Identification of Tea Plant Purple Acid Phosphatase Genes and Their Expression Responses to Excess Iron
Abstract
1. Introduction
2. Results
2.1. The Tea Plant Genome Encodes 19 Putative Purple Acid Phosphatases (PAPs)
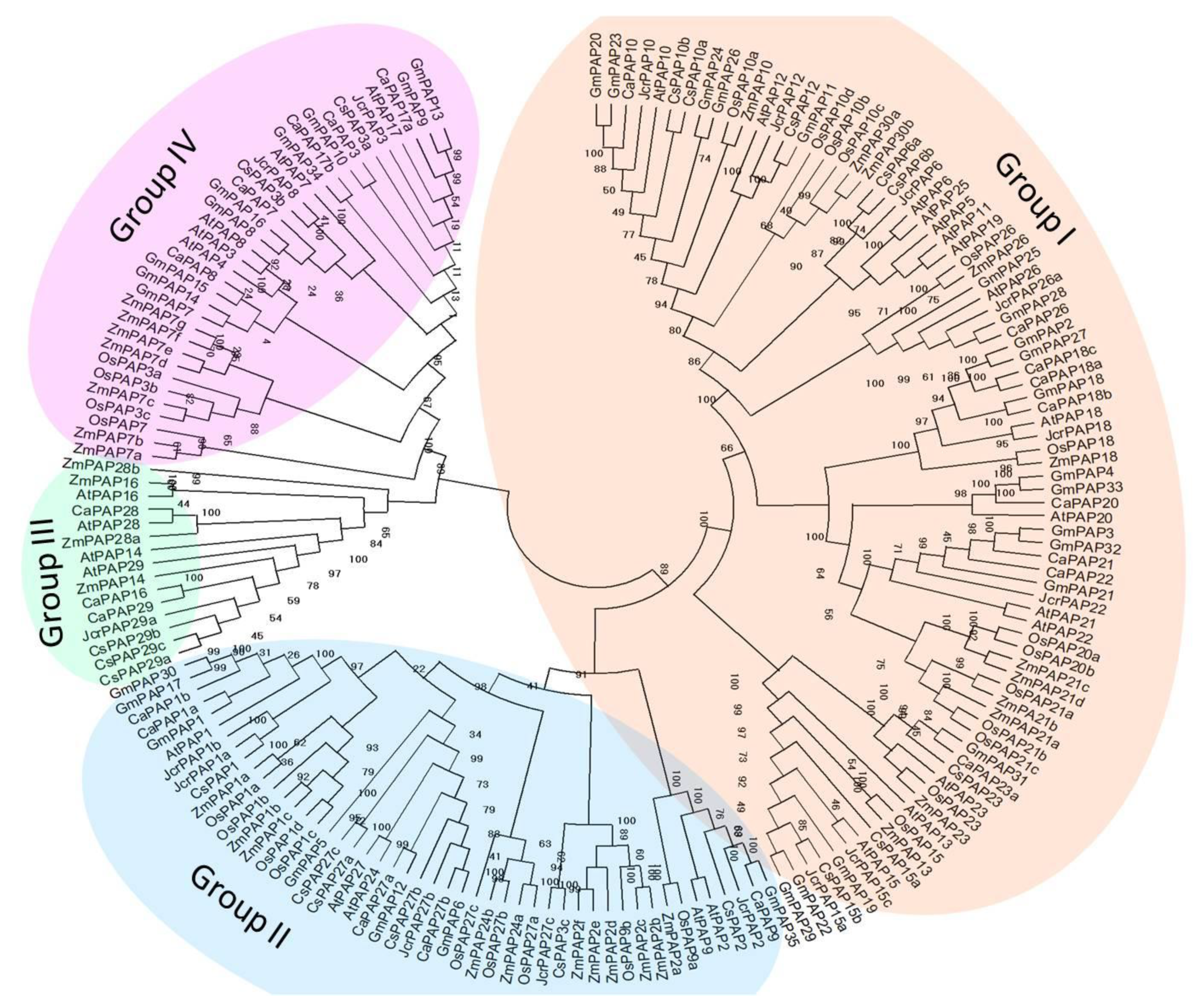
2.2. Phylogenetic Investigations of Tea Plant PAPs
