Different Effects of Cisplatin and Transplatin on the Higher-Order Structure of DNA and Gene Expression
Abstract
:1. Introduction
2. Results
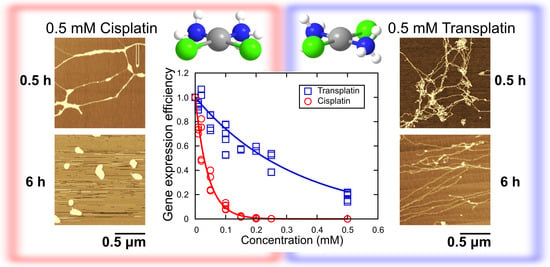
2.1. Luciferase Assay for Gene Expression and Higher-Order DNA Structure
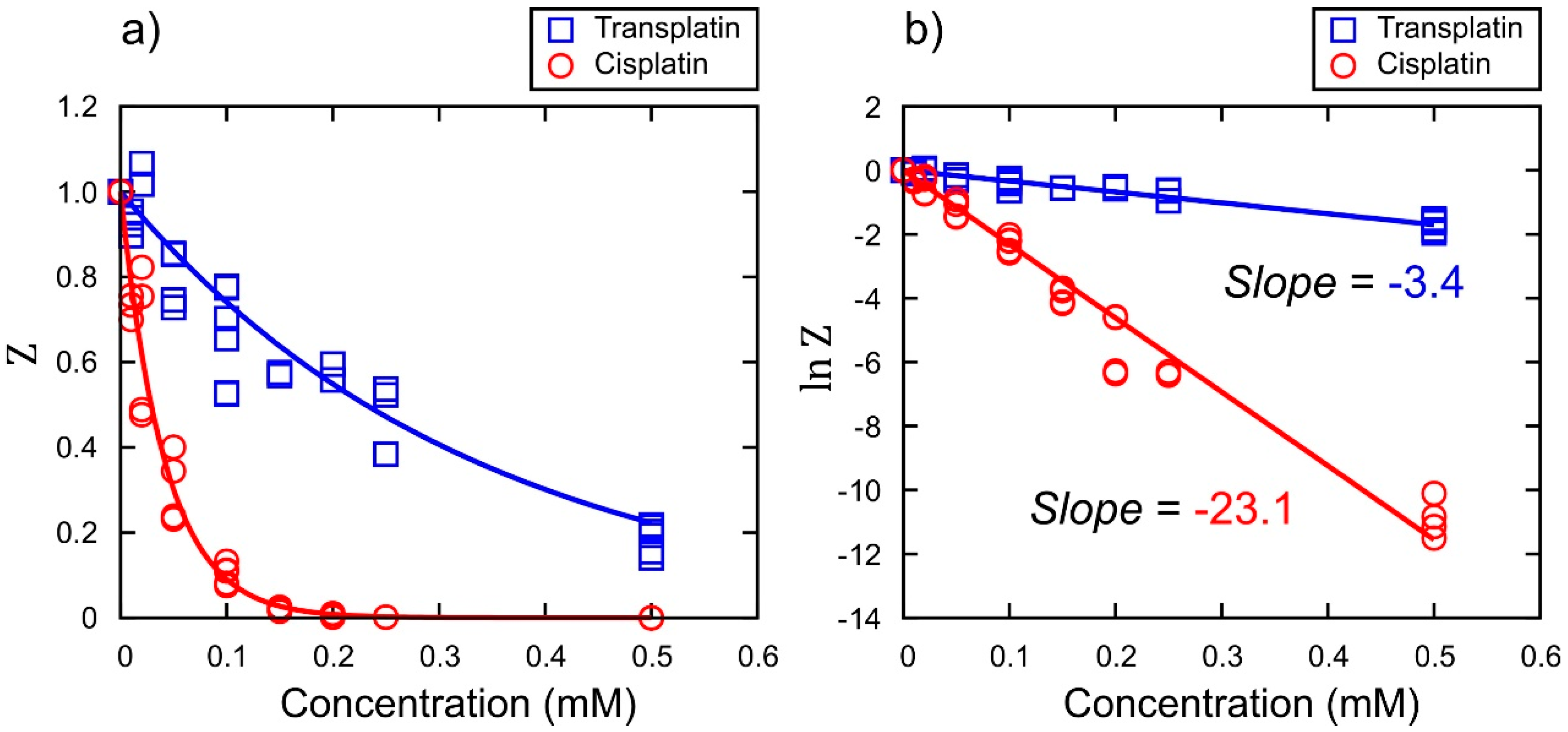
Inhibitory Effect on Gene Expression
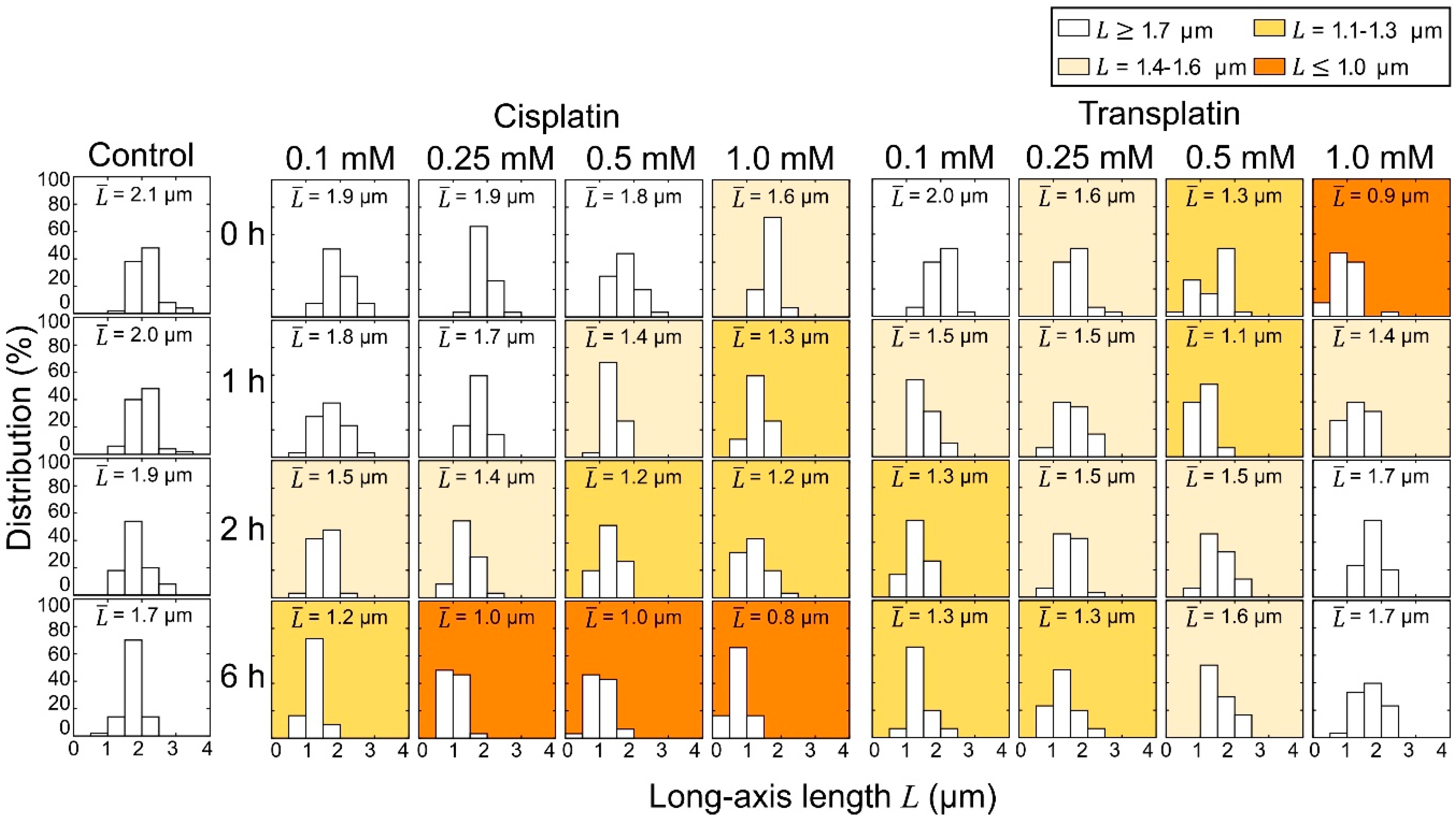
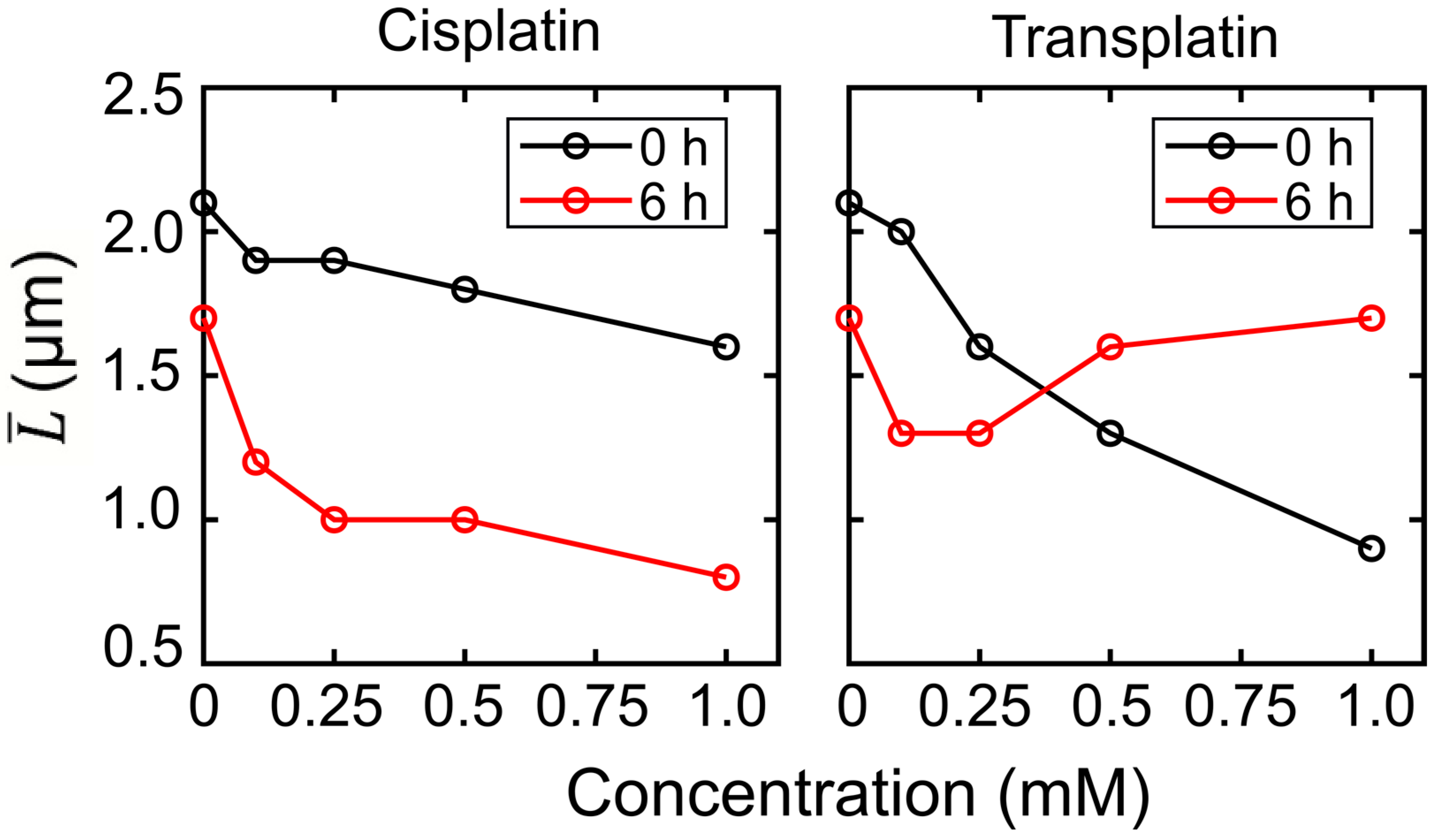
2.2. Higher-Order Conformational Change in T4 Phage DNA
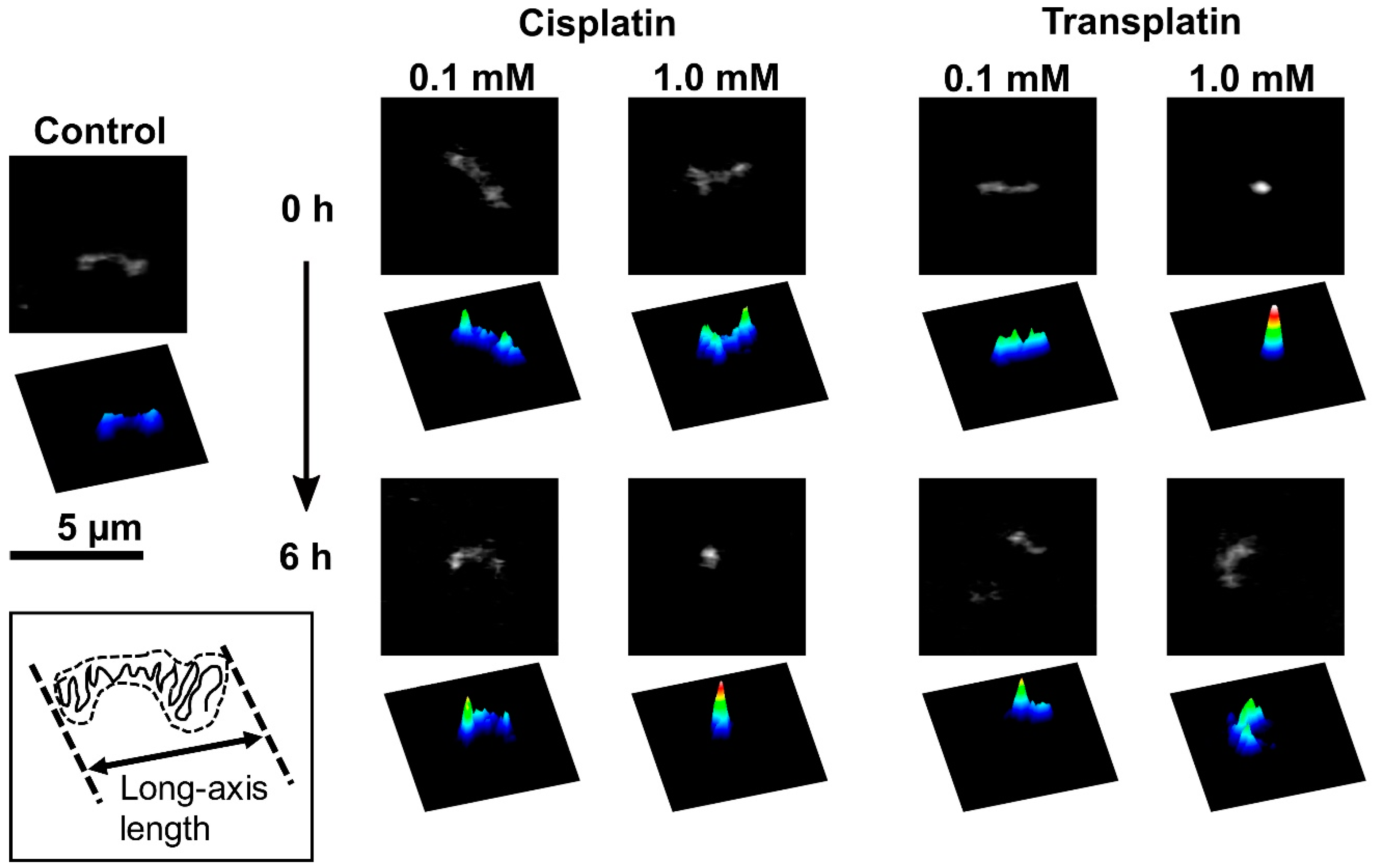
2.2.1. FM Observations
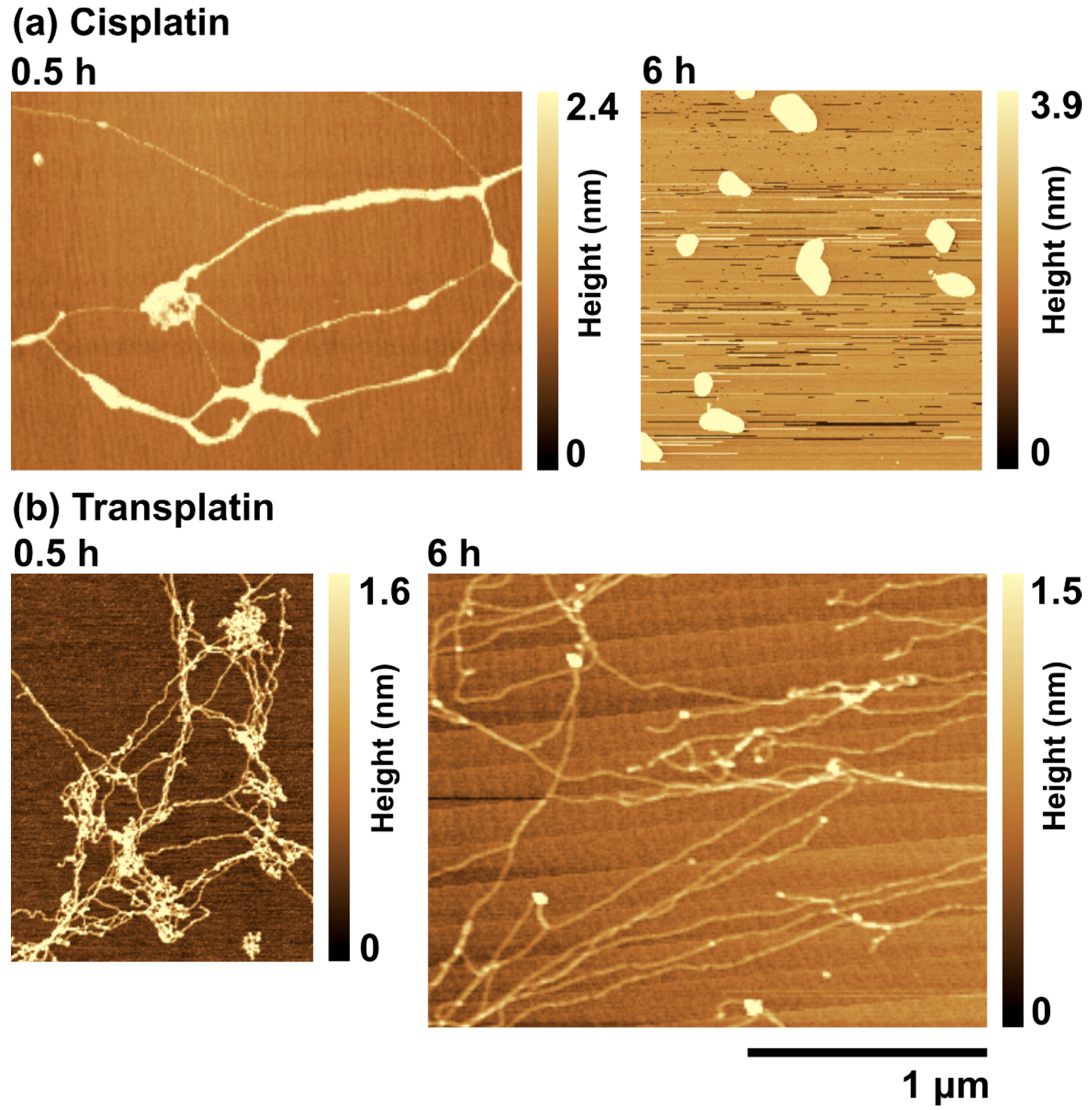
2.2.2. AFM Observations
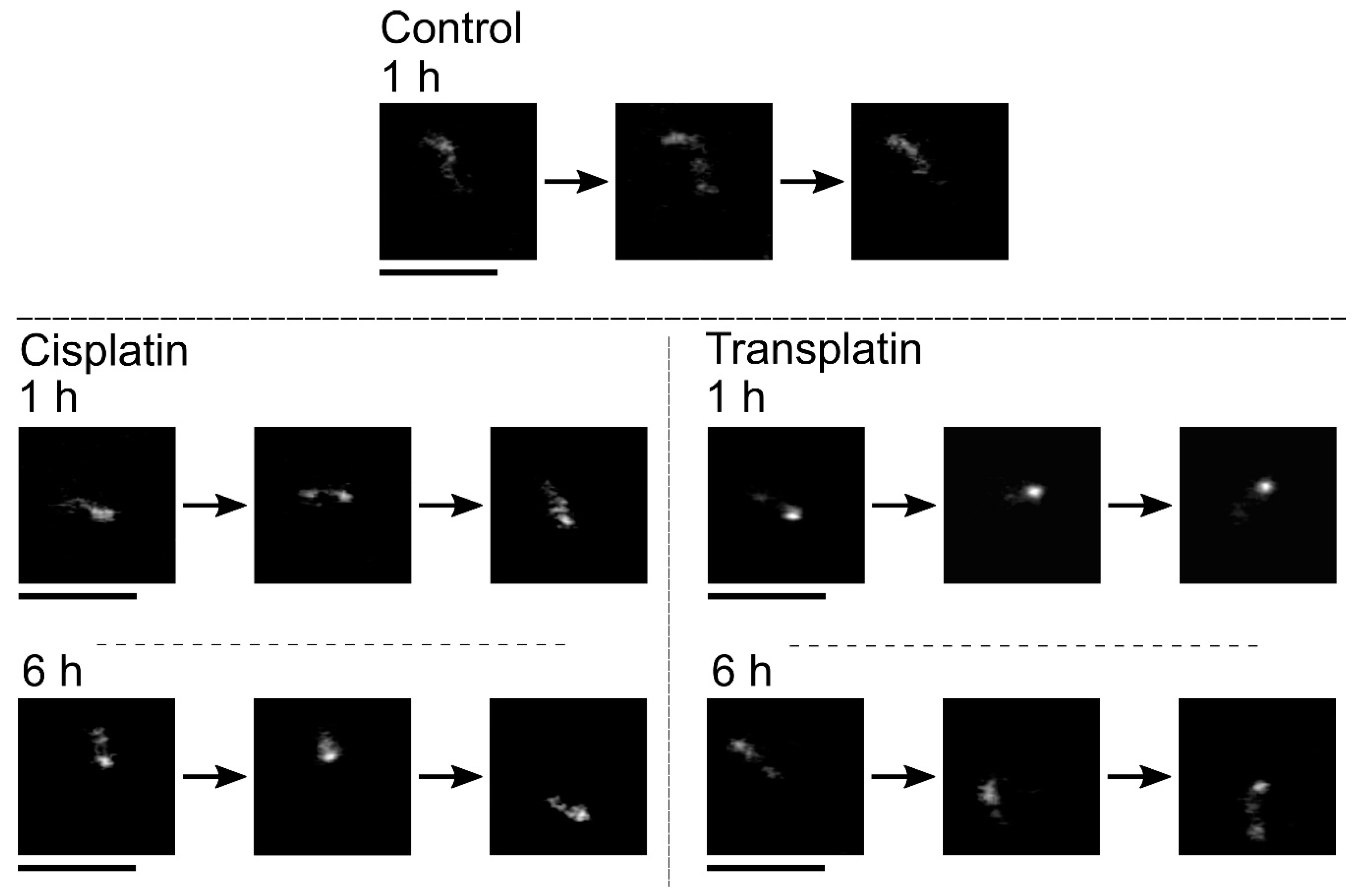
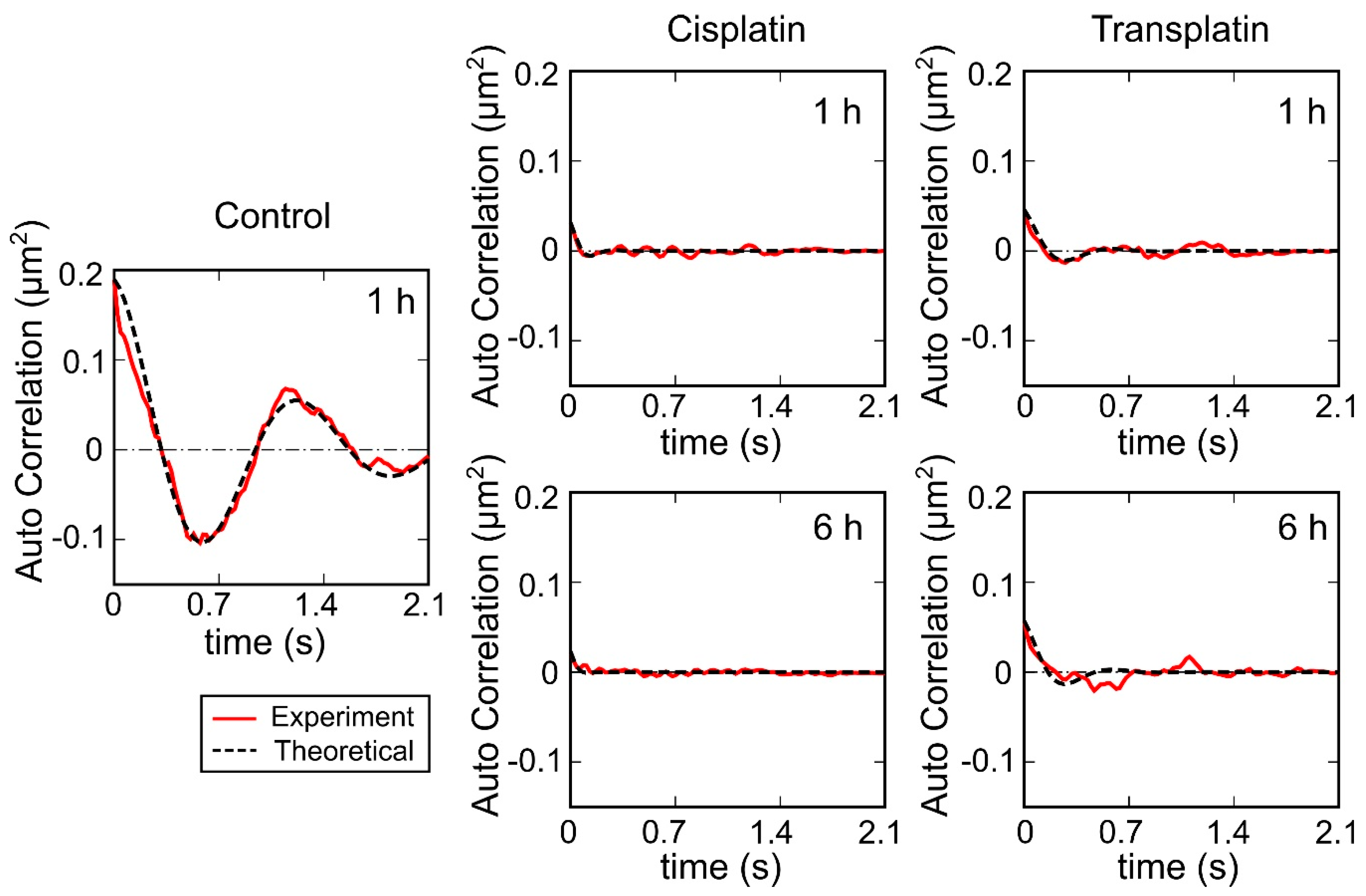
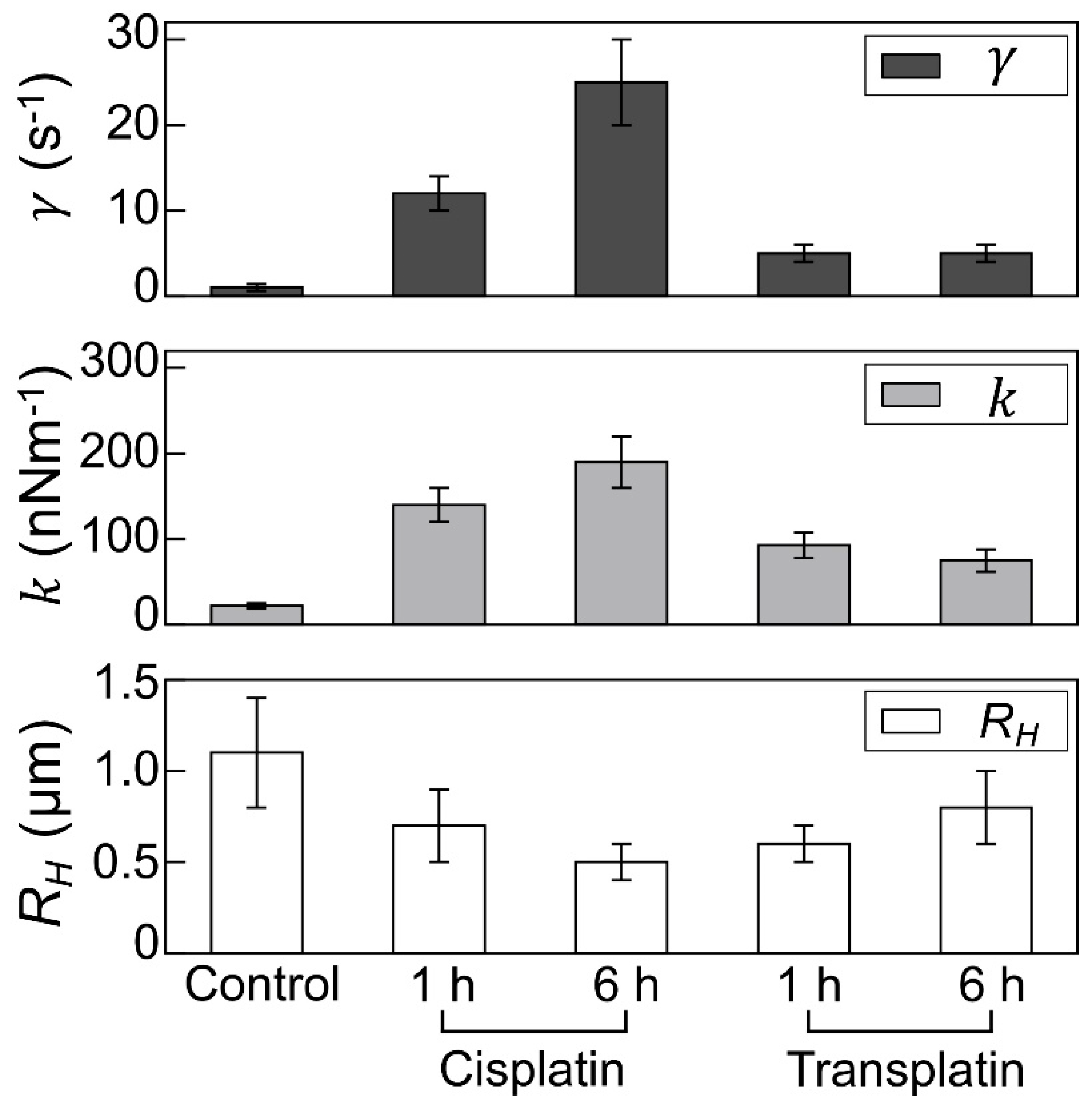
2.2.3. Visco-Elasticity and Hydrodynamic Radius RH of the DNA Molecule
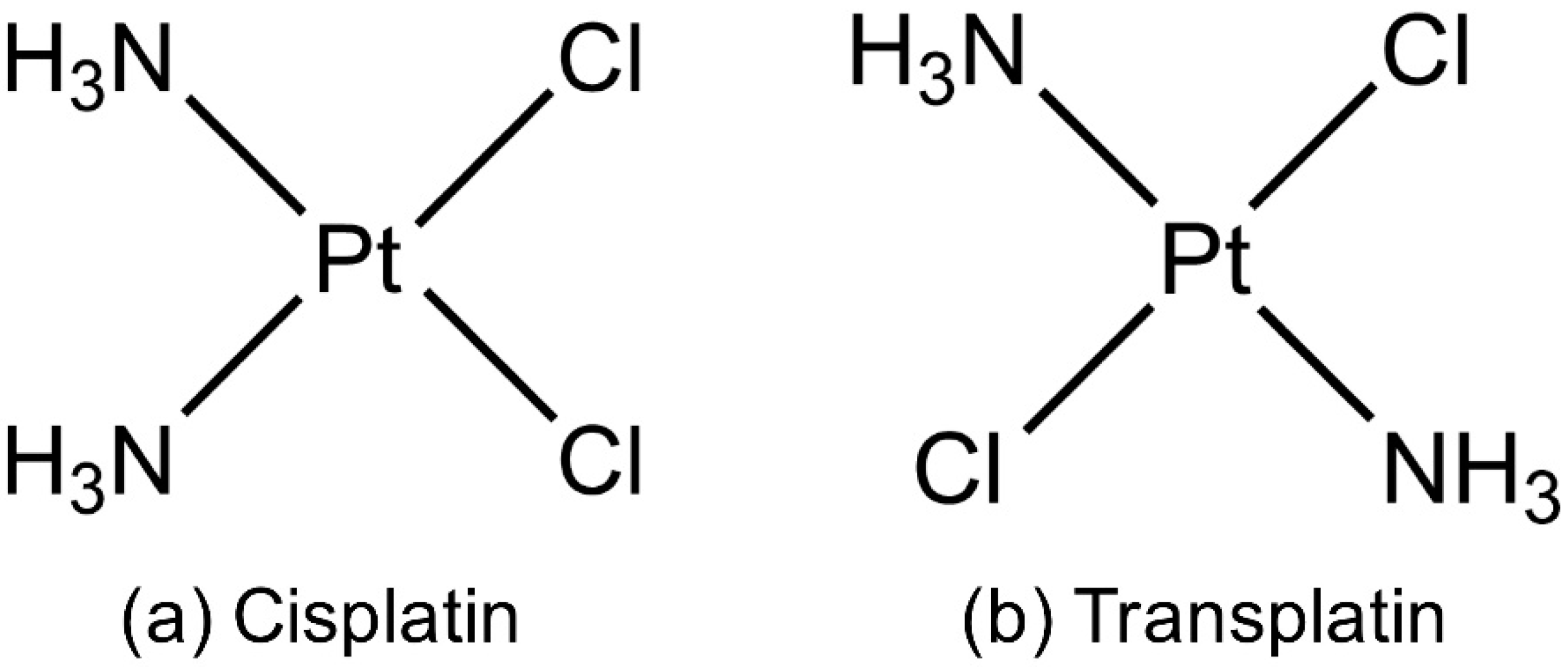
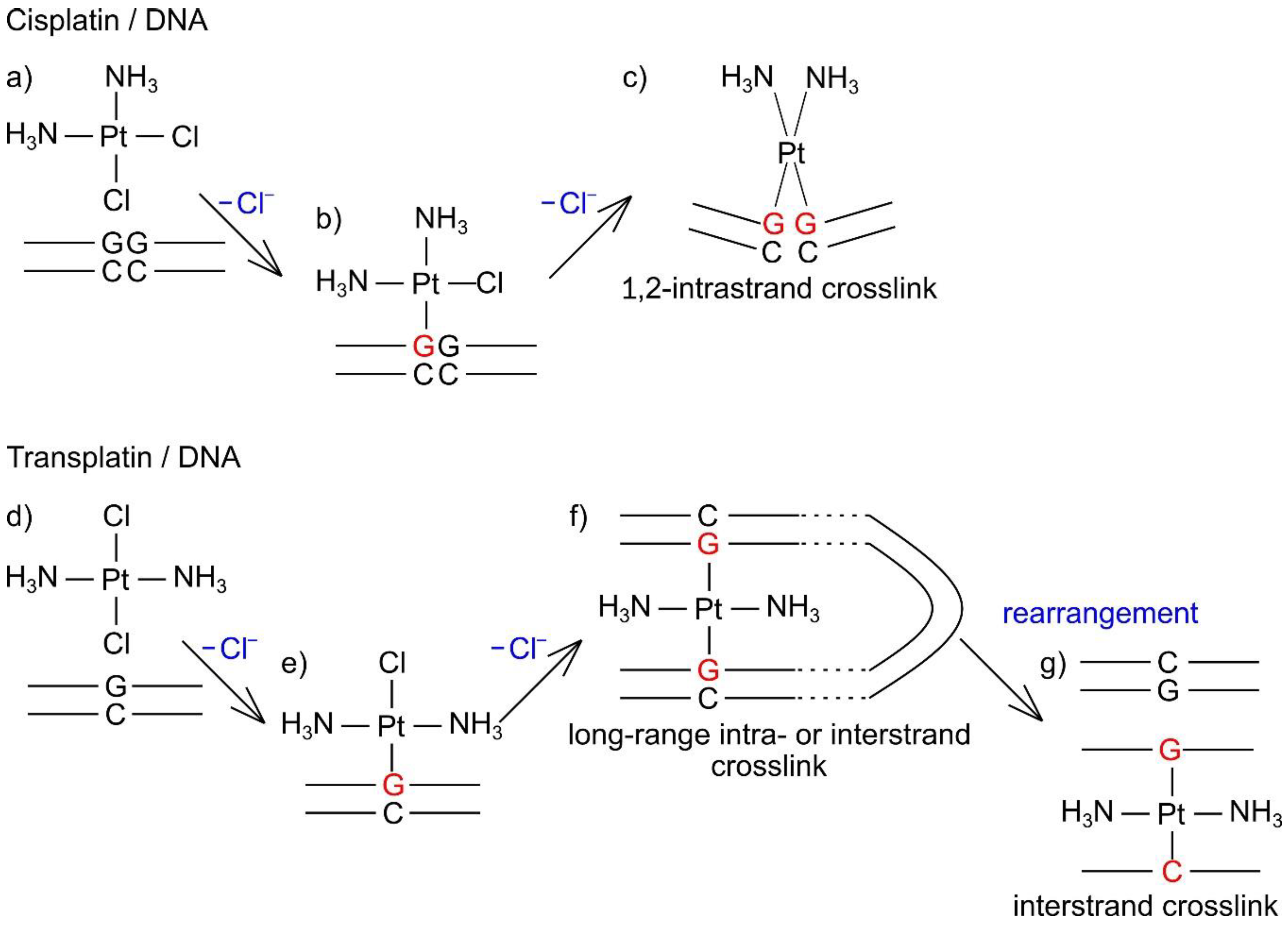
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Luciferase Assay for Gene Expression
4.2.2. FM Observations
4.2.3. AFM Measurements
4.2.4. Evaluation of Visco-Elasticity and Hydrodynamic Radius RH from the Analysis of DNA Molecule Fluctuations Observed by FM
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FM | Fluorescence microscopy |
| AFM | Atomic force microscopy |
References
- Rosenberg, B.; VanCamp, L.; Trosko, J.E.; Mansour, V.H. Platinum compounds: A new class of potent antitumour agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, B.; VanCamp, L. The successful regression of large solid sarcoma 180 tumors by platinum compounds. Cancer Res. 1970, 30, 1799–1802. [Google Scholar] [PubMed]
- Prestayko, A.W.; D’aoust, J.; Issell, B.; Crooke, S. Cisplatin (cis-diamminedichloroplatinum II). Cancer Treat. Rev. 1979, 6, 17–39. [Google Scholar] [CrossRef]
- Calvert, A.H.; Harland, S.J.; Newell, D.R.; Siddik, Z.H.; Jones, A.C.; McElwain, T.J.; Raju, S.; Wiltshaw, E.; Smith, I.E.; Baker, J.M.; et al. Early clinical studies with cis-diammine-1,1-cyclobutane dicarboxylate platinum II. Cancer Chemother. Pharmacol. 1982, 9, 140–147. [Google Scholar] [CrossRef]
- Sharma, H.; Thatcher, N.; Baer, J.; Zaki, A.; Smith, A.; McAucliffe, C.A.; Crowther, D.; Owens, S.; Fox, B.W. Blood clearance of radioactively labelled cis-diammine 1,1-cyclobutane dicarboxylate platinum (II) (CBDCA) in cancer patients. Cancer Chemother. Pharmacol. 1983, 11, 5–7. [Google Scholar] [CrossRef]
