Role of Metastasis-Related Genes in Cisplatin Chemoresistance in Gastric Cancer
Abstract
1. Introduction
2. Results
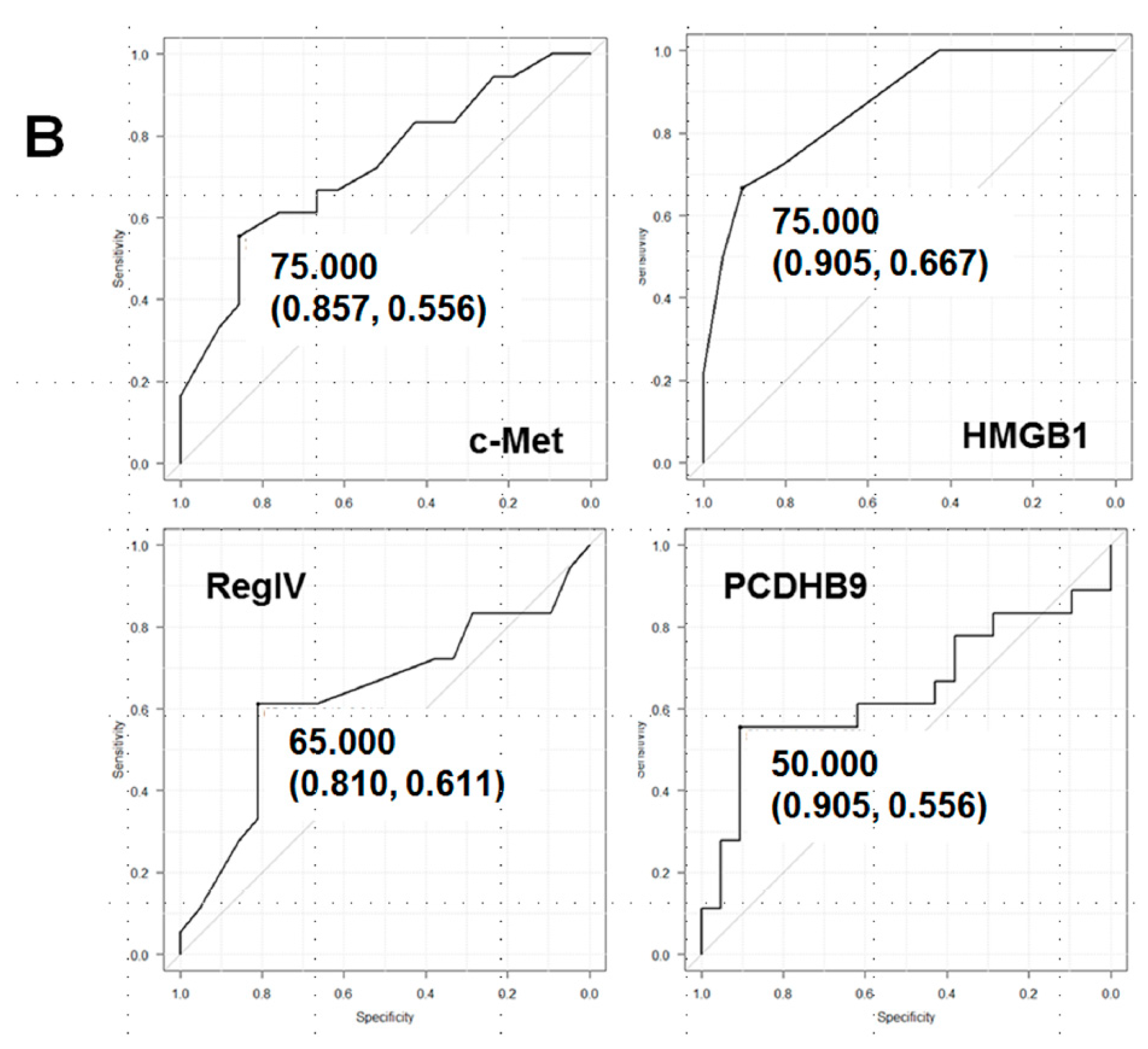
2.1. Expression of the Four Metastasis-Related Genes by Immunohistochemistry
| Parameter | Resistant | Sensitive | P2 | |
|---|---|---|---|---|
| Number | 39 | 18 | 21 | |
| Grade 1 | 0-Ia | Ib-2 | ||
| Sex (M:F) | 11:7 | 12:9 | NS | |
| Age (years) | 72 (58–91) | 75 (56–85) | NS | |
| Neoadjuvant | CDDP alone | 9 | 10 | NS |
| CDDP+5-FU | 9 | 11 | ||
| Histology 3 | tub1 | 7 | 6 | NS |
| tub2 | 5 | 6 | ||
| por1 | 2 | 3 | ||
| por2/sig | 4 | 6 | ||
| pT 3 | 3 | 3 | 3 | NS |
| 4a | 11 | 12 | ||
| 4b | 4 | 6 | ||
| pN 3 | 0 | 0 | 0 | NS |
| 1 | 7 | 11 | ||
| 2 | 10 | 7 | ||
| 3 | 1 | 3 | ||
| pM 3 | 0 | 9 | 10 | NS |
| 1 | 9 | 11 | ||
| pStage 3 | IIIA | 5 | 5 | NS |
| IIIB | 4 | 5 | ||
| IV | 9 | 11 | ||
| Positive Expression 4 | c-Met | 12 (67%) | 7 (33%) | NS |
| HMGB1 | 13 (72%) | 4 (19%) | 0.0013 | |
| REGIV | 11 (61%) | 4 (19%) | 0.01 | |
| PCDHB9 | 8 (44%) | 2 (10%) | 0.025 |

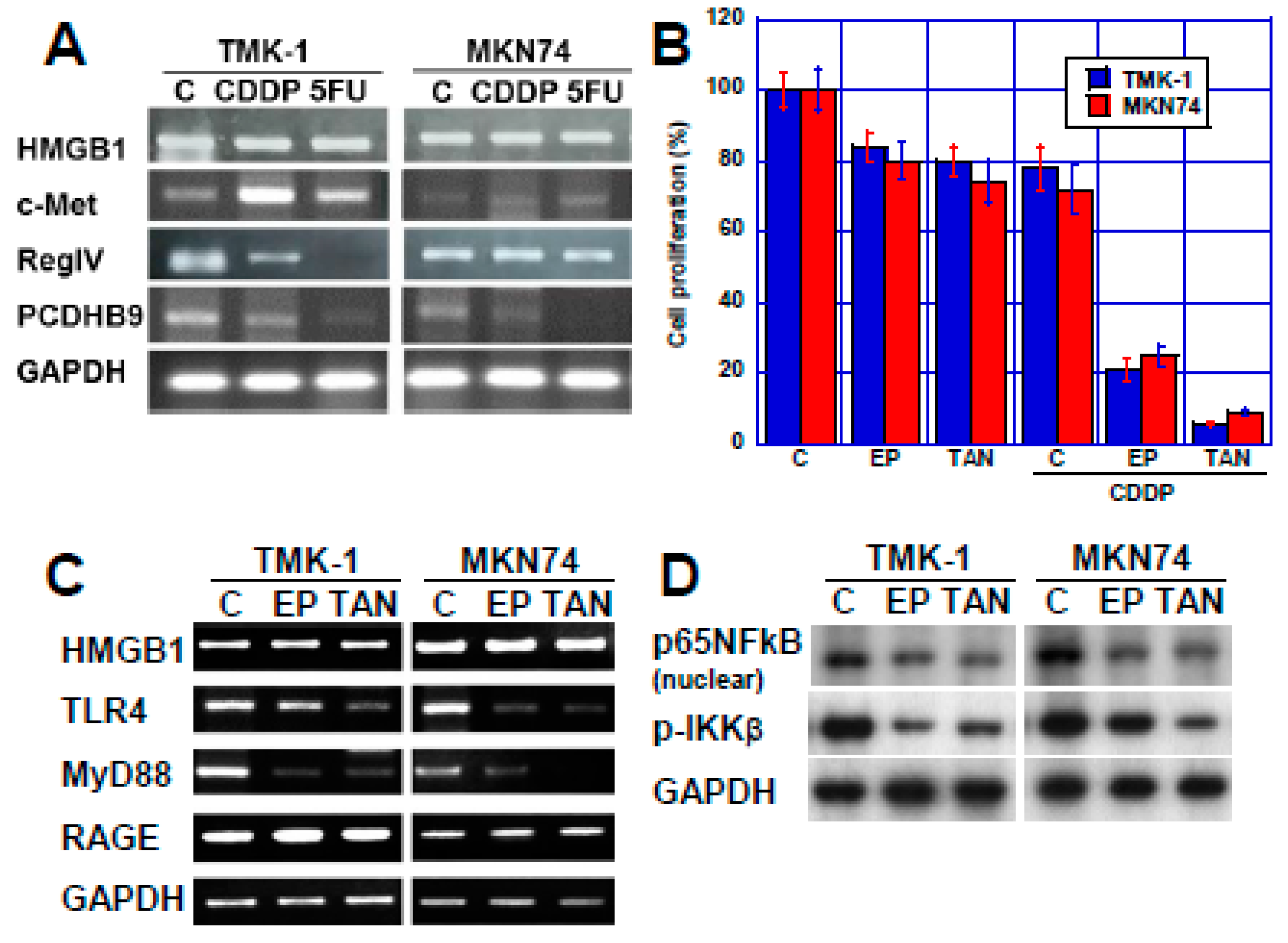
2.2. Effect of HMGB1 Inhibition on CDDP Sensitivity in Two Human Gastric Cancer Cell Lines
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Cells and Reagents
4.3. Cell Growth and Apoptosis
4.4. Immunohistochemistry
4.5. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.6. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2014 (ver. 4). Gastric Cancer 2017, 20, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef]
- Zhu, H.; Luo, H.; Zhang, W.; Shen, Z.; Hu, X.; Zhu, X. Molecular mechanisms of cisplatin resistance in cervical cancer. Drug Des. Dev. Ther. 2016, 10, 1885–1895. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Yasui, W.; Yokozaki, H.; Kitadai, Y.; Tahara, E. Aberrant expression of c-met mRNA in human gastric carcinomas. Int. J. Cancer 1993, 55, 72–75. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Yasui, W.; Kitadai, Y.; Yokozaki, H.; Ito, H.; Tahara, E. Frequent amplification of the c-met gene in scirrhous type stomach cancer. Biochem. Biophys. Res. Commun. 1992, 189, 227–232. