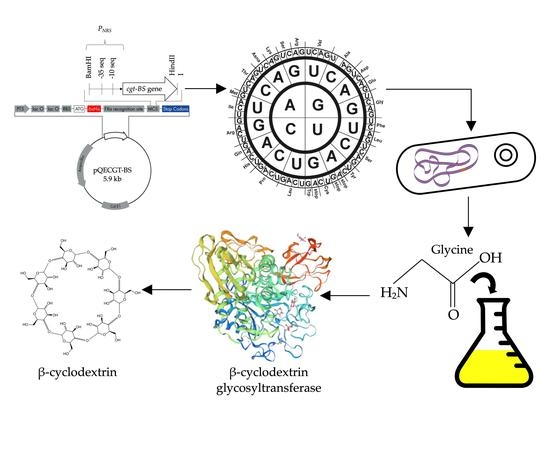
Combined Optimization of Codon Usage and Glycine Supplementation Enhances the Extracellular Production of a β-Cyclodextrin Glycosyltransferase from Bacillus sp. NR5 UPM in Escherichia coli
Abstract
1. Introduction
2. Results and Discussion
2.1. Construction of Recombinant E. coli for Expression of codon-Optimized cgt-BS Gene
2.2. Prediction of Theoretical Physicochemical Properties of β-Cyclodextrin Glycosyltransferase (CGT-BS)
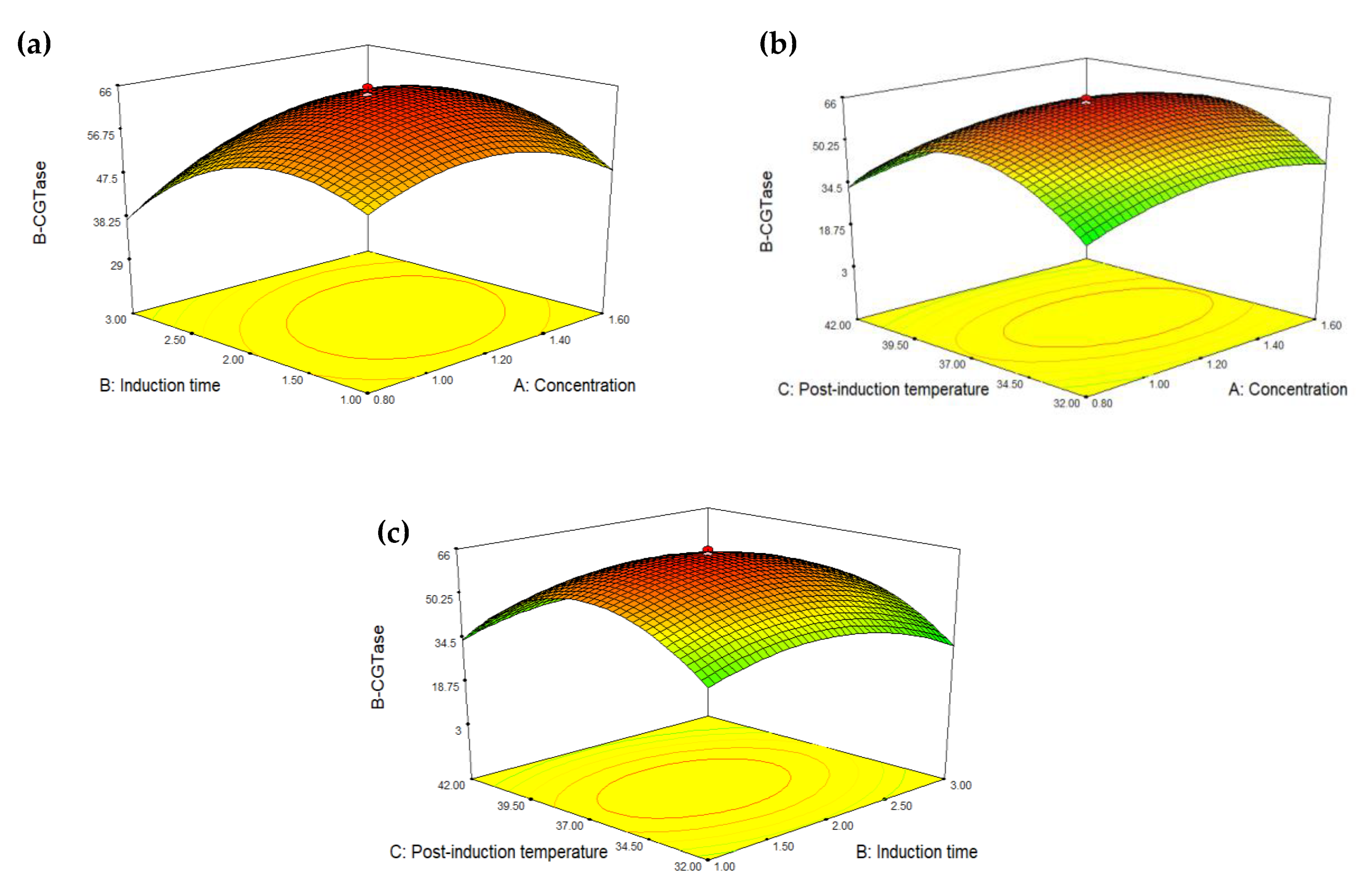
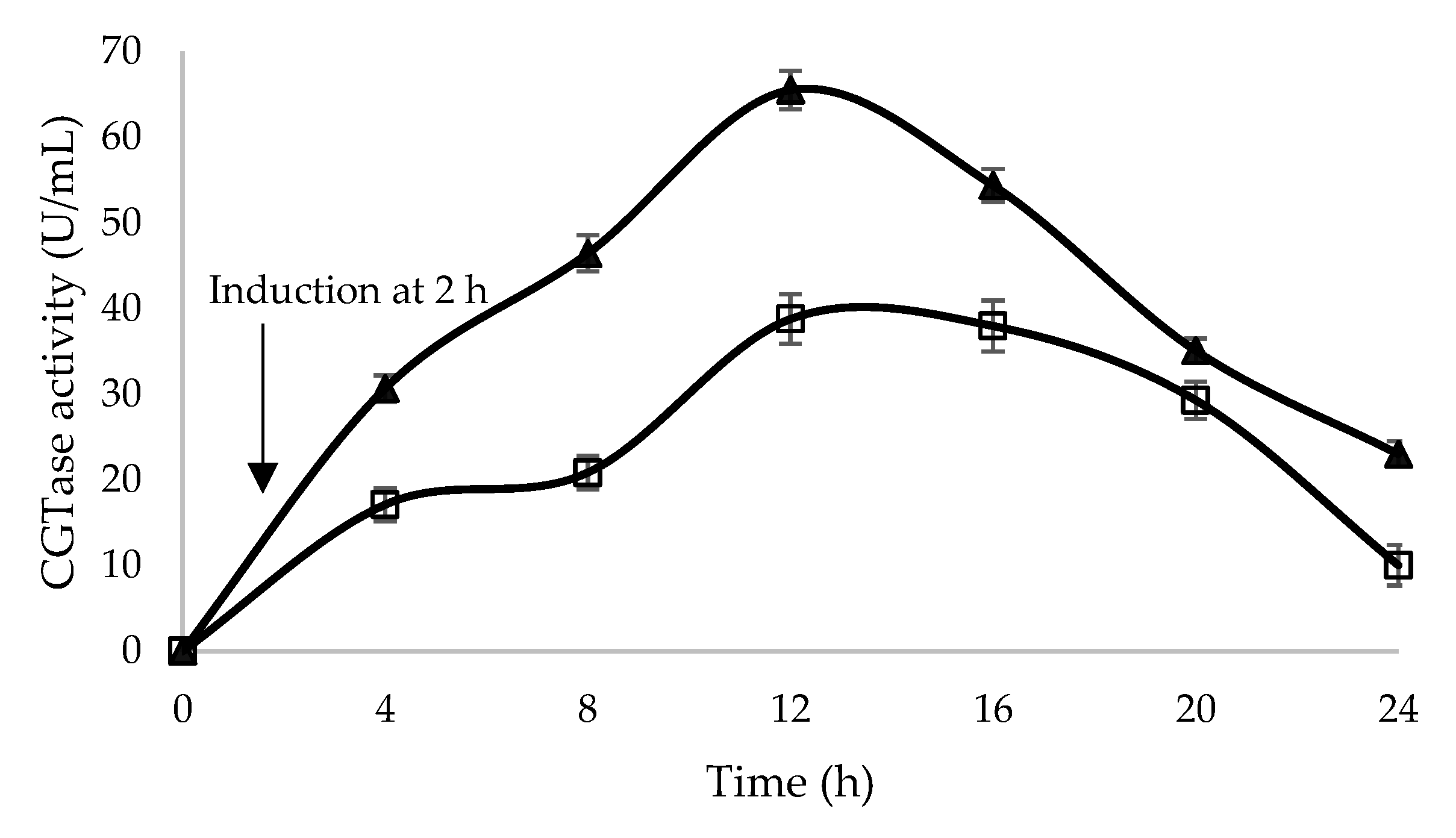
2.3. Optimization of Glycine Supplementation to Improve Extracellular Secretion of Recombinant β-CGTase
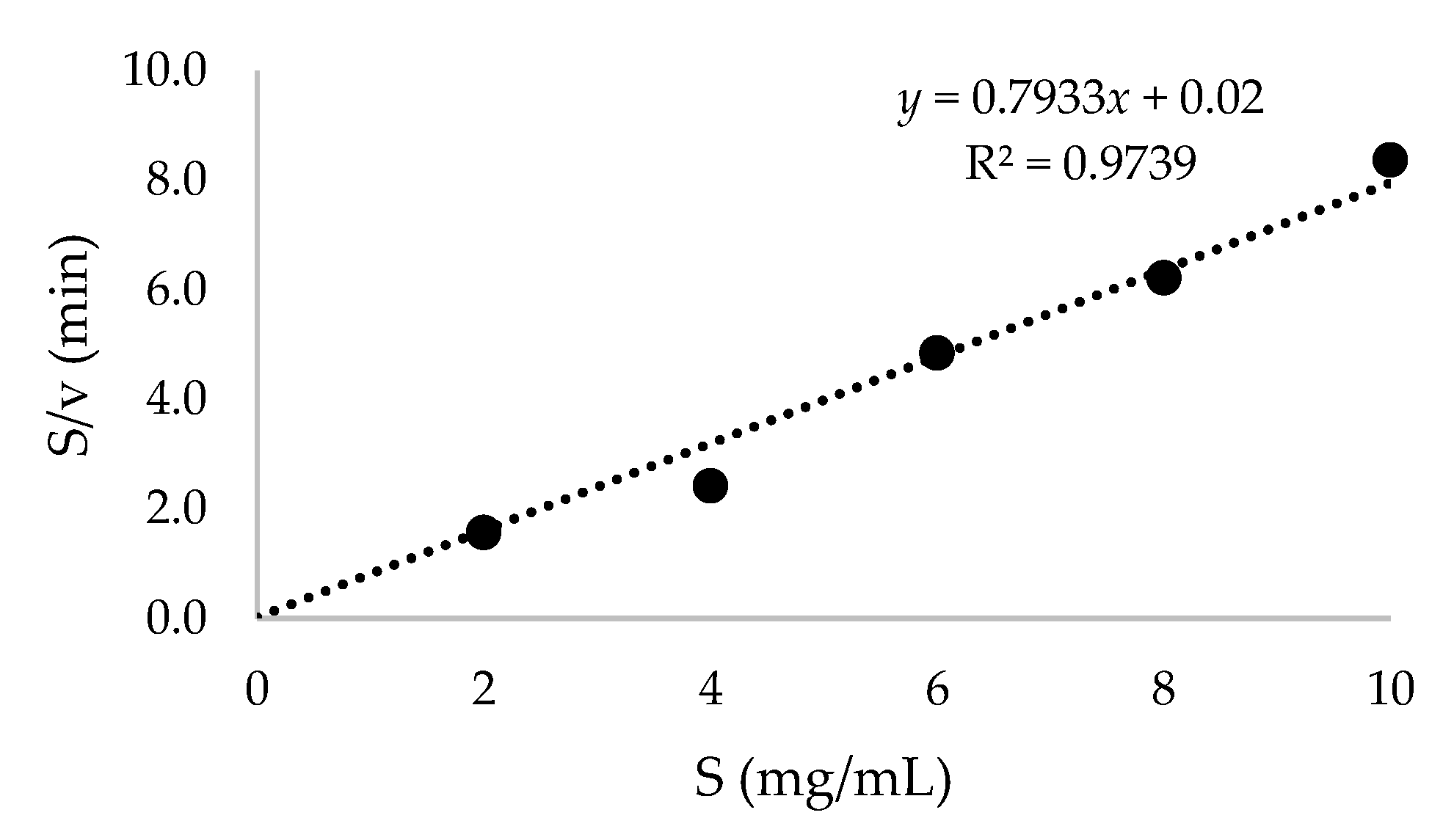
2.4. Purification of Recombinant β-CGTase and Its Kinetic Parameters
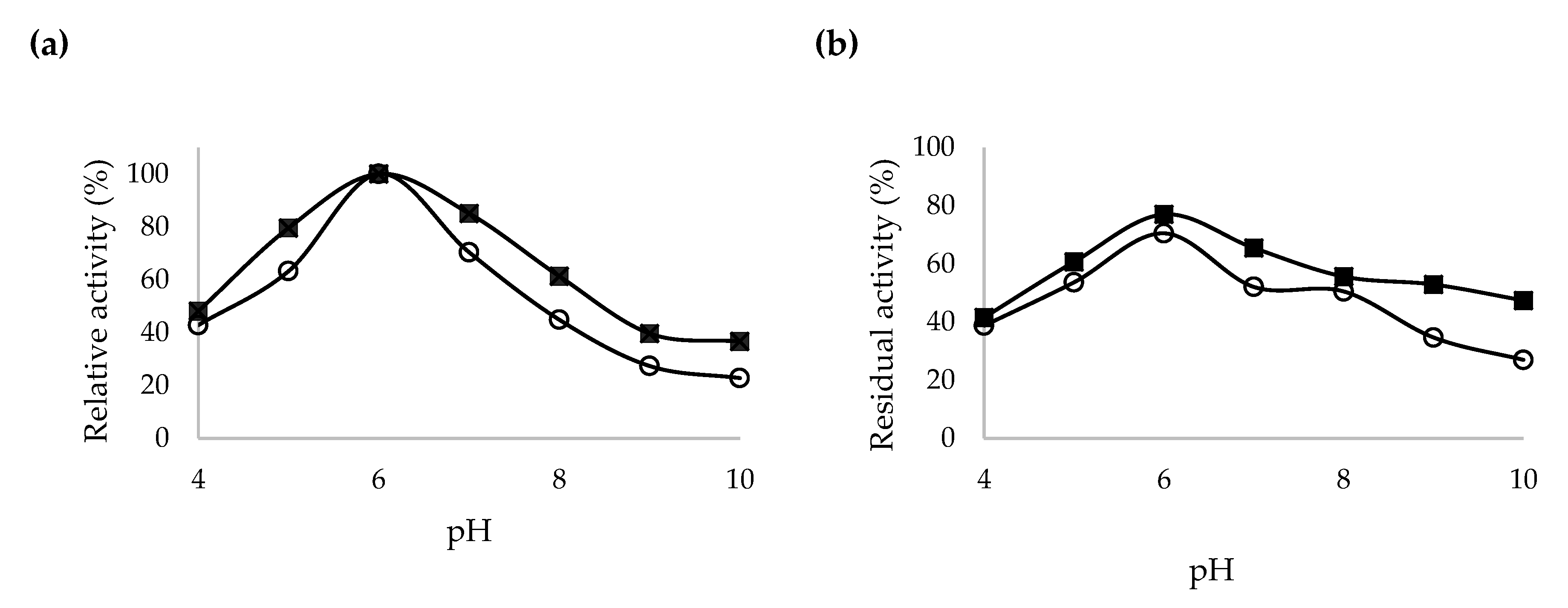
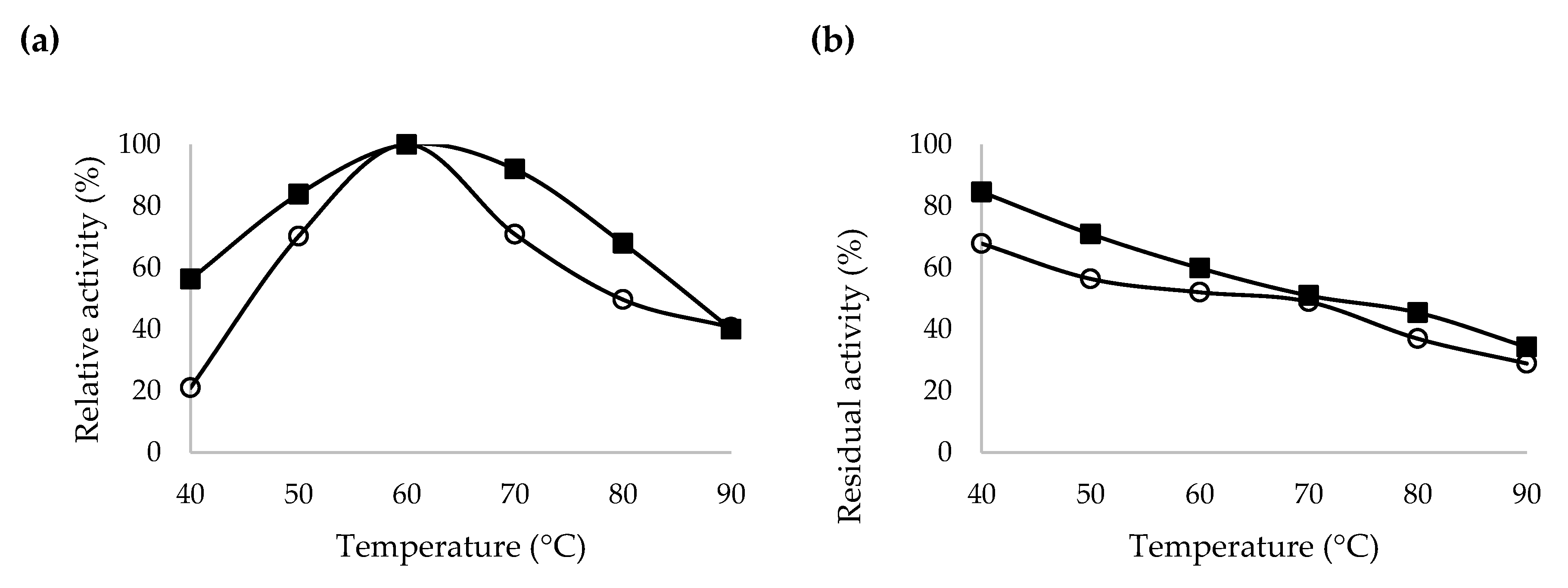
2.5. Characterization of Recombinant β-CGTase
3. Materials and Methods
3.1. Bacterial Strain and Plasmid
3.2. Codon Optimization of cgt-BS Gene
3.3. Construction of the Expression System
3.4. Prediction of Physicochemical Characteristics
3.5. Medium and Culture Conditions
3.6. Design of Experiments for Optimum Condition of Glycine Supplementation
3.7. Purification of Recombinant β-CGTase
3.8. Assay of β-CGTase Activity
3.9. Molecular Mass Determination of β-CGTase
3.10. Characterization of Crude and Purified Codon Optimized Recombinant β-CGTase
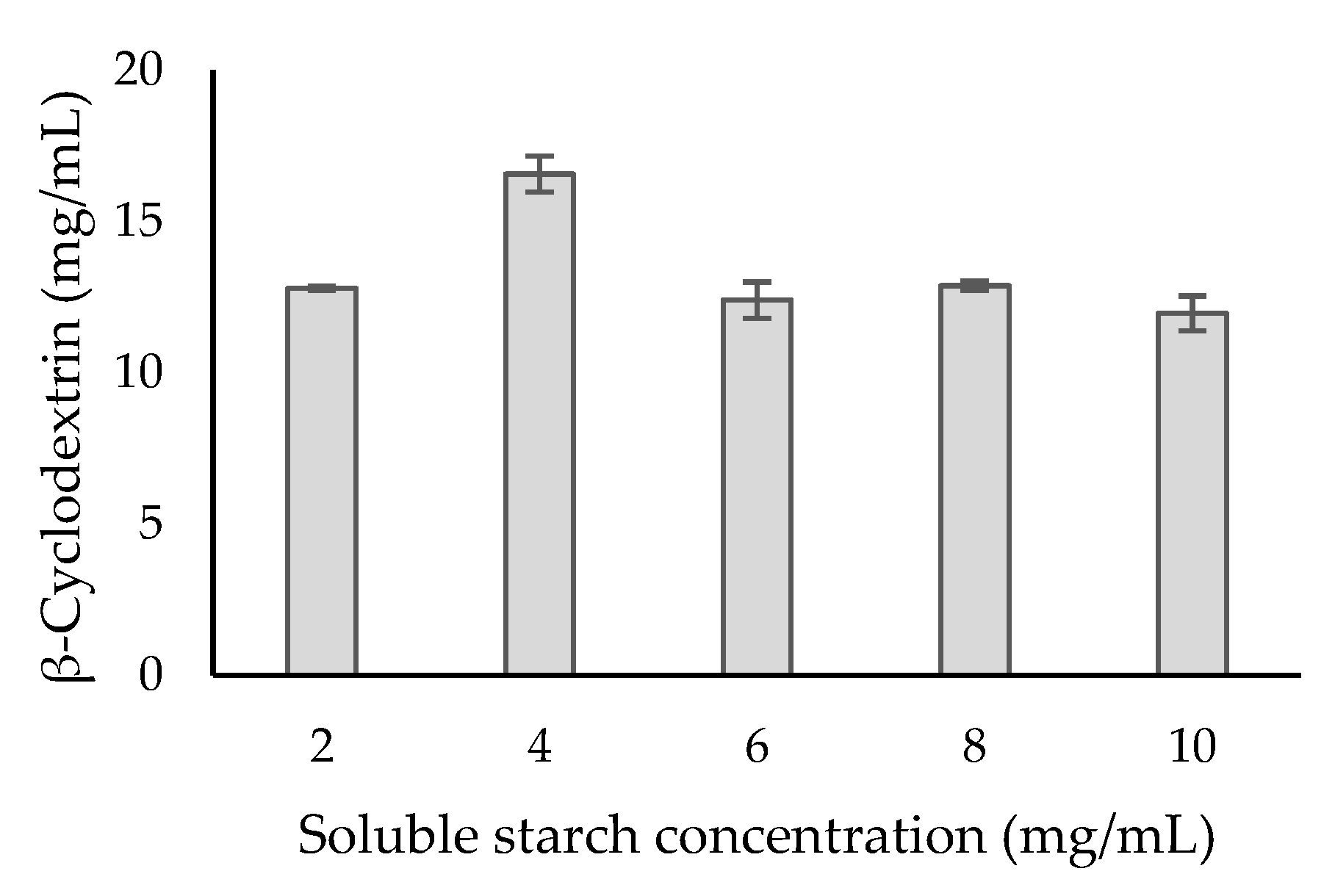
3.11. Cyclodextrin Production and Kinetic Parameters of Purified β-CGTase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van der Veen, B.A.; Uitdehaag, J.C.M.; Dijkstra, B.W.; Dijkhuizen, L. Engineering of cyclodextrin glycosyltransferase reaction and product specificity. Biochim. Biophys. Acta 2000, 1543, 336–360. [Google Scholar] [CrossRef]
- Liu, H.; Li, J.; Du, G.; Zhou, J.; Chen, J. Enhanced production of α-cyclodextrin glycosyltransferase in Escherichia coli by systematic codon usage optimization. J. Ind. Microbiol. Biotechnol. 2012, 39, 1841–1849. [Google Scholar] [CrossRef] [PubMed]
- Samperio, C.; Boyer, R.; Eigel, W.N., III; Holland, K.W.; McKinney, J.S.; O’Keefe, S.F.; Smith, R.; Marcy, J.E. Enhancement of plant essential oils’ aqueous solubility and stability using alpha and beta cyclodextrin. J. Agric. Food Chem. 2010, 58, 12950–12956. [Google Scholar] [CrossRef] [PubMed]