- Tashiro, T.; Kawada, Y.; Sakurai, Y.; Kidani, Y. Antitumor activity of a new platinum complex, oxalato (trans-l-1,2-diaminocyclohexane)platinum (II): New experimental data. Biomed. Pharmacol. 1989, 43, 251–260. [Google Scholar] [CrossRef]
- Cvitkovic, E. Ongoing and unsaid on oxaliplatin: The hope. Br. J. Cancer 1998, 77, 8–11. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, E.R.; Lippard, S.J. Structure, Recognition, and Processing of Cisplatin-DNA Adducts. Chem. Rev. 1999, 99, 2467–2498. [Google Scholar] [CrossRef]
- Reedijk, J. New clues for platinum antitumor chemistry: Kinetically controlled metal binding to DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 3611–3616. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, B. Some biological effects of platinum compounds. Platin. Met. Rev. 1971, 15, 42–51. [Google Scholar]
- Johnson, N.P.; Hoeschele, J.D.; Rahn, R.O.; O’Neill, J.P.; Hsie, A.W. Mutagenicity, cytotoxicity, and DNA binding of platinum (II)-chloroammines in Chinese hamster ovary cells. Cancer Res. 1980, 40, 1463–1468. [Google Scholar] [PubMed]
- Dedon, P.C.; Borch, R.F. Characterization of the reactions of platinum antitumor agents with biologic and nonbiologic sulfur-containing nucleophiles. Biochem. Pharmacol. 1987, 36, 1955–1964. [Google Scholar] [CrossRef]
- Żelazowski, A.J.; Garvey, J.S.; Hoeschele, J.D. In vivo and in vitro binding of platinum to metallothionein. Arch. Biochem. Biophys. 1984, 229, 246–252. [Google Scholar] [CrossRef]
- Ciccarelli, R.B.; Solomon, M.J.; Varshavsky, A.; Lippard, S.J. In vivo effects of cis- and trans-diamminedichloroplatinum(II) on SV40 chromosomes: Differential repair, DNA-protein cross-linking, and inhibition of replication. Biochemistry 1985, 24, 7533–7540. [Google Scholar] [CrossRef] [PubMed]
- Farrell, N.; Ha, T.T.B.; Souchard, J.P.; Wimmer, F.L.; Cros, S.; Johnson, N.P. Cytostatic Trans-Platinum(Ii) Complexes. J. Med. Chem. 1989, 32, 2240–2241. [Google Scholar] [CrossRef] [PubMed]
- Farrell, N.; Kelland, L.R.; Roberts, J.D.; Van Beusichem, M. Activation of the trans geometry in platinum antitumor complexes: A survey of the cytotoxicity of trans complexes containing planar ligands in murine L1210 and human tumor panels and studies on their mechanism of action. Cancer Res. 1992, 52, 5065–5072. [Google Scholar] [PubMed]
