Zygotic Vsx1 Plays a Key Role in Defining V2a Interneuron Sub-Lineage by Directly Repressing tal1 Transcription in Zebrafish
Abstract
1. Introduction
2. Results
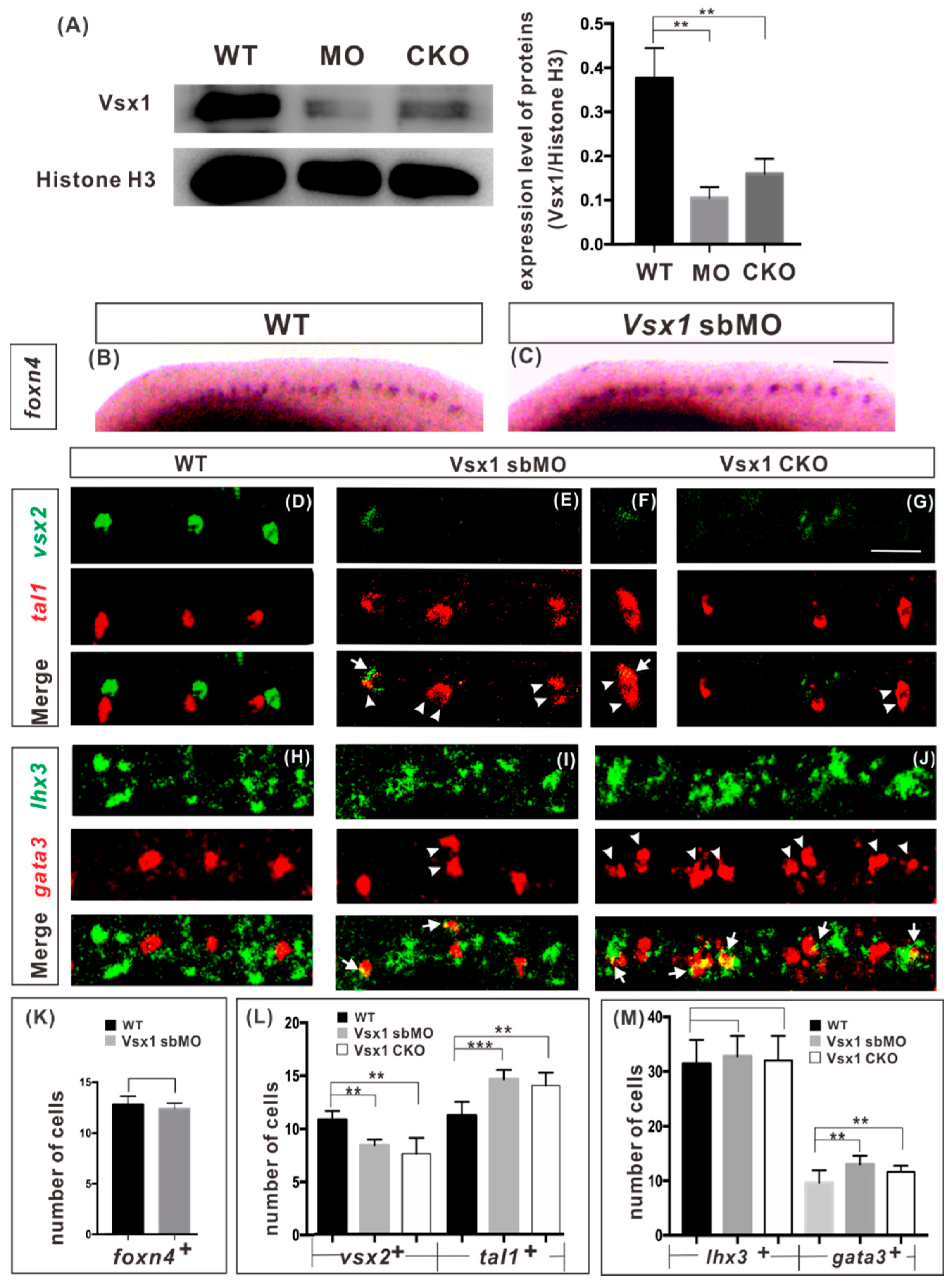
2.1. Zygotic Vsx1 Has no Impact on the Generation of both P2 Progenitors and Its Adjacent Neurons
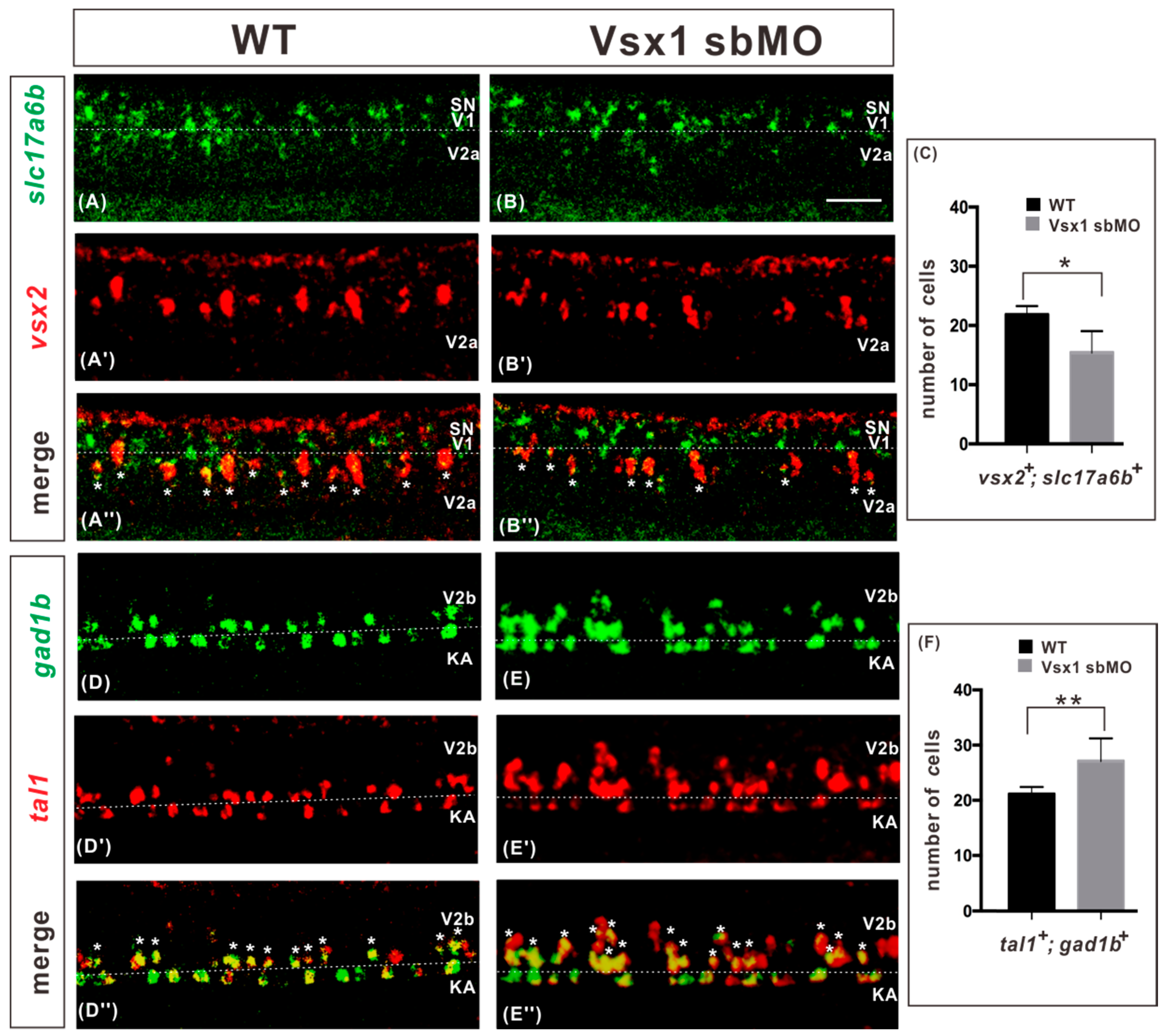
2.2. Vsx1 is Essential for Repressing Tal1 Expression in Presumptive V2a Cells and Defining V2a Sub-Lineage
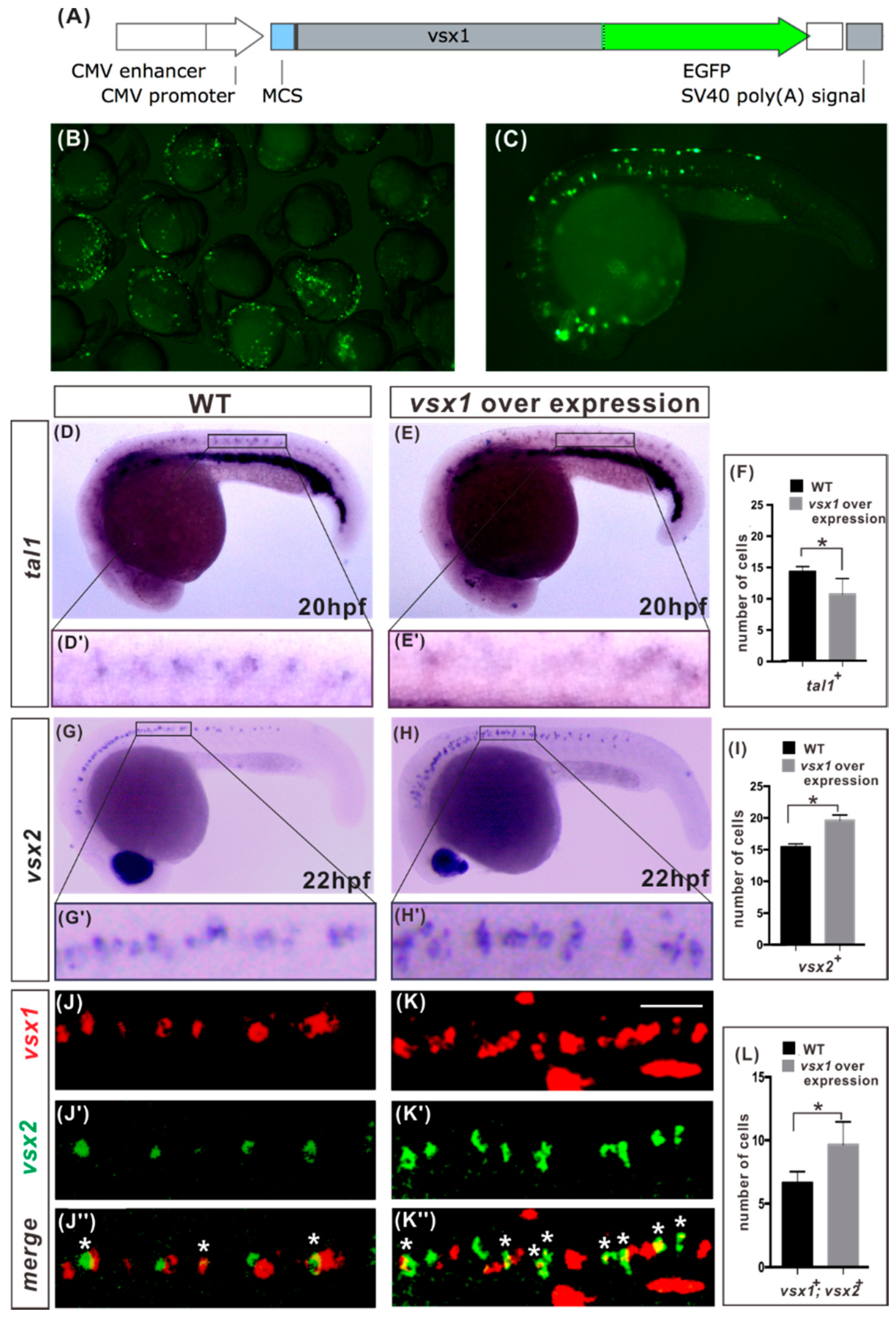
2.3. Vsx1 Overexpression can Repress tal1 Expression in Presumptive V2b Cells and Elicit Ectopic V2a Generation
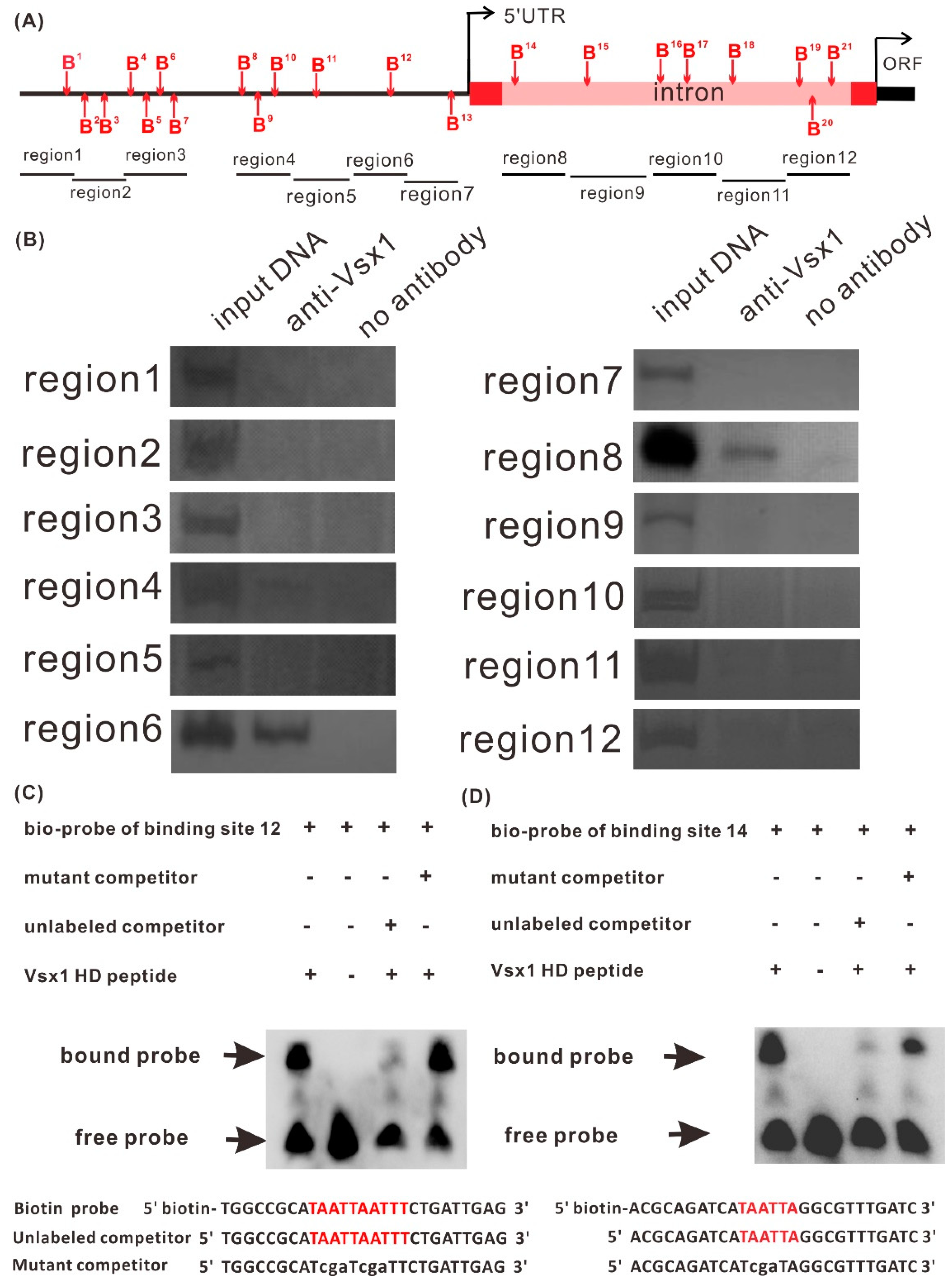
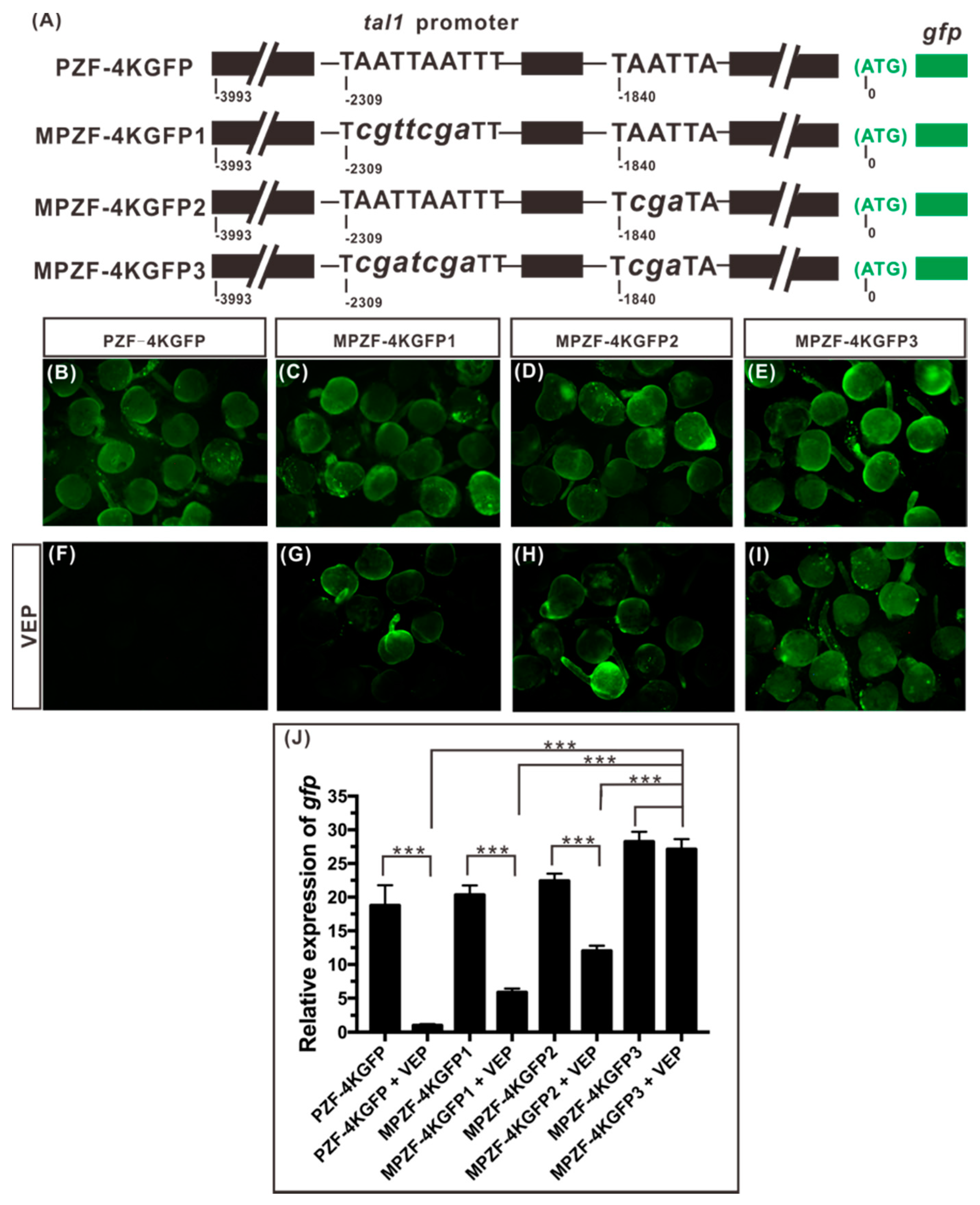
2.4. Vsx1 Can Directly Repress tal1 Transcription
2.5. Both Binding Sites, 12 and 14, Play a Role in Mediating the Repression of tal1 Transcription by Vsx1
3. Discussion
3.1. Vsx1 Plays a Role in Stimulating vsx2 Expression in V2a Interneuron Sub-Lineage in Zebrafish
3.2. Direct Repression of tal1 by Paired-Like Homeodomain Repressor might Be an Evolutionarily Conserved Mechanism of V2a and V2b Diversification
3.3. Notch-Delta Signaling may Activate Ubiquitin-Dependent Proteolysis of Vsx1 to Promote tal1 Transcription
4. Materials and Methods
4.1. Zebrafish Maintance and Embryo Acquisition
4.2. Morpholino Oligonucleotides
4.3. Transient Knockout of vsx1 by CRISPR/Cas9
4.4. Construction of vsx1–egfp Fusion Gene Expression Plasmids
4.5. Cloning of Proximal Promoter Sequence of Zebrafish tal1 and Construction of GFP Sensors
4.6. Microinjection
4.7. Western Blotting
4.8. Digoxigenin or Fluorescein-Labeled RNA Probe Synthesis