- Campos, E.V.R.; Proenç, P.L.F.; Oliveira, J.L.; Melville, C.C. Chitosan nanoparticles functionalized with β-cyclodextrin: A promising carrier for botanical pesticides. Sci. Rep. 2018. [Google Scholar] [CrossRef]
- Qiu, N.; Cheng, X.; Wang, G.; Wang, W.; Wen, J.; Zhang, Y.; Song, H.; Ma, L.; Wei, Y.; Peng, A.; et al. Inclusion complex of barbigerone with hydroxypropyl-β-cyclodextrin: Preparation and in vitro evaluation. Carbohydr. Polym. 2013, 1–30. [Google Scholar] [CrossRef]
- Xu, G.; Xie, X.; Qin, L.; Hu, X.; Zhang, D.; Xu, J.; Li, D.; Ji, X.; Huang, Y.; Tu, Y.; et al. Simple synthesis of a swellable porous β-cyclodextrin-based polymer for rapid removal of organic micro-pollutants from water. Green Chem. 2019, 1–33. [Google Scholar] [CrossRef]
- Li, Z.; Li, B.; Gu, Z.; Du, G.; Wu, J.; Chen, J. Extracellular expression and biochemical characterization of alpha-cyclodextrin glycosyltransferase from Paenibacillus macerans. Carbohydr. Res. 2010, 345, 886–892. [Google Scholar] [CrossRef]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli : Advances and challenges. Front Microbiol. 2014, 5, 1–17. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, H.; Du, G.; Li, J.; Chen, J. Production of α-cyclodextrin glycosyltransferase in Bacillus megaterium MS941 by systematic codon usage optimization. J. Agric. Food Chem. 2012, 60, 10285–10292. [Google Scholar] [CrossRef]
- Angov, E. Codon usage: Nature’s roadmap to expression and folding of proteins. Biotechnol. J. 2011, 6, 650–659. [Google Scholar] [CrossRef]
- Gasmi, N.; Fudalej, F.; Kallel, H. A molecular approach to optimize hIFN α2b expression and secretion in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2011, 89, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Scorer, C.A.; Carrier, M.J.; Rosenberger, R.F. Amino acid misincorporation during high-level expression of mouse epidermal growth factor in Escherichia coli. Nucleic. Acids Res. 1991, 19, 3511–3516. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koda, A.; Bogaki, T.; Minetoki, T.; Hirotsune, M. High expression of a synthetic gene encoding potato α-glucan phosphorylase in Aspergillus niger. J. Biosci. Bioeng. 