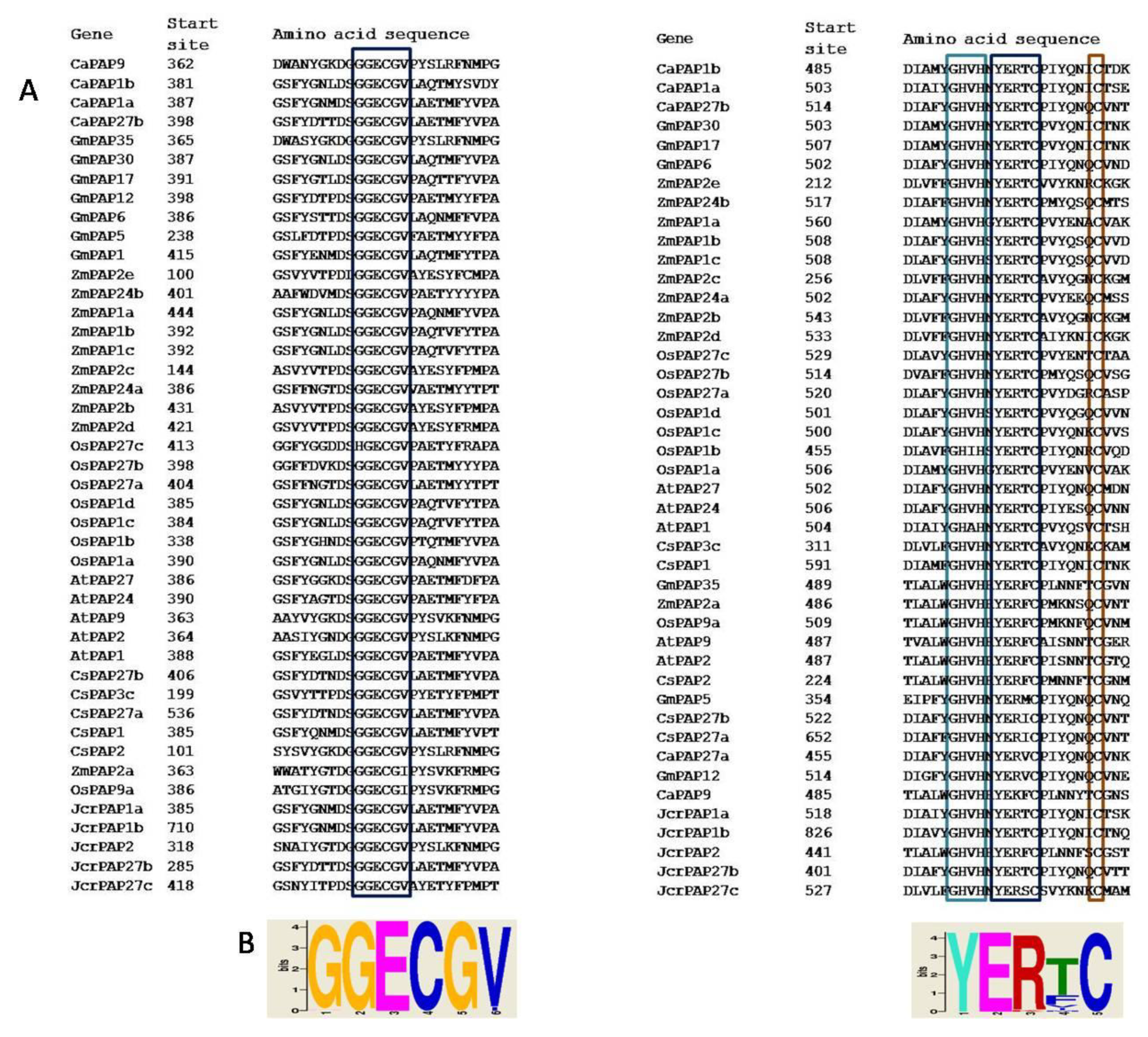
2.3. Group II PAPs Shared Two Cysteine-containing Conserved Blocks
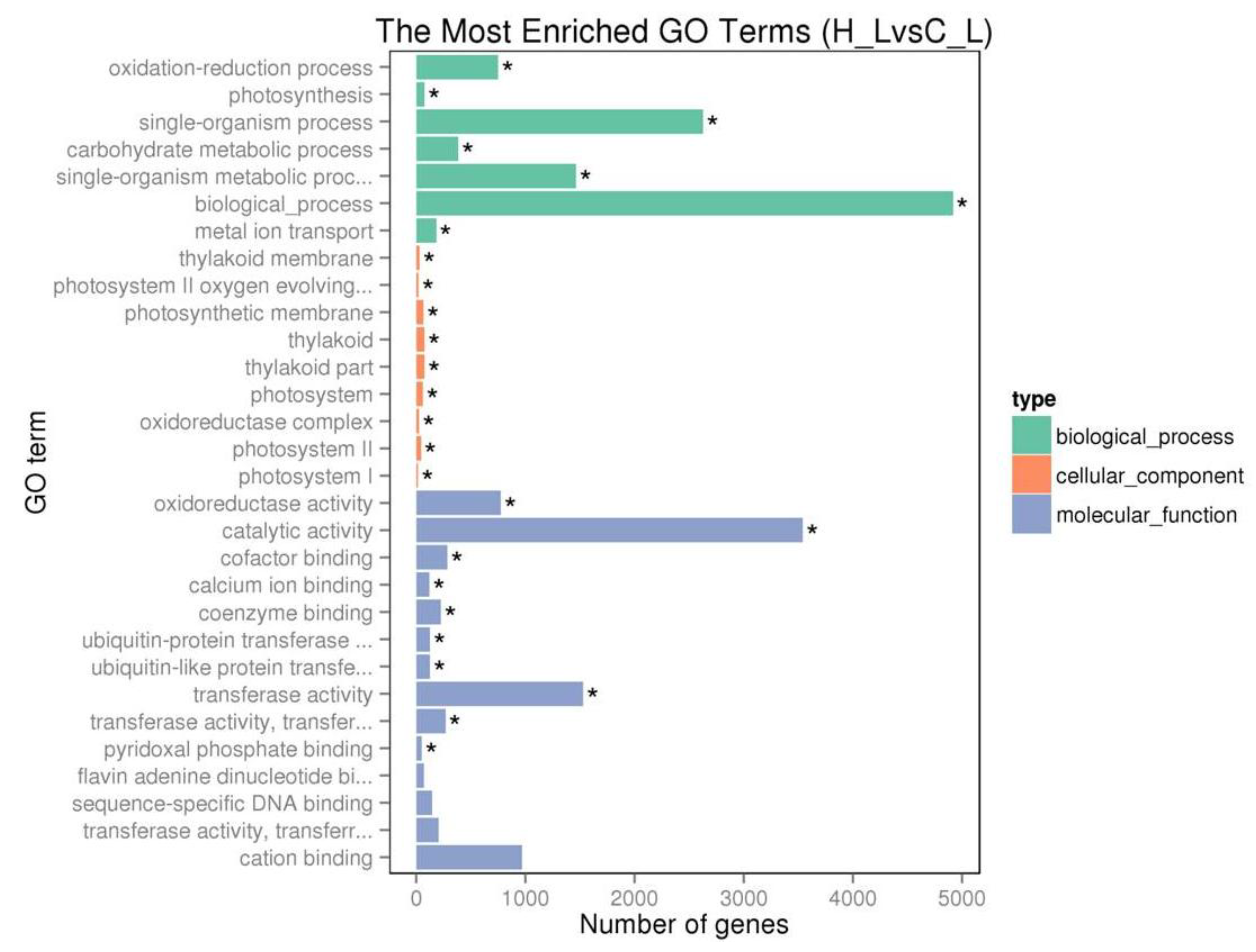
2.4. Transcriptome Analysis of Tea Plant
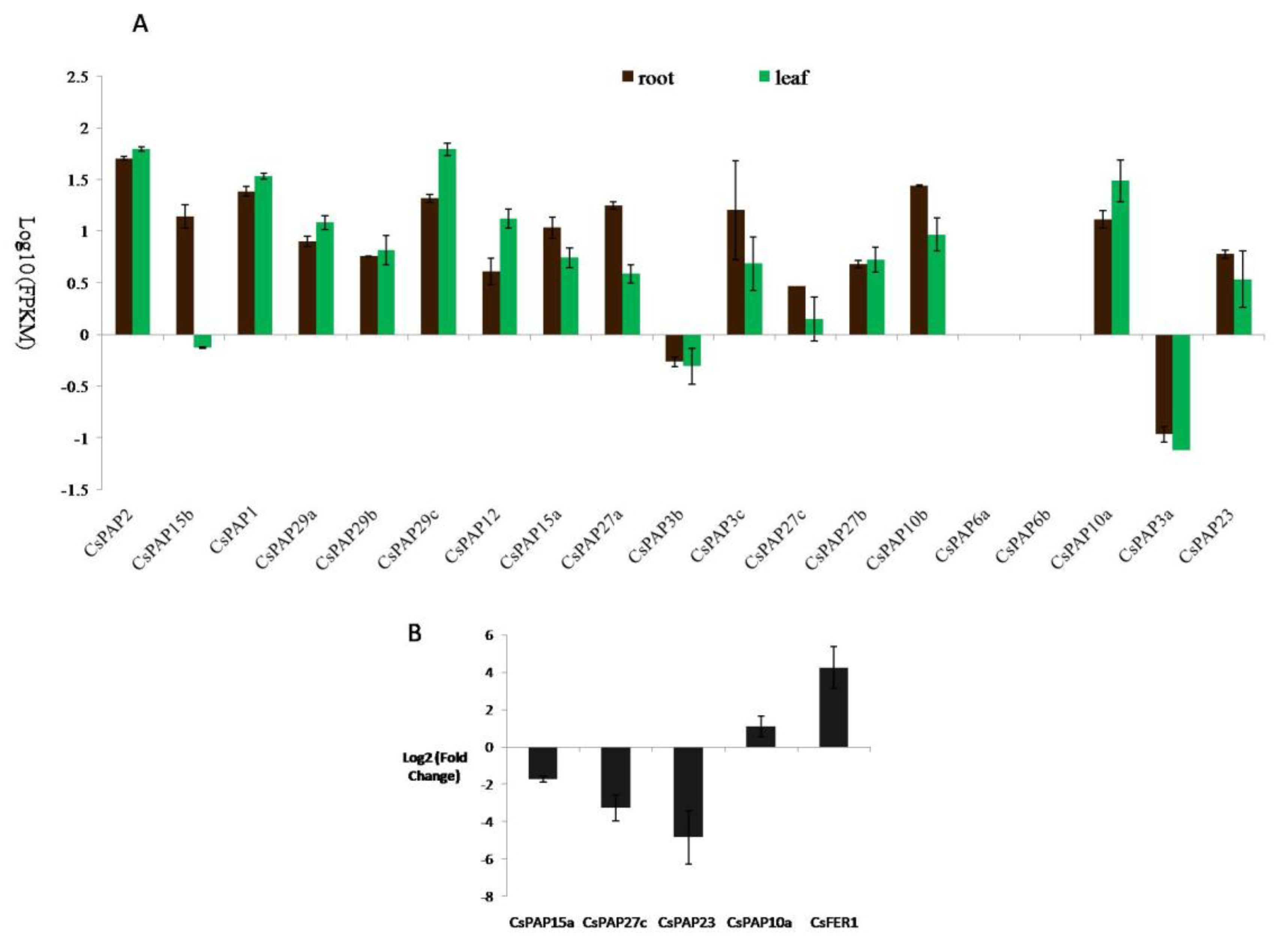
2.5. The Expression of Tea Plant PAP Genes and Their Responses to Iron Supply
