In Vivo Validation of the BIANCA Biophysical Model: Benchmarking against Rat Spinal Cord RBE Data
Abstract
1. Introduction
2. Results
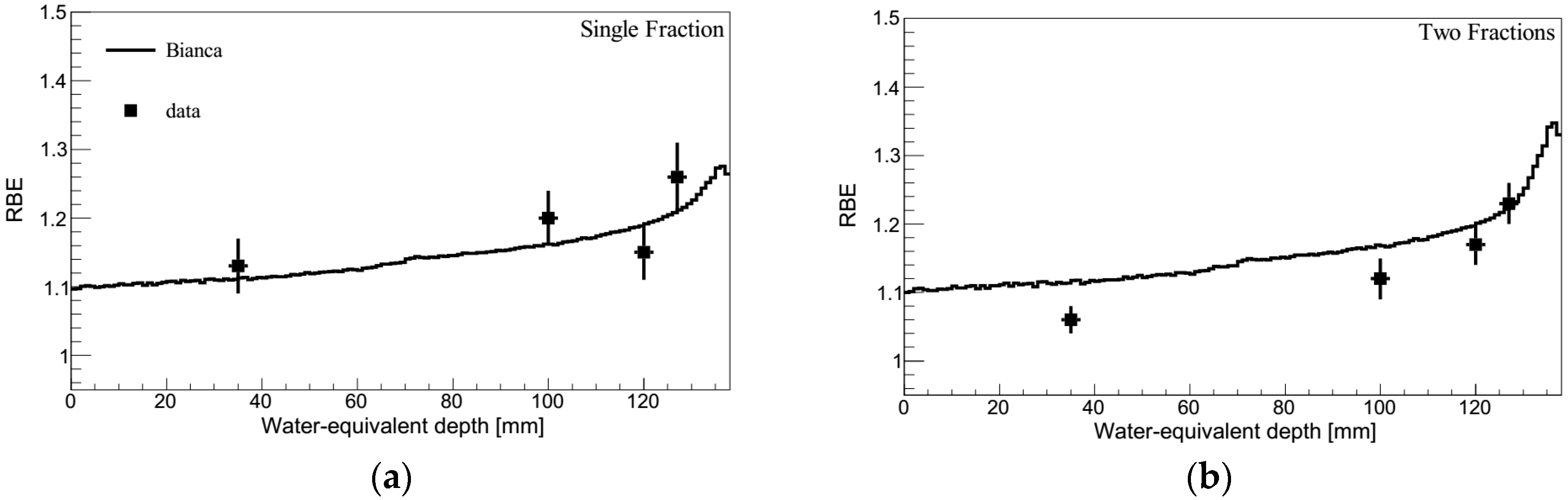
2.1. RBE-LET Relationship
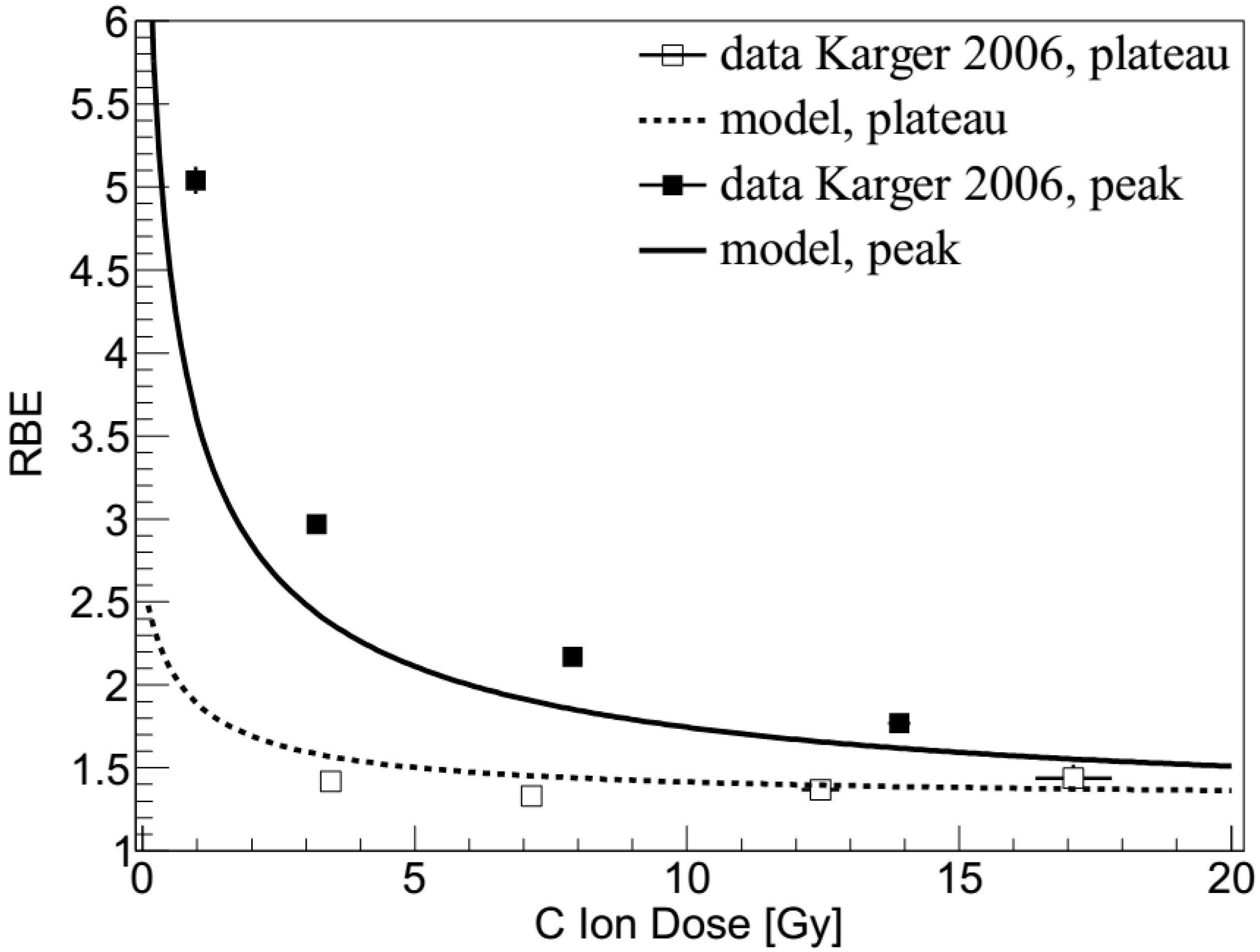
2.2. Carbon Ions: Dose Dependence
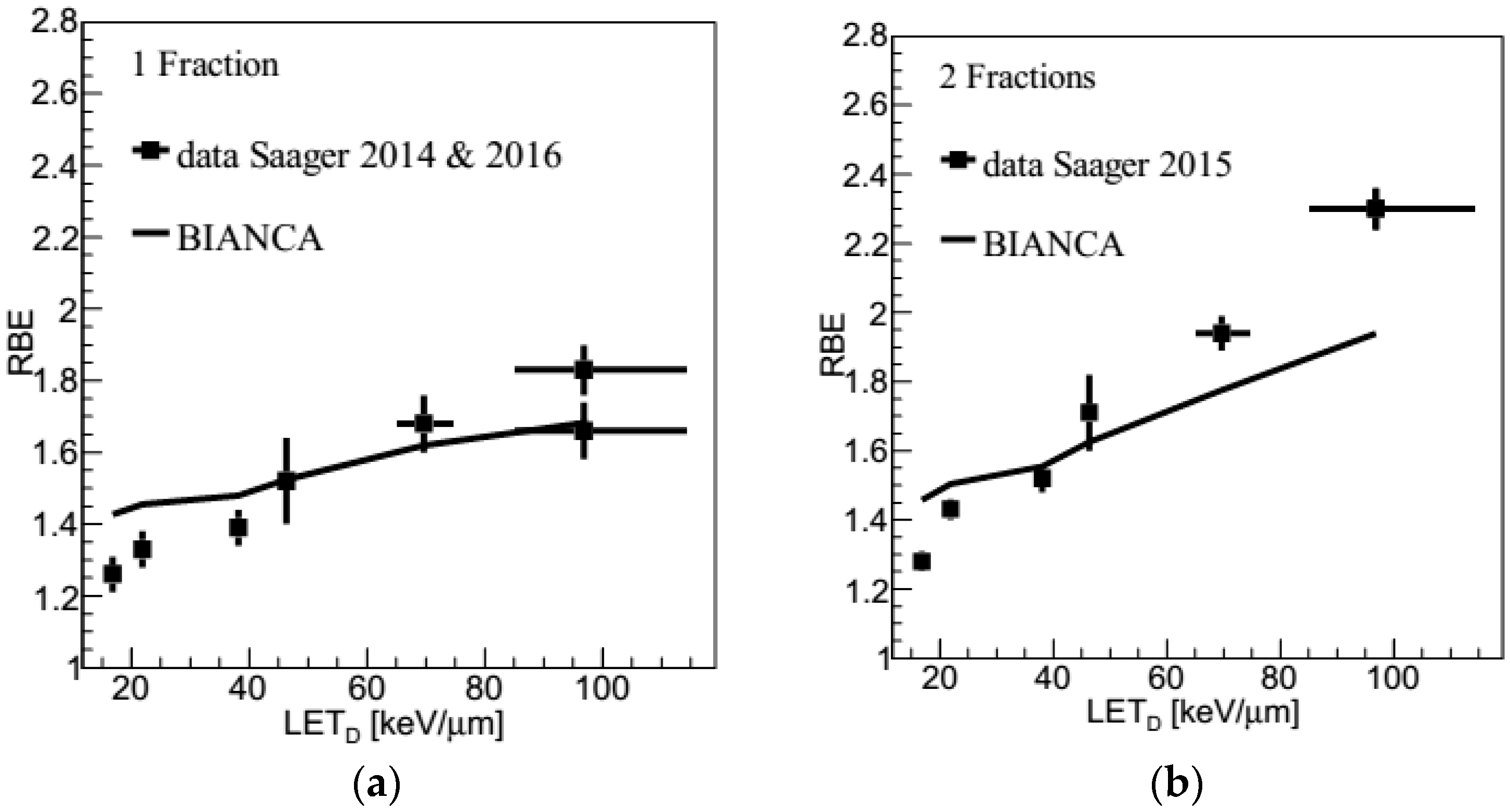
2.3. Carbon Ions: LET Dependence
2.4. Protons
3. Discussion
4. Materials and Methods
4.1. The BIANCA Model: Assumptions, Parameters and Structure
4.2. Construction of a Radiobiological Database and RBE Prediction
4.3. Interface between BIANCA and the FLUKA Monte Carlo Transport Code
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Georgakilas, A.G.; O’Neill, P.; Stewart, R.D. Induction and repair of clustered DNA lesions: What do we know so far? Radiat. Res. 2013, 180, 100–109. [Google Scholar] [CrossRef]
- Karger, C.P.; Peschke, P. RBE and related modeling in carbon-ion therapy. Phys. Med. Biol. 2017, 63, 01TR02. [Google Scholar] [CrossRef]
- Schuemann, J.; McNamara, A.L.; Warmenhoven, J.W.; Henthorn, N.T.; Kirkby, K.J.; Merchant, M.J.; Ingram, S.; Paganetti, H.; Held, K.D.; Faddegon, B.; et al. A new standard DNA damage data format (SDD). Radiat. Res. 2019, 191, 76–92. [Google Scholar] [CrossRef]
- Suit, H.; DeLaney, T.; Goldberg, S.; Paganetti, H.; Clasie, B.; Gerweck, L.; Niemierko, A.; Hall, E.; Flanz, J.; Hallman, J.; et al. Proton vs carbon ion beams in the definitive radiation treatment of cancer patients. Radiother. Oncol. 2010, 95, 3–22. [Google Scholar] [CrossRef]
- Scholz, M.; Kellerer, A.M.; Kraft-Weyrather, W.; Kraft, G. Computation of cell survival in heavy ion beams for therapy. The model and its approximation. Radiat. Environ. Biophys. 1997, 36, 59–66. [Google Scholar] [CrossRef]
- Elsässer, T.; Weyrather, W.K.; Friedrich, T.; Durante, M.; Iancu, G.; Krämer, M.; Kragl, G.; Brons, S.; Winter, M.; Weber, K.J.; et al. Quantification of the relative biological effectiveness for ion beam radiotherapy: Direct experimental comparison of proton and carbon ion beams and a novel approach for treatment planning. Int. J. Radiat. Oncol. Biol. Phys. 2010, 78, 1177–1183. [Google Scholar] [CrossRef]