- Aris, S.M.; Farrell, N.P. Towards antitumor active trans-platinum compounds. Eur. J. Inorg. Chem. 2009, 2009, 1293–1302. [Google Scholar] [CrossRef] [Green Version]
- Coluccia, M.; Nassi, A.; Loseto, F.; Boccarelli, A.; Mariggio, M.A.; Giordano, D.; Intini, F.P.; Caputo, P.; Natile, G. A Trans-Platinum Complex Showing Higher Antitumor-Activity Than the Cis Congeners. J. Med. Chem. 1993, 36, 510–512. [Google Scholar] [CrossRef]
- Coluccia, M.; Boccarelli, A.; Mariggio, M.A.; Cardellicchio, N.; Caputo, P.; Intini, F.P.; Natile, G. Platinum(II) complexes containing iminoethers: A trans platinum antitumour agent. Chem.-Biol. Interact. 1995, 98, 251–266. [Google Scholar] [CrossRef]
- Blommaert, F.A.; van Dijk-Knijnenburg, H.C.; Dijt, F.J.; den Engelse, L.; Baan, R.A.; Berends, F.; Fichtinger-Schepman, A.M. Formation of DNA adducts by the anticancer drug carboplatin: Different nucleotide sequence preferences in vitro and in cells. Biochemistry 1995, 34, 8474–8780. [Google Scholar] [CrossRef]
- Fichtinger-Schepman, A.M.; van der Veer, J.L.; den Hartog, J.H.; Lohman, P.H.; Reedijk, J. Adducts of the antitumor drug cis-diamminedichloroplatinum(II) with DNA: Formation, identification, and quantitation. Biochemistry 1985, 24, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Fichtinger-Schepman, A.M.; Lohman, P.H.; Berends, F.; Reedijk, J.; van Oosterom, A.T. Interactions of the antitumour drug cisplatin with DNA in vitro and in vivo. IARC Sci. Publ. 1986, 78, 83–99. [Google Scholar]
- Eastman, A.; Barry, M.A. Interaction of trans-diamminedichloroplatinum(II) with DNA: Formation of monofunctional adducts and their reaction with glutathione. Biochemistry 1987, 26, 3303–3307. [Google Scholar] [CrossRef] [PubMed]
- Brabec, V.; Leng, M. DNA interstrand cross-links of trans-diamminedichloroplatinum(II) are preferentially formed between guanine and complementary cytosine residues. Proc. Natl. Acad. Sci. USA 1993, 90, 5345–5349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofr, C.; Brabec, V. Thermal and thermodynamic properties of duplex DNA containing site-specific interstrand cross-link of antitumor cisplatin or its clinically ineffective trans isomer. J. Biol. Chem. 2001, 276, 9655–9661. [Google Scholar] [CrossRef] [Green Version]
- Kasparkova, J.; Brabec, V. Recognition of DNA interstrand cross-links of cis-diamminedichloroplatinum (II) and its trans isomer by DNA-binding proteins. Biochemistry 1995, 34, 12379–12387. [Google Scholar] [CrossRef]