4.9. Whole Mount In Situ Hybridization (WISH)
4.10. Cell Counts and Statistical Analyses
4.11. Chromatin Immuno Precipitation (ChIP)
4.12. Electrophoretic Mobility Shift Assay (EMSA)
4.13. Quantitative RT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| hpf | hours post fertilization |
| sbMO | splice-blocking Morpholino |
| WT | wide type |
| KD | knock down |
| CKO | chimeric knock out |
| WISH | whole mount in situ hybridization |
References
- Crone, S.A.; Quinlan, K.A.; Zagoraiou, L.; Droho, S.; Restrepo, C.E.; Lundfald, L.; Endo, T.; Setlak, J.; Jessell, T.M.; Kiehn, O.; et al. Genetic ablation of V2a ipsilateral interneurons disrupts left right locomotor coordination in mammalian spinal cord. Neuron 2008, 60, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Droho, S.; Crone, S.A.; Dietz, S.; Kwan, A.C.; Webb, W.W.; Sharma, K.; Harris-Warrick, R.M. Electrophysiological characterization of V2a interneurons and their locomotor-related activity in the neonatal mouse spinal cord. J. Neurosci. 2010, 30, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Ausborn, J.; Mahmood, R.; Manira, A.E. Decoding the rules of recruitment of excitatory interneurons in the adult zebrafish locomotor network. Proc. Natl. Acad. Sci. USA 2012, 109, E3631–E3639. [Google Scholar] [CrossRef] [PubMed]
- Eklöf-Ljunggren, E.; Haupt, S.; Ausborn, J.; Dehnisch, I.; Uhlén, P.; Higashima, S.; Manira, A.E. Origin of excitation underlying locomotion in the spinal circuit of zebrafish. Proc. Natl. Acad. Sci. USA 2012, 109, 5511–5516. [Google Scholar] [CrossRef]