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Yasui, W.; Yokozaki, H.; Akagi, M.; Akama, Y.; Kitahara, K.; Fujii, K.; Tahara, E. Frequent loss of heterozygosity of the long arm of chromosome 7 is closely associated with progression of human gastric carcinomas. Int. J. Cancer 1994, 59, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, H.; Luo, Y.; Kuniyasu, H. Non-histone nuclear factor HMGB1 as a therapeutic target in colorectal cancer. Expert Opin. Ther. Targets 2011, 15, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Kuniyasu, H.; Oue, N.; Wakikawa, A.; Shigeishi, H.; Matsutani, N.; Kuraoka, K.; Ito, R.; Yokozaki, H.; Yasui, W. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J. Pathol. 2002, 196, 163–170. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef]
- Luo, Y.; Chihara, Y.; Fujimoto, K.; Sasahira, T.; Kuwada, M.; Fujiwara, R.; Fujii, K.; Ohmori, H.; Kuniyasu, H. High mobility group box 1 released from necrotic cells enhances regrowth and metastasis of cancer cells that have survived chemotherapy. Eur. J. Cancer 2013, 49, 741–751. [Google Scholar] [CrossRef]
- Mitani, Y.; Oue, N.; Matsumura, S.; Yoshida, K.; Noguchi, T.; Ito, M.; Tanaka, S.; Kuniyasu, H.; Kamata, N.; Yasui, W. Reg IV is a serum biomarker for gastric cancer patients and predicts response to 5-fluorouracil-based chemotherapy. Oncogene 2007, 26, 4383–4393. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Oue, N.; Sasahira, T.; Yi, L.; Moriwaka, Y.; Shimomoto, T.; Fujii, K.; Ohmori, H.; Yasui, W. Reg IV enhances peritoneal metastasis in gastric carcinomas. Cell Prolif. 2009, 42, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Mukai, S.; Oue, N.; Oshima, T.; Imai, T.; Sekino, Y.; Honma, R.; Sakamoto, N.; Sentani, K.; Kuniyasu, H.; Egi, H.; et al. Overexpression of PCDHB9 promotes peritoneal metastasis and correlates with poor prognosis in patients with gastric cancer. J. Pathol. 2017, 243, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Japanese Gastric Cancer Association. Japanese classification of gastric carcinoma; 3rd ed. Gastric Cancer 2011, 14, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Huo, Y.; Zheng, H.; Zhao, J.; Jia, L.; Wang, P. Ethyl pyruvate suppresses the growth, invasion and migration and induces the apoptosis of nonsmall cell lung cancer cells via the HMGB1/RAGE axis and the NFκB/STAT3 pathway. Oncol. Rep. 2019, 42, 817–825. [Google Scholar] [PubMed]