- Kasparkova, J.; Pospisilova, S.; Brabec, V. Different recognition of DNA modified by antitumor cisplatin and its clinically ineffective trans isomer by tumor suppressor protein p53. J. Biol. Chem. 2001, 276, 16064–16069. [Google Scholar] [CrossRef] [Green Version]
- Kasparkova, J.; Thibault, T.; Kostrhunova, H.; Stepankova, J.; Vojtiskova, M.; Muchova, T.; Midoux, P.; Malinge, J.M.; Brabec, V. Different affinity of nuclear factor-kappa B proteins to DNA modified by antitumor cisplatin and its clinically ineffective trans isomer. FEBS J. 2014, 281, 1393–1408. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Komeda, S.; Uemura, M.; Kanbe, T.; Chikuma, M.; Yoshikawa, K.; Imanaka, T. Highly Efficient DNA Compaction Mediated by an In Vivo Antitumor-Active Tetrazolato-Bridged Dinuclear Platinum(II) Complex. Inorg. Chem. 2011, 50, 11729–11735. [Google Scholar] [CrossRef]
- Uemura, M.; Yoshikawa, Y.; Yoshikawa, K.; Sato, T.; Mino, Y.; Chikuma, M.; Komeda, S. Second- and higher-order structural changes of DNA induced by antitumor-active tetrazolato-bridged dinuclear platinum(II) complexes with different types of 5-substituent. J. Inorg. Biochem. 2013, 127, 169–174. [Google Scholar] [CrossRef]
- Komeda, S.; Yoneyama, H.; Uemura, M.; Muramatsu, A.; Okamoto, N.; Konishi, H.; Takahashi, H.; Takagi, A.; Fukuda, W.; Imanaka, T.; et al. Specific Conformational Change in Giant DNA Caused by Anticancer Tetrazolato-Bridged Dinuclear Platinum(II) Complexes: Middle-Length Alkyl Substituents Exhibit Minimum Effect. Inorg. Chem. 2017, 56, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Kida, N.; Katsuda, Y.; Yoshikawa, Y.; Komeda, S.; Sato, T.; Saito, Y.; Chikuma, M.; Suzuki, M.; Imanaka, T.; Yoshikawa, K. Characteristic effect of an anticancer dinuclear platinum(II) complex on the higher-order structure of DNA. J. Biol. Inorg. Chem. 2010, 15, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Carnerero, J.M.; Masuoka, S.; Baba, H.; Yoshikawa, Y.; Prado-Gotor, R.; Yoshikawa, K. Decorating a single giant DNA with gold nanoparticles. RSC Adv. 2018, 8, 26571–26579. [Google Scholar] [CrossRef] [Green Version]
- Araki, S.; Nakai, T.; Hizume, K.; Takeyasu, K.; Yoshikawa, K. Hydrodynamic radius of circular DNA is larger than that of linear DNA. Chem. Phys. Lett. 2006, 418, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, M.; Sakaguchi, T.; Kimura, H.; Doi, M.; Minagawa, K.; Matsuzawa, Y.; Yoshikawa, K. Direct observation of brownian motion of macromolecules by fluorescence microscope. J. Polym. Sci. B 1992, 30, 779–783. [Google Scholar] [CrossRef]