- Azim, E.; Jiang, J.; Alstermark, B.; Jessell, T.M. Skilled reaching relies on a V2a propriospinal internal copy circuit. Nature 2014, 508, 357–363. [Google Scholar] [CrossRef]
- Karunaratne, A.; Hargrave, M.; Poh, A.; Yamada, T. GATA proteins identify a novel ventral interneuron subclass in the developing chick spinal cord. Dev. Biol. 2002, 249, 30–43. [Google Scholar] [CrossRef]
- Li, S.; Misra, K.; Matise, M.P.; Xiang, M. Foxn4 acts synergistically with Mash1 to specify subtype identity of V2 interneurons in the spinal cord. Proc. Natl. Acad. Sci. USA 2005, 102, 10688–10693. [Google Scholar] [CrossRef]
- Smith, E.; Hargrave, M.; Yamada, T.; Begley, C.G. Little MH. Coexpression of TAL1 and GATA3 in the V2 interneurons of the developing mouse spinal cord. Dev. Dyn. 2002, 224, 231–237. [Google Scholar] [CrossRef]
- Zhou, Y.; Yamamoto, M.; Engel, J.D. GATA2 is required for the generation of V2 interneurons. Development 2000, 127, 3829–3838. [Google Scholar]
- Peng, C.Y.; Yajima, H.; Burns, C.E.; Zon, L.I.; Sisodia, S.S.; Pfaff, S.L.; Sharma, K. Notch and MAML signaling drives Tal1-dependent interneuron diversity in the spinal cord. Neuron 2007, 53, 813–827. [Google Scholar] [CrossRef]
- Batista, M.F.; Jacobstein, J.; Lewis, K.E. Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev. Biol. 2008, 322, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Satou, C.; Higashijima, S. V2a and V2b neurons are generated by the final divisions of pair-producing progenitors in the zebrafish spinal cord. Development 2008, 135, 3001–3005. [Google Scholar] [CrossRef] [PubMed]