2005, 100, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Komar, A.A.; Lesnik, T.; Reiss, C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999, 462, 387–391. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Cheng, J.; Wu, D.; Chen, S.; Chen, J.; Wu, J. High-level extracellular production of α-cyclodextrin glycosyltransferase with recombinant Escherichia coli BL21 (DE3). J. Agric. Food Chem. 2011, 59, 3797–3802. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Li, Z.; Chen, S.; Wu, D.; Wu, J.; Chen, J. Enhanced secretion of recombinant α-cyclodextrin glucosyltransferase from E. coli by medium additives. Process. Biochem. 2010, 45, 880–886. [Google Scholar] [CrossRef]
- Tesfai, B.T.; Wu, D.; Chen, S.; Chen, J.; Wu, J. Strategies for enhancing extracellular secretion of recombinant cyclodextrin glucanotransferase in E. coli. Appl. Biochem. Biotechnol. 2012, 167, 897–908. [Google Scholar] [CrossRef]
- Nik-Pa, N.I.M.; Abd-Aziz, S.; Ibrahim, M.F.; Alitheen, N.B.M.; Ramli, N. Improved extracellular secretion of β-cyclodextrin glycosyltransferase from Escherichia coli by glycine supplementation without apparent cell lysis. Asia Pac. J. Mol. Biol. Biotechnol. 2019, 27, 93–102. [Google Scholar] [CrossRef]
- Li, B.; Wang, L.; Su, L.; Chen, S.; Li, Z.; Chen, J.; Wu, J. Glycine and Triton X-100 enhanced secretion of recombinant α-CGTase mediated by OmpA signal peptide in Escherichia coli. Biotechnol. Bioprocess. Eng. 2012, 17, 1128–1134. [Google Scholar] [CrossRef]
- Anumalla, B.; Prabhu, N.P. Counteracting effect of charged amino acids against the destabilization of proteins by arginine. Appl. Biochem. Biotechnol. 2019, 189, 541–555. [Google Scholar] [CrossRef]
- Smith, M.S.; Billings, W.M.; Whitby, F.G.; Miller, M.B.; Price, J.L. Enhancing a long-range salt bridge with intermediate aromatic and nonpolar amino acids. Org. Biomol. Chem. 2017, 15, 5882–5886. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Lee, S.Y. Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl. Microbiol. Biotechnol. 