 1 (CTR1) expression by mouse testicular germ cells, but not Sertoli cells, is essential for functional spermatogenesis. PLoS ONE 2019, 14, e0215522. [Google Scholar] [CrossRef]
- Gebuhr, T.C.; Kovalev, G.I.; Bultman, S.; Godfrey, V.; Su, L.; Magnuson, T. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J. Exp. Med. 2003, 198, 1937–1949. [Google Scholar] [CrossRef]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef]
- Garcia, T.; Hofmann, M.C. Isolation of undifferentiated and early differentiating type A spermatogonia from Pou5f1-GFP reporter mice. Methods Mol. Biol. 2012, 825, 31–44. [Google Scholar]
- Meng, J.; Holdcraft, R.W.; Shima, J.E.; Griswold, M.D.; Braun, R.E. Androgens regulate the permeability of the blood-testis barrier. Proc. Natl. Acad. Sci. USA 2005, 102, 16696–16700. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Wang, P.; Liang, D.; Wang, Y. BRG1 Is Dispensable for Sertoli Cell Development and Functions in Mice. Int. J. Mol. Sci. 2020, 21, 4358. https://doi.org/10.3390/ijms21124358
Wang S, Wang P, Liang D, Wang Y. BRG1 Is Dispensable for Sertoli Cell Development and Functions in Mice. International Journal of Molecular Sciences. 2020; 21(12):4358. https://doi.org/10.3390/ijms21124358
Chicago/Turabian StyleWang, Shuai, Pengxiang Wang, Dongli Liang, and Yuan Wang. 2020. "BRG1 Is Dispensable for Sertoli Cell Development and Functions in Mice" International Journal of Molecular Sciences 21, no. 12: 4358. https://doi.org/10.3390/ijms21124358
APA StyleWang, S., Wang, P., Liang, D., & Wang, Y. (2020). BRG1 Is Dispensable for Sertoli Cell Development and Functions in Mice. International Journal of Molecular Sciences, 21(12), 4358. https://doi.org/10.3390/ijms21124358