- Muroyama, Y.; Fujiwara, Y.; Orkin, S.H.; Rowitch, D.H. Specification of astrocytes by bHLH protein TAL1 in a restricted region of the neural tube. Nature 2005, 438, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Del Barrio, M.G.; Taveira-Marques, R.; Muroyama, Y.; Yuk, D.; Li, S.; Wines-Samuelson, M.; Shen, J.; Smith, H.K.; Xiang, M.; Rowitch, D.; et al. A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development 2007, 134, 3427–3436. [Google Scholar] [CrossRef] [PubMed]
- Andrzejczuk, L.A.; Banerjee, S.; England, S.J.; Voufo, C.; Kamara, K.; Lewis, K.E. Tal1, Gata2a, and Gata3 have distinct functions in the development of V2b and cerebrospinal fluid-contacting KA spinal neurons. Front. Neurosci. 2018, 12, 170. [Google Scholar] [CrossRef] [PubMed]
- Clovis, Y.M.; Seo, S.Y.; Kwon, J.; Rhee, J.C.; Yeo, S.; Lee, J.W.; Lee, S.; Lee, S.K. Vsx2 Consolidates V2a Interneuron Identity through Two Distinct Gene Repression Modes. Cell Rep. 2016, 16, 1642–1652. [Google Scholar] [CrossRef]
- Tanabe, Y.; William, C.; Jessell, T.M. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell 1998, 95, 67–80. [Google Scholar] [CrossRef]
- Thaler, J.P.; Lee, S.K.; Jurata, L.W.; Gill, G.N.; Pfaff, S.L. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 2002, 110, 237–249. [Google Scholar] [CrossRef]
- Passini, M.A.; Kurtzman, A.L.; Canger, A.K.; Asch, W.S.; Wray, G.A.; Raymond, P.A.; Schechter, N. Cloning of zebrafish vsx1, expression of a paired-like homeobox gene during CNS development. Dev. Genet. 1998, 23, 128–141. [Google Scholar] [CrossRef]
- Chen, C.M.; Cepko, C.L. Expression of Vsx2 and Vsx2-1 in the developing chicken retina. Mech. Dev. 2000, 90, 293–297. [Google Scholar] [CrossRef]