2004, 64, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gu, Z.; Wang, M.; Du, G.; Wu, J.; Chen, J. Delayed supplementation of glycine enhances extracellular secretion of the recombinant alpha-cyclodextrin glycosyltransferase in Escherichia coli. Appl Microbiol Biotechnol 2010, 85, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, J.; Liu, X.; Pan, X.; Hou, J.; Ran, C.; Zhou, Z. Improving extracellular production of Serratia marcescens lytic polysaccharide monooxygenase CBP21 and Aeromonas veronii B565 chitinase Chi92 in Escherichia coli and their synergism. AMB Express 2017, 7, 170. [Google Scholar] [CrossRef]
- Lo, P.K.; Hassan, O.; Ahmad, A.; Mahadi, N.M.; Illias, R.M. Excretory over-expression of Bacillus sp. G1 cyclodextrin glucanotransferase (CGTase) in Escherichia coli: Optimization of the cultivation conditions by response surface methodology. Enzyme Microb. Technol. 2007, 40, 1256–1263. [Google Scholar] [CrossRef]
- Donovan, R.S.; Robinson, C.W.; Click, B.R. Review: Optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter. J. Ind. Microbiol. 1996, 16, 145–154. [Google Scholar] [CrossRef]
- Sanden, A.M.; Prytz, I.; Tubulekas, I.; Forberg, C.; Le, H.; Hektor, A.; Neubauer, P.; Pragai, Z.; Harwood, C.; Ward, A.; et al. Limiting factors in Escherichia coli Fed-batch production of recombinant proteins. Biotechnol. Bioeng. 2003, 81, 158–166. [Google Scholar] [CrossRef]
- Duan, X.; Chen, S.; Chen, J.; Wu, J. Enhancing the cyclodextrin production by synchronous utilization of isoamylase and α-CGTase. Appl. Microbiol. Biotechnol. 2013, 97, 3467–3674. [Google Scholar] [CrossRef]
- Farewell, A.; Neidhardt, F.C. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J. Bacteriol. 1998, 180, 4704–4710. [Google Scholar] [CrossRef]
- Ramli, N.; Abd-aziz, S.; Hassan, M.A.; Alitheen, N.; Kamaruddin, K. Potential cyclodextrin glycosyltransferase producer from locally isolated bacteria. Afr. J. Biotechnol. 2010, 9, 7317–7321. [Google Scholar] [CrossRef]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biocehmistry 2015, 59, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Li, J.; Shin, H.; Du, G.; Chen, J.; Liu, L. Efficient expression of cyclodextrin glycosyltransferase from Geobacillus stearothermophilus in Escherichia coli by promoter engineering and downstream box evolution. J. Biotechnol. 2018, 266, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Rakmai, J.; Cheirsilp, B. Continuous production of β-cyclodextrin by cyclodextrin glycosyltransferase immobilized in mixed gel beads: Comparative study in continuous stirred tank reactor and packed bed reactor. Biochem. Eng. J. 2016, 105, 107–113. [Google Scholar] [CrossRef]