- Li, X.; Wu, Y.; Zhang, W.; Gong, J.; Cheng, Y. Pre-conditioning with tanshinone IIA attenuates the ischemia/reperfusion injury caused by liver grafts via regulation of HMGB1 in rat Kupffer cells. Biomed. Pharmacother 2017, 89, 1392–1400. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, X.; Liu, L.; Cui, L.; Yang, R.; Li, M.; Du, W. Tanshinone II A down-regulates HMGB1, RAGE, TLR4, NF-κB expression, ameliorates BBB permeability and endothelial cell function, and protects rat brains against focal ischemia. Brain Res. 2010, 1321, 143–151. [Google Scholar] [CrossRef]
- Liao, T.T.; Yang, M.H. Revisiting epithelial-mesenchymal transition in cancer metastasis: The connection between epithelial plasticity and stemness. Mol. Oncol. 2017, 11, 792–804. [Google Scholar] [CrossRef]
- Chang, J.C. Cancer stem cells: Role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine 2016, 95 (Suppl. S1), S20–S25. [Google Scholar] [CrossRef]
- Sharma, A.K.; Sharma, V.R.; Gupta, G.K.; Ashraf, G.M.; Kamal, M.A. Advanced Glycation End Products (AGEs), Glutathione and Breast Cancer: Factors, Mechanism and Therapeutic Interventions. Curr. Drug. Metab. 2019, 20, 65–71. [Google Scholar] [CrossRef]
- Wu, K.; Zhang, H.; Fu, Y.; Zhu, Y.; Kong, L.; Chen, L.; Zhao, F.; Yu, L.; Chen, X. TLR4/MyD88 signaling determines the metastatic potential of breast cancer cells. Mol. Med. Rep. 2018, 18, 3411–3420. [Google Scholar] [CrossRef] [PubMed]
- Fages, C.; Nolo, R.; Huttunen, H.J.; Eskelinen, E.; Rauvala, H. Regulation of cell migration by amphoterin. J. Cell Sci. 2000, 113 Pt 4, 611–620. [Google Scholar]
- Huttunen, H.J.; Fages, C.; Rauvala, H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-B require the cytoplasmic domain of the receptor but different downstream signaling pathways. J. Biol. Chem. 1999, 274, 19919–19924. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Qi, X.; Zhang, X.; Gao, D.; Fang, K.; Guo, Z.; Li, L. Med19 is involved in chemoresistance by mediating autophagy through HMGB1 in breast cancer. J. Cell Biochem. 2019, 120, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Mandke, P.; Vasquez, K.M. Interactions of high mobility group box protein 1 (HMGB1) with nucleic acids: Implications in DNA repair and immune responses. DNA Repair 2019, 83, 102701. [Google Scholar] [CrossRef]