- D’Autilia, S.; Decembrini, S.; Casarosa, S.; He, R.Q.; Barsacchi, G.; Cremisi, F.; Andreazzoli, M. Cloning and developmental expression of the Xenopus homeobox gene Xvsx1. Dev. Genes Evol. 2006, 216, 829–834. [Google Scholar] [CrossRef]
- Francius, C.; Hidalgo-Figueroa, M.; Debrulle, S.; Pelosi, B.; Rucchin, V.; Ronellenfitch, K.; Panayiotou, E.; Makrides, N.; Misra, K.; Harris, A.; et al. Vsx1 Transiently defines an early intermediate V2 interneuron precursor compartment in the mouse developing spinal cord. Front. Mol. Neurosci. 2016, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xu, X.; Zhao, S.; Ma, S.; Sun, L.; Liu, Z.; Luo, C. Maternal control of axial-paraxial mesoderm patterning via direct transcriptional repression in zebrafish. Dev. Biol. 2014, 386, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; He, Y.; Sun, L.; Ma, S.; Luo, C. Maternal Vsx1 plays an essential role in regulating prechordal mesendoderm and forebrain formation in zebrafish. Dev. Biol. 2014, 394, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.M.; Yun, S.; Veien, E.S.; Wu, Y.Y.; Chow, R.L.; Dorsky, R.L.; Levine, E.M. Negative regulation of Vsx1 by its paralog Chx10/Vsx2 is conserved in the vertebrate retina. Brain Res. 2008, 1192, 99–113. [Google Scholar] [CrossRef]
- Jessell, T.M. Neuronal specification in the spinal cord, inductive signals and transcriptional codes. Nat. Rev. Gen. 2000, 1, 20–29. [Google Scholar] [CrossRef]
- Lewis, K.E. How do genes regulate simple behaviours? Understanding how different neurons in the vertebrate spinal cord are genetically specified. Philos. Trans. R Soc. Lond. B Biol. Sci. 2006, 361, 45–66. [Google Scholar] [CrossRef]