- Jamieson, E.R.; Jacobson, M.P.; Barnes, C.M.; Chow, C.S.; Lippard, S.J. Structural and kinetic studies of a cisplatin-modified DNA icosamer binding to HMG1 domain B. J. Biol. Chem. 1999, 274, 12346–12354. [Google Scholar] [CrossRef] [Green Version]
- Sandman, K.E.; Marla, S.S.; Zlokarnik, G.; Lippard, S.J. Rapid fluorescence-based reporter-gene assays to evaluate the cytotoxicity and antitumor drug potential of platinum complexes. Chem. Biol. 1999, 6, 541–551. [Google Scholar] [CrossRef] [Green Version]
- Komeda, S.; Yoneyama, H.; Uemura, M.; Tsuchiya, T.; Hoshiyama, M.; Sakazaki, T.; Hiramoto, K.; Harusawa, S. Synthesis and structure-activity relationships of tetrazolato-bridged dinuclear platinum(II) complexes: A small modification at tetrazole C5 markedly influences the in vivo antitumor efficacy. J. Inorg. Biochem. 2019, 192, 82–86. [Google Scholar] [CrossRef]
- Onoa, G.B.; Cervantes, G.; Moreno, V.; Prieto, M.J. Study of the interaction of DNA with cisplatin and other Pd (II) and Pt (II) complexes by atomic force microscopy. Nucleic Acids Res. 1998, 26, 1473–1480. [Google Scholar] [CrossRef] [Green Version]
- Malina, J.; Farrell, N.P.; Brabec, V. DNA condensing effects and sequence selectivity of DNA binding of antitumor noncovalent polynuclear platinum complexes. Inorg. Chem. 2014, 53, 1662–1671. [Google Scholar] [CrossRef]
- Lippert, B. Trans-diammineplatinum (II): What makes it different from cis-DDP? Coordination chemistry of a neglected relative of cisplatin and its interaction with nucleic acids. Met. Ions Biol. Syst. 1996, 33, 105–142. [Google Scholar] [PubMed]
- Bancroft, D.P.; Lepre, C.A.; Lippard, S.J. Platinum-195 NMR kinetic and mechanistic studies of cis- and trans-diamminedichloroplatinum(II) binding to DNA. J. Am. Chem. Soc. 1990, 112, 6860–6871. [Google Scholar] [CrossRef]
- Miller, S.E.; Gerard, K.J.; House, D.A. The hydrolysis products of cis-diamminedichloroplatinum (II) 6. A kinetic comparison of the cis-and trans-isomers and other cis-di (amine) di (chloro) platinum (II) compounds. Inorg. Chim. Acta 1991, 190, 135–144. [Google Scholar] [CrossRef]