- Ong, R.M.; Goh, K.M.; Mahadi, N.M.; Hassan, O.; Rahman, R.N.Z.R.A.; Illias, R.M. Cloning, extracellular expression and characterization of a predominant β-CGTase from Bacillus sp. G1 in E. coli. J. Ind. Microbiol. Biotechnol. 2008, 35, 1705–1714. [Google Scholar] [CrossRef] [PubMed]
- Ramli, N.; Abd-Aziz, S.; Alitheen, N.B.; Hassan, M.A.; Maeda, T. Improvement of cyclodextrin glycosyltransferase gene expression in Escherichia coli by insertion of regulatory sequences involved in the promotion of RNA transcription. Mol. Biotechnol. 2013, 54, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Kato, T.; Nakamura, N.; Horikoshi, K. Spectrophotometric determination of cyclization activity of β-cyclodextrin-forming cyclomaltodextrin glucanotransferase. J. Jpn. Soc. Starch. Sci. 1987, 34, 45–48. [Google Scholar] [CrossRef]
- Lowry, O.H.; Randall, R.J.; Rosebrough, N.J.; Farr, A.L. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Rahman, K.; Illias, R.M.; Hassan, O.; Mahmood, N.A.N.; Rashid, N.A.A. Molecular cloning of a cyclodextrin glucanotransferase gene from alkalophilic Bacillus sp. TS1-1 and characterization of the recombinant enzyme. Enzyme Microb. Technol. 2006, 39, 74–84. [Google Scholar] [CrossRef]
- Martins, R.F.; Hatti-Kaul, R. A new cyclodextrin glycosyltransferase from an alkaliphilic Bacillus agaradhaerens isolate: Purification and characterisation. Enzyme Microb. Technol. 2002, 30, 116–124. [Google Scholar] [CrossRef]
| Amino Acid | Codon | Relative Frequency | Frequency | Amino Acid | Codon | Relative Frequency | Frequency | ||
|---|---|---|---|---|---|---|---|---|---|
| E. coli | cgt-BS | cocgt-BS | E. coli | cgt-BS | cocgt-BS | ||||
| Ala (A) | GCA | 0.22 | 11 | 7 | Leu (L) | CUA | 0.03 | 5 | 0 |
| GCC | 0.25 | 8 | 9 | CUC | 0.10 | 4 | 6 | ||
| GCG | 0.34 | 5 | 16 | CUG | 0.55 | 2 | 27 | ||
| GCU | 0.19 | 9 | 1 | CUU | 0.10 | 9 | 5 | ||
| Arg (R) | AGA | 0.04 | 3 | 0 | UUA | 0.11 | 31 | 13 | |
| AGG | 0.03 | 1 | 0 | UUG | 0.11 | 9 | 9 | ||