- Gerber, V.; Yang, L.; Takamiya, M.; Ribes, V.; Gourain, V.; Peravali, R.; Stegmaier, J.; Mikut, R.; Reischl, M.; Ferg, M.; et al. The HMG box transcription factors Sox1a and Sox1b specify a new class of glycinergic interneuron in the spinal cord of zebrafish embryos. Development 2019, 146. [Google Scholar] [CrossRef]
- Shi, Z.; Trenholm, S.; Zhu, M.; Buddingh, S.; Star, E.N.; Awatramani, G.B.; Chow, R.L. Vsx1 regulates terminal differentiation of type 7 ON bipolar cells. J. Neurosci. 2011, 31, 13118–13127. [Google Scholar] [CrossRef]
- Levine, E.M.; Hitchcock, P.; Glascow, E.; Schechter, N. Restricted expression of a new paired-class homeobox gene in normal and regenerating adult goldfish retina. J. Comp. Neurol. 1994, 348, 596–606. [Google Scholar] [CrossRef]
- Levine, E.M.; Passini, M.A.; Hitchcock, P.; Glascow, E.; Schechter, N. Vsx-1 and Vsx-2, Two Vsx2-like homeobox genes expressed in overlapping domains in the adult goldfish retina. J. Comp. Neurol. 1997, 387, 439–448. [Google Scholar] [CrossRef]
- Percin, E.F.; Ploder, L.A.; Yu, J.J.; Arici, K.; Horsford, D.J.; Rutherford, A.; Bapat, B.; Cox, D.W.; Duncan, A.M.; Kalnins, V.I.; et al. Human microphthalmia associated with mutations in the retinal homeobox gene VSX2. Nat. Genet. 2000, 25, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Dorval, K.M.; Bobechko, B.P.; Fujieda, H.; Chen, S.; Zack, D.J.; Bremner, R. VSX2 targets a subset of photoreceptor genes. J. Biol. Chem. 2006, 281, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Dorval, K.M.; Bobechko, B.P.; Ahmad, K.F.; Bremner, R. Transcriptional activity of the paired-like homeodomain proteins VSX2 and VSX1. J. Biol. Chem. 2005, 280, 10100–10108. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Levine, E.M. Vsx2 controls eye organogenesis and retinal progenitor identity via homeodomain and non-homeodomain residues required for high affinity DNA binding. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef]
- Debrulle, S.; Baudouin, C.; Hidalgo-Figueroa, M.; Pelosi, B.; Francius, C.; Rucchin, V.; Ronellenfitch, K.; Chow, R.L.; Tissir, F.; Lee, S.-K.; et al. Vsx1 and Chx10 paralogs sequentially secure V2 interneuron identity during spinal cord development. Cellular and Molecular Life Sciences. Cel. Mol. Life Sci. 2019, 1–15. [Google Scholar]
- Itoh, M.; Kim, C.; Palardy, G.; Oda, T.; Jiang, Y.J.; Maust, D.; Yeo, S.Y.; Lorick, K.; Wright, G.J.; Ariza-McNaughton, L.; et al. Mind bomb is an ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell. 2003, 4, 67–82. [Google Scholar] [CrossRef]
- Kang, K.; Lee, D.; Hong, S.; Park, S.; Song, M. The E3 Ligase Mind Bomb-1 (Mib1) Modulates Delta-Notch Signaling to Control Neurogenesis and Gliogenesis in the Developing Spinal Cord. J. Biol. Chem. 2013, 288, 2580–2592. [Google Scholar] [CrossRef]
- Kurtzman, A.L.; Gregori, L.; Haas, A.L.; Schechter, N. Ubiquitination and degradation of the zebrafish paired-liked homeobox protein Vsx1. J. Neurochem. 2000, 75, 48–55. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Vejnar, C.E.; Moreno-Mateos, M.A.; Cifuentes, D.; Bazzini, A.A.; Giraldez, A.J. Optimized CRISPR–Cas9 System for Genome Editing in Zebrafish. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.S.; Lam, I.I.; Clay, H.; Duong, D.N.; Deo, R.C.; Coughlin, S.R. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev. Cell. 2018, 46, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Lauter, G.; Söll, I.; Hauptmann, G. Sensitive whole-mount fluorescent in situ hybridization in zebrafish using enhanced tyramide signal amplification. J. Meth. Mol. Biol. 2014, 1082, 175. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Xu, H.; Zhao, W.; Zheng, J.; Sun, L.; Luo, C. Zygotic Vsx1 Plays a Key Role in Defining V2a Interneuron Sub-Lineage by Directly Repressing tal1 Transcription in Zebrafish. Int. J. Mol. Sci. 2020, 21, 3600. https://doi.org/10.3390/ijms21103600
Zhang Q, Xu H, Zhao W, Zheng J, Sun L, Luo C. Zygotic Vsx1 Plays a Key Role in Defining V2a Interneuron Sub-Lineage by Directly Repressing tal1 Transcription in Zebrafish. International Journal of Molecular Sciences. 2020; 21(10):3600. https://doi.org/10.3390/ijms21103600
Chicago/Turabian StyleZhang, Qi, Haomang Xu, Wei Zhao, Jianbo Zheng, Lei Sun, and Chen Luo. 2020. "Zygotic Vsx1 Plays a Key Role in Defining V2a Interneuron Sub-Lineage by Directly Repressing tal1 Transcription in Zebrafish" International Journal of Molecular Sciences 21, no. 10: 3600. https://doi.org/10.3390/ijms21103600
APA StyleZhang, Q., Xu, H., Zhao, W., Zheng, J., Sun, L., & Luo, C. (2020). Zygotic Vsx1 Plays a Key Role in Defining V2a Interneuron Sub-Lineage by Directly Repressing tal1 Transcription in Zebrafish. International Journal of Molecular Sciences, 21(10), 3600. https://doi.org/10.3390/ijms21103600