- Arpalahti, J.; Mikola, M.; Mauristo, S. Kinetics and mechanism of the complexation of cis-diamminedichloroplatinum (II) with the purine nucleoside inosine in aqueous solution. Inorg. Chem. 1993, 32, 3327–3332. [Google Scholar] [CrossRef]
- Mikola, M.; Arpalahti, J. Kinetics and mechanism of the complexation of trans-diamminedichloroplatinum (II) with the purine nucleoside inosine in aqueous solution. Inorg. Chem. 1994, 33, 4439–4445. [Google Scholar] [CrossRef]
- Bierbach, U.; Qu, Y.; Hambley, T.W.; Peroutka, J.; Nguyen, H.L.; Doedee, M.; Farrell, N. Synthesis, structure, biological activity, and DNA binding of platinum (II) complexes of the type trans-[PtCl2 (NH3) L](L= planar nitrogen base). Effect of L and cis/trans isomerism on sequence specificity and unwinding properties observed in globally platinated DNA. Inorg. Chem. 1999, 38, 3535–3542. [Google Scholar]
- Navarro, M.; Castro, W.; Higuera-Padilla, A.R.; Sierraalta, A.; Abad, M.J.; Taylor, P.; Sánchez-Delgado, R.A. Synthesis, characterization and biological activity of trans-platinum (II) complexes with chloroquine. J. Inorg. Biochem. 2011, 105, 1684–1691. [Google Scholar] [CrossRef] [Green Version]
- Mello, J.A.; Lippard, S.J.; Essigmann, J.M. DNA adducts of cis-diamminedichloroplatinum(II) and its trans isomer inhibit RNA polymerase II differentially in vivo. Biochemistry 1995, 34, 14783–14791. [Google Scholar] [CrossRef]
- Hou, X.-M.; Zhang, X.-H.; Wei, K.-J.; Ji, C.; Dou, S.-X.; Wang, W.-C.; Li, M.; Wang, P.-Y. Cisplatin induces loop structures and condensation of single DNA molecules. Nucleic Acids Res. 2009, 37, 1400–1410. [Google Scholar] [CrossRef]
- Liu, Z.; Tan, S.; Zu, Y.; Fu, Y.; Meng, R.; Xing, Z. The interactions of cisplatin and DNA studied by atomic force microscopy. Micron 2010, 41, 833–839. [Google Scholar] [CrossRef]
- Lima, C.; Caquito, J.; de Oliveira, R.; Rocha, M. Pixantrone anticancer drug as a DNA ligand: Depicting the mechanism of action at single molecule level. Eur. Phys. J. E 2019, 42, 130. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.; Smith, S.B.; Liphardt, J.; Smith, D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000, 10, 279–285. [Google Scholar] [CrossRef]
- Wang, M.D.; Yin, H.; Landick, R.; Gelles, J.; Block, S.M. Stretching DNA with optical tweezers. Biophys. J. 1997, 72, 1335–1346. [Google Scholar] [CrossRef] [Green Version]