- Stros, M.; Cherny, D.; Jovin, T.M. HMG1 protein stimulates DNA end joining by promoting association of DNA molecules via their ends. Eur. J. Biochem. 2000, 267, 4088–4097. [Google Scholar] [CrossRef]
- Hughes, E.N.; Engelsberg, B.N.; Billings, P.C. Purification of nuclear proteins that bind to cisplatin-damaged DNA. Identity with high mobility group proteins 1 and 2. J. Biol. Chem. 1992, 267, 13520–13527. [Google Scholar] [PubMed]
- Calandrini, V.; Rossetti, G.; Arnesano, F.; Natile, G.; Carloni, P. Computational metallomics of the anticancer drug cisplatin. J. Inorg. Biochem. 2015, 153, 231–238. [Google Scholar] [CrossRef]
- Yusein-Myashkova, S.; Ugrinova, I.; Pasheva, E. Non-histone protein HMGB1 inhibits the repair of damaged DNA by cisplatin in NIH-3T3 murine fibroblasts. BMB Rep. 2016, 49, 99–104. [Google Scholar] [CrossRef]
- He, Y.; Ding, Y.; Wang, D.; Zhang, W.; Chen, W.; Liu, X.; Qin, W.; Qian, X.; Chen, H.; Guo, Z. HMGB1 bound to cisplatin-DNA adducts undergoes extensive acetylation and phosphorylation in vivo. Chem. Sci. 2015, 6, 2074–2078. [Google Scholar] [CrossRef]
- Patrick, S.M.; Henkels, K.M.; Turchi, J.J. High-mobility group 1 protein inhibits helicase catalyzed displacement of cisplatin-damaged DNA. Biochim. Biophys. Acta 1997, 1354, 279–290. [Google Scholar] [CrossRef]
- Ugrinova, I.; Zlateva, S.; Pashev, I.G.; Pasheva, E.A. Native HMGB1 protein inhibits repair of cisplatin-damaged nucleosomes in vitro. Int. J. Biochem. Cell Biol. 2009, 41, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lippard, S.J. Redox state-dependent interaction of HMGB1 and cisplatin-modified DNA. Biochemistry 2011, 50, 2567–2574. [Google Scholar] [CrossRef] [PubMed]