| CGA | 0.05 | 5 | 0 | Lys (K) | AAA | 0.76 | 22 | 27 | |
| CGC | 0.37 | 4 | 12 | AAG | 0.24 | 10 | 5 | ||
| CGG | 0.08 | 1 | 2 | Met (M) | AUG | 1.00 | 15 | 15 | |
| CGU | 0.42 | 8 | 8 | Phe (F) | UUC | 0.49 | 11 | 14 | |
| Asn (N) | AAC | 0.61 | 34 | 41 | UUU | 0.51 | 24 | 21 | |
| AAU | 0.39 | 36 | 29 | Pro (P) | CCA | 0.20 | 7 | 4 | |
| Asp (D) | GAC | 0.41 | 11 | 20 | CCC | 0.10 | 3 | 2 | |
| GAU | 0.59 | 37 | 28 | CCG | 0.55 | 2 | 18 | ||
| Cys (C) | UGC | 0.57 | 1 | 4 | CCU | 0.16 | 7 | 5 | |
| UGU | 0.43 | 3 | 0 | Ser (S) | AGC | 0.27 | 16 | 19 | |
| Gln (Q) | CAA | 0.31 | 22 | 10 | AGU | 0.13 | 11 | 8 | |
| CAG | 0.69 | 10 | 22 | UCA | 0.12 | 16 | 14 | ||
| Glu (E) | GAA | 0.70 | 22 | 21 | UCC | 0.17 | 6 | 9 | |
| GAG | 0.30 | 9 | 10 | UCG | 0.13 | 4 | 14 | ||
| Gly (G) | GGA | 0.09 | 13 | 3 | UCU | 0.19 | 14 | 3 | |
| GGC | 0.40 | 17 | 22 | Thr (T) | ACA | 0.30 | 17 | 11 | |
| GGG | 0.13 | 13 | 7 | ACC | 0.43 | 14 | 26 | ||
| GGU | 0.38 | 16 | 27 | ACG | 0.23 | 14 | 13 | ||
| His (H) | CAC | 0.48 | 7 | 7 | ACU | 0.21 | 11 | 6 | |
| CAU | 0.52 | 10 | 10 | Trp (V) | UGG | 1.00 | 15 | 15 | |
| Ile (I) | AUA | 0.07 | 12 | 0 | Tyr (Y) | UAC | 0.47 | 20 | 19 |
| AUC | 0.46 | 13 | 30 | UAU | 0.53 | 28 | 29 | ||
| AUU | 0.47 | 24 | 19 | Val (V) | GUA | 0.17 | 18 | 10 | |
| GUC | 0.20 | 11 | 18 | ||||||
| GUG | 0.34 | 5 | 11 | ||||||
| GUU | 0.29 | 19 | 14 | ||||||
| Description | Accession Number | Theoretical MW (Da) | pI | R+ | R− | EC a (M−1·cm−1) | EC b (M−1·cm−1) | II | Stability | AI | GRAVY | TNA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus sp. NR5 UPM cyclodextrin glycosyltransferase (cgt) gene * | HQ876173.1 | 80,622.34 | 4.81 | 48 | 76 | 142,810 | 142,560 | 24.57 | stable | 75.36 | −0.405 | 11,108 |
| B. circulans strain 251 cgt gene | X78145.1 | 77,309.28 | 6.08 | 52 | 58 | 120,795 | 120,670 | 22.72 | stable | 74.05 | −0.250 | 10,710 |
| B. licheniformis cgtA gene for cyclomaltodextrin glucanotransferase (CGTase) | X15752.1 | 78,002.68 | 5.57 | 48 | 59 | 124,805 | 124,680 | 18.17 | stable | 71.75 | −0.243 | 10,785 |
| Bacillus sp. (strain no. 38-2) cyclomaltodextrin glucanotransferase (CGTase) gene | M19880.1 | 78,249.29 | 5.41 | 49 | 63 | 133,285 | 133,160 | 25.23 | stable | 75.76 | −0.286 | 10,826 |