- Dutta, S.; Rivetti, C.; Gassman, N.R.; Young, C.G.; Jones, B.T.; Scarpinato, K.; Guthold, M. Analysis of single, cisplatin-induced DNA bends by atomic force microscopy and simulations. J. Mol. Recognit. 2018, 31, e2731. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; van Boom, S.S.G.E.; Reedijk, J.; van Boom, J.H.; Wang, A.H.J. Structure and isomerization of an intrastrand cisplatin-cross-linked octamer DNA duplex by NMR analysis. Biochemistry 1995, 34, 12912–12920. [Google Scholar] [CrossRef] [PubMed]
- Van Boom, S.S.; Yang, D.; Reedijk, J.; van der Marel, G.A.; Wang, A.H. Structural effect of intra-strand cisplatin-crosslink on palindromic DNA sequences. J. Biomol. Struct. Dyn. 1996, 13, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Shellard, S.A.; Fichtinger-Schepman, A.M.; Lazo, J.S.; Hill, B.T. Evidence of differential cisplatin-DNA adduct formation, removal and tolerance of DNA damage in three human lung carcinoma cell lines. Anti-Cancer Drugs 1993, 4, 491–500. [Google Scholar] [CrossRef]
- Takahara, P.M.; Rosenzweig, A.C.; Frederick, C.A.; Lippard, S.J. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature 1995, 377, 649–652. [Google Scholar] [CrossRef]
- Coste, F.; Malinge, J.M.; Serre, L.; Shepard, W.; Roth, M.; Leng, M.; Zelwer, C. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 A resolution: Hydration at the platinated site. Nucleic Acids Res. 1999, 27, 1837–1846. [Google Scholar] [CrossRef] [Green Version]
- Eastman, A.; Jennerwein, M.M.; Nagel, D.L. Characterization of bifunctional adducts produced in DNA by trans-diamminedichloroplatinum(II). Chem.-Biol. Interact. 1988, 67, 71–80. [Google Scholar] [CrossRef]
- Paquet, F.; Boudvillain, M.; Lancelot, G.; Leng, M. NMR solution structure of a DNA dodecamer containing a transplatin interstrand GN7-CN3 cross-link. Nucleic Acids Res. 1999, 27, 4261–4268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brabec, V.; Sip, M.; Leng, M. DNA conformational change produced by the site-specific interstrand cross-link of trans-diamminedichloroplatinum (II). Biochemistry 1993, 32, 11676–11681. [Google Scholar] [CrossRef] [PubMed]
- Brabec, V.; Reedijk, J.; Leng, M. Sequence-dependent distortions induced in DNA by monofunctional platinum (II) binding. Biochemistry 1992, 31, 12397–12402. [Google Scholar] [CrossRef] [PubMed]
- Kasparkova, J.; Marini, V.; Bursova, V.; Brabec, V. Biophysical studies on the stability of DNA intrastrand cross-links of transplatin. Biophys. J. 2008, 95, 4361–4371. [Google Scholar] [CrossRef] [Green Version]