- Amornsupak, K.; Insawang, T.; Thuwajit, P.; O-Charoenrat, P.; Eccles, S.A.; Thuwajit, C. Cancer-associated fibroblasts induce high mobility group box 1 and contribute to resistance to doxorubicin in breast cancer cells. BMC Cancer 2014, 14, 955. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Zhou, F.; Yang, H.; Wei, Y.; Gong, J.; Mei, Z.; Wu, L.; Yu, H.; Zhou, Y. Downregulation of high mobility group box 1 modulates telomere homeostasis and increases the radiosensitivity of human breast cancer cells. Int. J. Oncol. 2015, 46, 1051–1058. [Google Scholar] [CrossRef]
- Zhao, X.L.; Lin, Y.; Jiang, J.; Tang, Z.; Yang, S.; Lu, L.; Liang, Y.; Liu, X.; Tan, J.; Hu, X.G.; et al. High-mobility group box 1 released by autophagic cancer-associated fibroblasts maintains the stemness of luminal breast cancer cells. J. Pathol. 2017, 243, 376–389. [Google Scholar] [CrossRef]
- Ochiai, A.; Yasui, W.; Tahara, E. Growth-promoting effect of gastrin on human gastric carcinoma cell line TMK-1. Jpn. J. Cancer Res. 1985, 76, 1064–1071. [Google Scholar]
- Kuniyasu, H.; Yano, S.; Sasaki, T.; Sasahira, T.; Sone, S.; Ohmori, H. Colon cancer cell-derived high mobility group 1/amphoterin induces growth inhibition and apoptosis in macrophages. Am. J. Pathol. 2005, 166, 751–760. [Google Scholar] [CrossRef]
- Huang, J.C.; Zamble, D.B.; Reardon, J.T.; Lippard, S.J.; Sancar, A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc. Natl. Acad. Sci. USA 1994, 91, 10394–10398. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 2013, 48, 452–458. [Google Scholar] [CrossRef]

| Genes | Coefficient | 95% Interval | Standard Error | t-Statistic | p-Value |
|---|---|---|---|---|---|
| Intercept | −1.072 | −1.6362–−0.5089 | 0.2772 | −3.8677 | 0.00052 |
| HMGB1 | 0.0169 | 0.0083–0.0255 | 0.0042 | 4.0011 | 0.00035 |
| c-MET | 0.0011 | −0.0042–0.0065 | 0.0026 | 0.4308 | 0.66935 |
| PCDHB9 | 0.0045 | −0.0007–0.0010 | 0.0025 | 1.7666 | 0.08627 |
| REGIV | 0.0034 | −0.0019–0.0087 | 0.0026 | 1.3162 | 0.19689 |
| Biopsy No.1 | Grade 2 | Effect | c-Met | HMGB1 | RegIV | PCDHB9 |
|---|---|---|---|---|---|---|
| Cutoff 3 | 75 | 75 | 65 | 50 | ||
| 1 | 1a | Resistant | 904 | 95 | 75 | 45 |
| 2 | 1a | Resistant | 80 | 90 | 80 | 55 |
| 3 | 1a | Resistant | 75 | 90 | 70 | 40 |
| 4 | 1a | Resistant | 60 | 90 | 65 | 55 |
| 5 | 1a | Resistant | 70 | 60 | 50 | 65 |