- Inaniwa, T.; Furukawa, T.; Kase, Y.; Matsufuji, N.; Toshito, T.; Matsumoto, Y.; Furusawa, Y.; Noda, K. Treatment planning for a scanned carbon beam with a modified microdosimetric kinetic model. Phys. Med. Biol. 2010, 55, 6721–6737. [Google Scholar] [CrossRef]
- Inaniwa, T.; Kanematsu, N.; Matsufuji, N.; Kanai, T.; Shirai, T.; Noda, K.; Tsuji, H.; Kamada, T.; Tsujii, H. Reformulation of a clinical-dose system for carbon-ion radiotherapy treatment planning at the National Institute of Radiological Sciences, Japan. Phys. Med. Biol. 2015, 60, 3271–3286. [Google Scholar] [CrossRef]
- Kanai, T.; Furusawa, Y.; Fukutsu, K.; Itsukaichi, H.; Eguchi-Kasai, K.; Ohara, H. Irradiation of mixed beam and design of spread-out Bragg peak for heavy-ion radiotherapy. Radiat. Res. 1997, 147, 78–85. [Google Scholar] [CrossRef]
- Karger, C.P.; Peschke, P.; Sanchez-Brandelik, R.; Scholz, M.; Debus, J. Radiation tolerance of the rat spinal cord after 6 and 18 fractions of photons and carbon ions: Experimental results and clinical implications. Int. J. Radiat. Oncol. Biol. Phys. 2006, 66, 1488–1497. [Google Scholar] [CrossRef]
- van der Kogel, A.J.; Barendsen, G.W. Late effects of spinal cord irradiation with 300 kV X rays and 15 MeV neutrons. Br. J. Radiol. 1974, 47, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Saager, M.; Glowa, C.; Peschke, P.; Brons, S.; Scholz, M.; Huber, P.E.; Debus, J.; Karger, C.P. Carbon ion irradiation of the rat spinal cord: Dependence of the relative biological effectiveness on linear energy transfer. Int. J. Radiat. Oncol. Biol. Phys. 2014, 90, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Saager, M.; Glowa, C.; Peschke, P.; Brons, S.; Grün, R.; Scholz, M.; Huber, P.E.; Debus, J.; Karger, C.P. Split dose carbon ion irradiation of the rat spinal cord: Dependence of the relative biological effectiveness on dose and linear energy transfer. Radiother. Oncol. 2015, 117, 358–363. [Google Scholar] [CrossRef]