- Giraud-Panis, M.-J.; Leng, M. Transplatin-modified oligonucleotides as modulators of gene expression. Pharmacol. Ther. 2000, 85, 175–181. [Google Scholar] [CrossRef]
- Dalbiès, R.; Payet, D.; Leng, M. DNA double helix promotes a linkage isomerization reaction in trans-diamminedichloroplatinum (II)-modified DNA. Proc. Natl. Acad. Sci. USA 1994, 91, 8147–8151. [Google Scholar] [CrossRef] [Green Version]
- Boudvillain, M.; Dalbies, R.; Aussourd, C.; Leng, M. Intrastrand cross-links are not formed in the reaction between transplatin and native DNA: Relationm with the clinical inefficency of transplatin. Nucleic Acids Res. 1995, 23, 2381–2388. [Google Scholar] [CrossRef] [Green Version]
- Campbell, M.A.; Miller, P.S. Transplatin-conjugated triplex-forming oligonucleotides form adducts with both strands of DNA. Bioconjugate Chem. 2009, 20, 2222–2230. [Google Scholar] [CrossRef] [Green Version]
- Colombier, C.; Lippert, B.; Leng, M. Interstrand cross-linking reaction in triplexes containing a monofunctional transplatin-adduct. Nucleic Acids Res. 1996, 24, 4519–4524. [Google Scholar] [CrossRef] [Green Version]
- Alderden, R.A.; Hall, M.D.; Hambley, T.W. The discovery and development of cisplatin. J. Chem. Educ. 2006, 83, 728. [Google Scholar] [CrossRef]
- Shimizu, Y.; Yoshikawa, Y.; Kenmotsu, T.; Komeda, S.; Yoshikawa, K. Conformational transition of DNA by dinuclear Pt(II) complexes causes cooperative inhibition of gene expression. Chem. Phys. Lett. 2017, 678, 123–129. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kishimoto, T.; Yoshikawa, Y.; Yoshikawa, K.; Komeda, S. Different Effects of Cisplatin and Transplatin on the Higher-Order Structure of DNA and Gene Expression. Int. J. Mol. Sci. 2020, 21, 34. https://doi.org/10.3390/ijms21010034
Kishimoto T, Yoshikawa Y, Yoshikawa K, Komeda S. Different Effects of Cisplatin and Transplatin on the Higher-Order Structure of DNA and Gene Expression. International Journal of Molecular Sciences. 2020; 21(1):34. https://doi.org/10.3390/ijms21010034
Chicago/Turabian StyleKishimoto, Toshifumi, Yuko Yoshikawa, Kenichi Yoshikawa, and Seiji Komeda. 2020. "Different Effects of Cisplatin and Transplatin on the Higher-Order Structure of DNA and Gene Expression" International Journal of Molecular Sciences 21, no. 1: 34. https://doi.org/10.3390/ijms21010034