| Bacillus sp. strain Y112 cyclodextrin glycosyltransferase precursor (cgtase) gene | KX579963.2 | 80,168.90 | 4.23 | 46 | 111 | 129,735 | 129,610 | 30.92 | stable | 73.39 | −0.483 | 10,983 |
| Bacillus sp. BL-31 cyclodextrin glucanotransferase (cgt) gene | EF363797.1 | 80,096.84 | 4.24 | 46 | 110 | 129,735 | 129,610 | 30.65 | stable | 73.39 | −0.478 | 10,974 |
| Run | Concentration (mM) | Induction Time (h) | Post-Induction Temperature (°C) | β-CGTase Activity (U/mL) | |
|---|---|---|---|---|---|
| Experimental | Predicted | ||||
| 1 | 1.60 | 1.00 | 42.0 | 23.47 | 23.47 |
| 2 | 1.60 | 3.00 | 32.0 | 34.23 | 32.38 |
| 3 | 1.20 | 2.00 | 37.0 | 64.76 | 64.17 |
| 4 | 1.20 | 2.00 | 37.0 | 63.88 | 64.17 |
| 5 | 1.60 | 3.00 | 42.0 | 24.37 | 25.77 |
| 6 | 1.20 | 2.00 | 37.0 | 62.72 | 64.17 |
| 7 | 1.20 | 2.00 | 37.0 | 65.73 | 64.17 |
| 8 | 1.20 | 3.68 | 37.0 | 29.96 | 30.70 |
| 9 | 1.20 | 2.00 | 37.0 | 63.97 | 64.17 |
| 10 | 1.60 | 1.00 | 32.0 | 30.18 | 31.67 |
| 11 | 1.20 | 2.00 | 37.0 | 63.83 | 64.17 |
| 12 | 0.80 | 3.00 | 32.0 | 16.61 | 17.19 |
| 13 | 1.20 | 0.32 | 37.0 | 42.88 | 41.31 |
| 14 | 0.80 | 3.00 | 42.0 | 17.57 | 16.66 |
| 15 | 1.20 | 2.00 | 28.6 | 8.47 | 9.11 |
| 16 | 0.80 | 1.00 | 32.0 | 32.92 | 32.10 |
| 17 | 1.20 | 2.00 | 45.4 | 3.11 | 1.65 |
| 18 | 1.87 | 2.00 | 37.0 | 47.64 | 47.31 |
| 19 | 0.80 | 1.00 | 42.0 | 27.55 | 29.98 |
| 20 | 0.53 | 2.00 | 37.0 | 40.50 | 40.01 |
| Source | Sum of Squares | Degree of Freedom (df) | Mean Square | F Value | p-Value (Probability > F) | Significant Term Based on p-Value |
|---|---|---|---|---|---|---|
| Model | 7839.26 | 9 | 871.03 | 329.64 | <0.0001 | Significant |
| A | 64.26 | 1 | 64.26 | 24.32 | 0.0006 | |
| B | 135.82 | 1 | 135.82 | 51.40 | <0.0001 | |
| C | 64.98 | 1 | 64.98 | 24.59 | 0.0006 | |
| AB | 121.96 | 1 | 121.96 | 46.16 | <0.0001 | |
| AC | 18.47 | 1 | 18.47 | 6.99 | 0.0246 | |
| BC | 1.27 | 1 | 1.27 | 0.48 | 0.5044 | |
| A2 | 758.03 | 1 | 758.03 | 286.88 | <0.0001 | |
| B2 | 1428.85 | 1 | 1428.85 | 540.75 | <0.0001 | |
| C2 | 6229.12 | 1 | 6229.12 | 2357.42 | <0.0001 | |
| Residual | 26.42 | 10 | 2.64 | |||
| Lack of fit | 21.29 | 5 | 4.26 | 4.14 | 0.0724 | Not significant |
| Pure error | 5.14 | 5 | 1.03 | |||