- Saager, M.; Glowa, C.; Peschke, P.; Brons, S.; Grün, R.; Scholz, M.; Huber, P.E.; Debus, J.; Karger, C.P. The relative biological effectiveness of carbon ion irradiations of the rat spinal cord increases linearly with LET up to 99 keV/μm. Acta Oncol. 2016, 55, 1512–1515. [Google Scholar] [CrossRef]
- Saager, M.; Peschke, P.; Brons, S.; Debus, J.; Karger, C.P. Determination of the proton RBE in the rat spinal cord: Is there an increase towards the end of the spread-out Bragg peak? Radiother. Oncol. 2018, 128, 115–120. [Google Scholar] [CrossRef]
- Carante, M.P.; Aimè, C.; Tello Cajiao, J.J.; Ballarini, F. BIANCA, a biophysical model of cell survival and chromosome damage by protons, C-ions and He-ions at energies and doses used in hadrontherapy. Phys. Med. Biol. 2018, 63, 075007. [Google Scholar] [CrossRef]
- Carante, M.P.; Tello Cajiao, J.J.; Ballarini, F. Predicting biological effects along hadrontherapy dose profiles by the BIANCA biophysical model. Radiat. Prot. Dosim. 2019, 183, 111–115. [Google Scholar] [CrossRef]
- Carante, M.P.; Aricò, G.; Ferrari, A.; Kozlowska, K.; Mairani, A.; Ballarini, F. First benchmarking of the BIANCA model for cell survival predictions in a clinical hadron therapy scenario. Phys. Med. Biol. 2019, 64, 215008. [Google Scholar] [CrossRef]
- Ferrari, A.; Sala, P.; Fassò, A.; Ranft, J. FLUKA: A Multi-Particle Transport Code Technical Report CERN-2005-10, INFN/TC 05/11, SLAC-R-773 CERN, INFN; SLAC: Menlo Park, CA, USA, 2005. [Google Scholar]
- Bohlen, T.T.; Cerutti, F.; Chin, M.; Fassò, A.; Ferrari, A.; Ortega, P.; Mairani, A.; Sala, P.; Smirnov, G.; Vlachoudis, V. The FLUKA code: Developments and challenges for high energy and medical applications. Nucl. Data Sheets 2014, 120, 211–214. [Google Scholar] [CrossRef]
- Battistoni, G.; Bauer, J.; Boehlen, T.T.; Cerutti, F.; Chin, M.P.W.; Dos Santos Augusto, R.; Ferrari, A.; Ortega, P.G.; Kozłowska, W.; Magro, G.; et al. The FLUKA Code: An Accurate Simulation Tool for Particle Therapy. Front. Oncol. 2016, 6, 116. [Google Scholar] [CrossRef]