| 6 | 1b | Effective | 60 | 50 | 70 | 55 |
| 7 | 1b | Effective | 50 | 55 | 75 | 30 |
| 8 | 2 | Effective | 50 | 70 | 50 | 35 |
| 9 | 2 | Effective | 65 | 70 | 55 | 55 |
| 10 | 2 | Effective | 75 | 60 | 50 | 30 |
| Predictive value | 7/10 | 9/10 | 7/10 | 6/10 | ||
| Gene | Accession No. | Forward | Reverse |
|---|---|---|---|
| c-met | NM_001127500.3 | CAGGCAGTGCAGCATGTAGT | GATGATTCCCTCGGTCAGAA |
| hmgb1 | CR456863.1 | ATATGGCAAAAGCGGACAAG | GCAACATCACCAATGGACAG |
| regIV | NM_001159352.1 | TGCTCCTGGATGGTTTTACC | TATCGGCTGGCTTCTCTGAT |
| pcdhb9 | NM_019119.4 | CACTGAGACAGATGGGCTGA | GCCTTTGTCTTGGAAAGCTG |
| tlr4 | AB445638.1 | CCTGTCCCTGAACCCTATGA | CCAGAACCAAACGATGGACT |
| myd88 | U84408.1 | GGATGGTGGTGGTTGTCTCT | AGGATGCTGGGGAACTCTTT |
| rage | AB036432.1 | GCTGTCAGCATCAGCATCAT | ATTCAGTTCTGCACGCTCCT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishiguchi, Y.; Oue, N.; Fujiwara-Tani, R.; Sasaki, T.; Ohmori, H.; Kishi, S.; Mori, S.; Mori, T.; Ikeda, N.; Matsumoto, S.; et al. Role of Metastasis-Related Genes in Cisplatin Chemoresistance in Gastric Cancer. Int. J. Mol. Sci. 2020, 21, 254. https://doi.org/10.3390/ijms21010254
Nishiguchi Y, Oue N, Fujiwara-Tani R, Sasaki T, Ohmori H, Kishi S, Mori S, Mori T, Ikeda N, Matsumoto S, et al. Role of Metastasis-Related Genes in Cisplatin Chemoresistance in Gastric Cancer. International Journal of Molecular Sciences. 2020; 21(1):254. https://doi.org/10.3390/ijms21010254
Chicago/Turabian StyleNishiguchi, Yukiko, Naohide Oue, Rina Fujiwara-Tani, Takamitsu Sasaki, Hitoshi Ohmori, Shingo Kishi, Shiori Mori, Takuya Mori, Naoya Ikeda, Sohei Matsumoto, and et al. 2020. "Role of Metastasis-Related Genes in Cisplatin Chemoresistance in Gastric Cancer" International Journal of Molecular Sciences 21, no. 1: 254. https://doi.org/10.3390/ijms21010254
APA StyleNishiguchi, Y., Oue, N., Fujiwara-Tani, R., Sasaki, T., Ohmori, H., Kishi, S., Mori, S., Mori, T., Ikeda, N., Matsumoto, S., Wakatsuki, K., Luo, Y., Yasui, W., Sho, M., & Kuniyasu, H. (2020). Role of Metastasis-Related Genes in Cisplatin Chemoresistance in Gastric Cancer. International Journal of Molecular Sciences, 21(1), 254. https://doi.org/10.3390/ijms21010254