| Cor total | 7865.69 | 19 | ||||
| R2 | 0.9966 | |||||
| Adjusted R2 | 0.9936 |
| Purification Step | Volume (mL) | Total Activity (U/mL) | Total Protein (mg/mL) | Specific Activity (U/mg) | Purification Fold | Purification Yield (%) |
|---|---|---|---|---|---|---|
| Crude | 1000 | 77.1 | 16.2 | 4.8 | 1.0 | 100.0 |
| Diafiltration | 250 | 65.6 | 10.6 | 6.2 | 1.3 | 85.2 |
| Affinity Chromatography | 9 | 19.1 | 0.2 | 95.6 | 20.0 | 24.8 |
| Ultrafiltration | 3 | 11.0 | 0.1 | 113.4 | 23.8 | 14.3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nik-Pa, N.I.M.; Sobri, M.F.M.; Abd-Aziz, S.; Ibrahim, M.F.; Kamal Bahrin, E.; Mohammed Alitheen, N.B.; Ramli, N. Combined Optimization of Codon Usage and Glycine Supplementation Enhances the Extracellular Production of a β-Cyclodextrin Glycosyltransferase from Bacillus sp. NR5 UPM in Escherichia coli. Int. J. Mol. Sci. 2020, 21, 3919. https://doi.org/10.3390/ijms21113919
Nik-Pa NIM, Sobri MFM, Abd-Aziz S, Ibrahim MF, Kamal Bahrin E, Mohammed Alitheen NB, Ramli N. Combined Optimization of Codon Usage and Glycine Supplementation Enhances the Extracellular Production of a β-Cyclodextrin Glycosyltransferase from Bacillus sp. NR5 UPM in Escherichia coli. International Journal of Molecular Sciences. 2020; 21(11):3919. https://doi.org/10.3390/ijms21113919
Chicago/Turabian StyleNik-Pa, Nik Ida Mardiana, Mohamad Farhan Mohamad Sobri, Suraini Abd-Aziz, Mohamad Faizal Ibrahim, Ezyana Kamal Bahrin, Noorjahan Banu Mohammed Alitheen, and Norhayati Ramli. 2020. "Combined Optimization of Codon Usage and Glycine Supplementation Enhances the Extracellular Production of a β-Cyclodextrin Glycosyltransferase from Bacillus sp. NR5 UPM in Escherichia coli" International Journal of Molecular Sciences 21, no. 11: 3919. https://doi.org/10.3390/ijms21113919
APA StyleNik-Pa, N. I. M., Sobri, M. F. M., Abd-Aziz, S., Ibrahim, M. F., Kamal Bahrin, E., Mohammed Alitheen, N. B., & Ramli, N. (2020). Combined Optimization of Codon Usage and Glycine Supplementation Enhances the Extracellular Production of a β-Cyclodextrin Glycosyltransferase from Bacillus sp. NR5 UPM in Escherichia coli. International Journal of Molecular Sciences, 21(11), 3919. https://doi.org/10.3390/ijms21113919