- Kozłowska, W.S.; Böhlen, T.T.; Cuccagna, C.; Ferrari, A.; Fracchiolla, F.; Magro, G.; Mairani, A.; Schwarz, M.; Vlachoudis, V.; Georg, D. FLUKA particle therapy tool for Monte Carlo independent calculation of scanned proton and carbon ion beam therapy. Phys. Med. Biol. 2019, 64, 075012. [Google Scholar] [CrossRef]
- Debus, J.; Scholz, M.; Haberer, T.; Peschke, P.; Jäkel, O.; Karger, C.P.; Wannenmacher, M. Radiation Tolerance of the Rat Spinal Cord after Single and Split Doses of Photons and Carbon Ions. Radiat. Res. 2003, 160, 536–542. [Google Scholar] [CrossRef]
- International Commission on Radiation Units and Measurements. Prescribing, Recording, and Reporting Proton-Beam Therapy; Report 78; International Commission on Radiation Units and Measurements: Bethesda, MD, USA, 2007. [Google Scholar]
- Paganetti, H. Relative biological effectiveness (RBE) values for proton beam therapy. Variations as a function of biological endpoint, dose, and linear energy transfer. Phys. Med. Biol. 2014, 59, R419–R472. [Google Scholar] [CrossRef]
- Mohan, R.; Grosshans, D. Proton therapy—Present and future. Adv. Drug Deliv. Rev. 2017, 109, 26–44. [Google Scholar] [CrossRef]
- Carante, M.P.; Ballarini, F. Calculating Variations in Biological Effectiveness for a 62 MeV Proton Beam. Front. Oncol. 2016, 6, 76. [Google Scholar]
- Ballarini, F.; Carante, M.P. Chromosome aberrations and cell death by ionizing radiation: Evolution of a biophysical model. Radiat. Phys. Chem. 2016, 128, 18–25. [Google Scholar] [CrossRef]
- Tello, J.J.; Carante, M.P.; Bernal, M.; Ballarini, F. Proximity effects in chromosome aberration induction by low-LET ionizing radiation. DNA Repair 2017, 58, 38–46. [Google Scholar] [CrossRef]
- Tello, J.J.; Carante, M.P.; Bernal, M.; Ballarini, F. Proximity effects in chromosome aberration induction: Dependence on radiation quality, cell type and dose. DNA Repair 2018, 64, 45–52. [Google Scholar] [CrossRef]
- Ballarini, F.; Altieri, S.; Bortolussi, S.; Carante, M.P.; Giroletti, E.; Protti, N. The BIANCA model/code of radiation-induced cell death: Application to human cells exposed to different radiation types. Radiat. Environ. Biophys. 2014, 53, 525–533. [Google Scholar] [CrossRef]
- Carante, M.P.; Altieri, S.; Bortolussi, S.; Postuma, I.; Protti, N.; Ballarini, F. Modelling radiation-induced cell death: Role of different levels of DNA damage clustering. Radiat. Environ. Biophys. 2015, 54, 305–316. [Google Scholar] [CrossRef]
- Carrano, A.V. Chromosome aberrations and radiation-induced cell death. I. Transmission and survival parameters of aberrations. Mutat. Res. 1973, 17, 341–353. [Google Scholar] [CrossRef]
- Cornforth, M.; Bedford, J. A quantitative comparison of potentially lethal damage repair and the rejoining of interphase chromosome breaks in low passage normal human fibroblasts. Radiat. Res. 1987, 111, 385–405. [Google Scholar] [CrossRef]
- Carante, M.P.; Ballarini, F. Modelling cell death for cancer hadrontherapy. AIMS Biophys. 2017, 4, 465–490. [Google Scholar] [CrossRef]
- Chatterjee, A.; Schaefer, H.J. Microdosimetric structure of heavy ion tracks in tissue. Radiat. Environ. Biophys. 1976, 13, 215–227. [Google Scholar] [CrossRef]
- Kato, T.A.; Tsuda, A.; Uesaka, M.; Fujimori, A.; Kamada, T.; Tsujii, H.; Okayasu, R. In vitro characterization of cells derived from chordoma cell line U-CH1 following treatment with X-rays, heavy ions and chemotherapeutic drugs. Radiat. Oncol. 2011, 6, 116. [Google Scholar] [CrossRef]
- Fujisawa, H.; Genik, P.C.; Kitamura, H.; Fujimori, A.; Uesaka, M.; Kato, T.A. Comparison of human chordoma cell-kill for 290 MeV/n carbon ions versus 70 MeV protons in vitro. Radiat. Oncol. 2013, 8, 91. [Google Scholar] [CrossRef]
- Henderson, F.C.; McCool, K.; Seigle, J.; Jean, W.; Harter, W.; Gagnon, G.J. Treatment of chordomas with CyberKnife: Georgetown University experience and treatment recommendations. Neurosurgery 2009, 64, A44–A53. [Google Scholar] [CrossRef]
- Stavreva, N.; Stavrev, P.; Warkentin, B.; Fallone, B.G. Derivation of the expressions for γ50 and D50 for different individual TCP and NTCP models. Phys. Med. Biol. 2002, 47, 3591–3604. [Google Scholar] [CrossRef]
- Nahum, A.E.; Tait, D.M. Maximizing Local Control by Customized Dose Prescription for Pelvic Tumours. In Tumor Response Monitoring and Treatment Planning; Breit, A., Heuck, A., Lukas, P., Kneschaurek, P., Mayr, M., Eds.; Springer: Berlin/Heidelberg, Germany, 1992. [Google Scholar]
- Mairani, A.; Brons, S.; Cerutti, F.; Fassò, A.; Ferrari, A.; Kramer, A.; Parodi, K.; Scholz, M.; Sommerer, F. The FLUKA Monte Carlo code coupled with the local effect model for biological calculations in carbon ion therapy. Phys. Med. Biol. 2010, 55, 4273–4289. [Google Scholar] [CrossRef]
- Zaider, M.; Rossi, H.H. The Synergistic Effects of Different Radiations. Radiat. Res. 1980, 83, 732–739. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carante, M.P.; Aricò, G.; Ferrari, A.; Karger, C.P.; Kozlowska, W.; Mairani, A.; Sala, P.; Ballarini, F. In Vivo Validation of the BIANCA Biophysical Model: Benchmarking against Rat Spinal Cord RBE Data. Int. J. Mol. Sci. 2020, 21, 3973. https://doi.org/10.3390/ijms21113973
Carante MP, Aricò G, Ferrari A, Karger CP, Kozlowska W, Mairani A, Sala P, Ballarini F. In Vivo Validation of the BIANCA Biophysical Model: Benchmarking against Rat Spinal Cord RBE Data. International Journal of Molecular Sciences. 2020; 21(11):3973. https://doi.org/10.3390/ijms21113973
Chicago/Turabian StyleCarante, Mario P., Giulia Aricò, Alfredo Ferrari, Christian P. Karger, Wioletta Kozlowska, Andrea Mairani, Paola Sala, and Francesca Ballarini. 2020. "In Vivo Validation of the BIANCA Biophysical Model: Benchmarking against Rat Spinal Cord RBE Data" International Journal of Molecular Sciences 21, no. 11: 3973. https://doi.org/10.3390/ijms21113973
APA StyleCarante, M. P., Aricò, G., Ferrari, A., Karger, C. P., Kozlowska, W., Mairani, A., Sala, P., & Ballarini, F. (2020). In Vivo Validation of the BIANCA Biophysical Model: Benchmarking against Rat Spinal Cord RBE Data. International Journal of Molecular Sciences, 21(11), 3973. https://doi.org/10.3390/ijms21113973