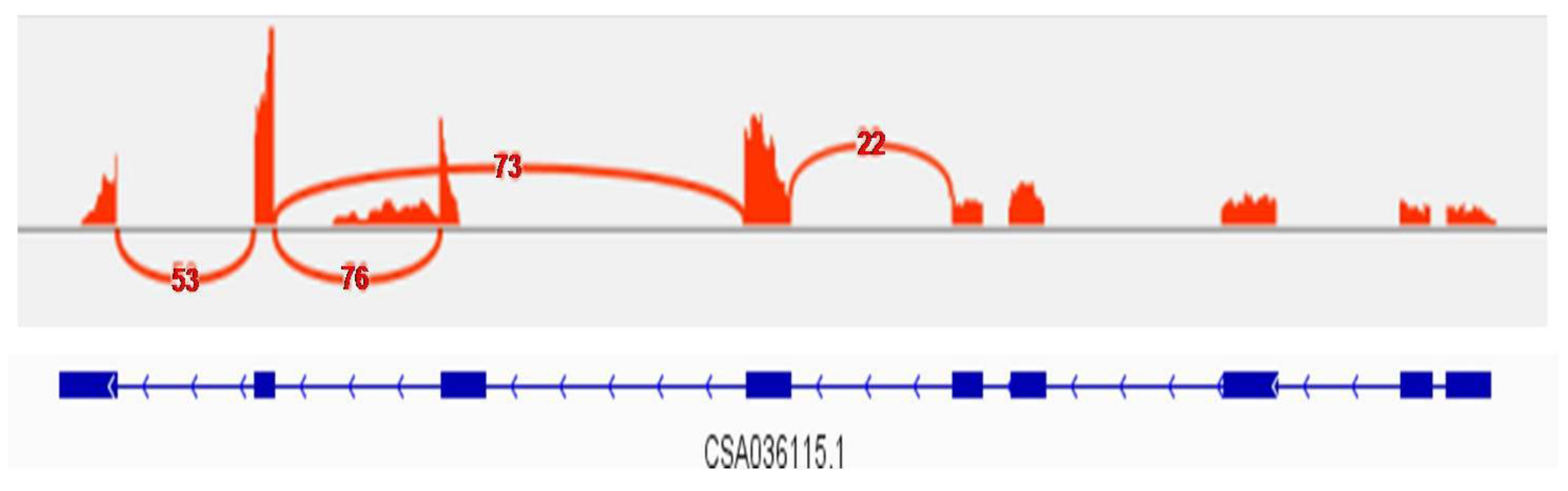
2.6. CsPAP23 Produced Splice Variants
3. Discussion
4. Materials and Methods
4.1. Database Search of PAP Genes in the Tea Plant Genome
4.2. Clustering and Bioinformatics Analysis of Tea Plant PAPs
4.3. Plant Materials and Treatments
4.4. RNA-seq Analysis of Tea Plants Grown Under Normal and High-Iron Conditions
4.5. Quantification of Gene Expression Level and Alternative Splicing(AS) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Schenk, G.; Guddat, L.W.; Ge, Y.; Carrington, L.E.; Hume, D.A.; Hamilton, S.; de Jersey, J. Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 2000, 250, 117–125. [Google Scholar] [CrossRef][Green Version]
- Schenk, G.; Mitić, N.; Hanson, G.R.; Comba, P. Purple acid phosphatase: A journey into the function and mechanism of a colorful enzyme. Coordin. Chem. Rev. 2013, 257, 473–482. [Google Scholar] [CrossRef]
- Bhadouria, J.; Singh, A.P.; Mehra, P.; Verma, L.; Srivastawa, R.; Parida, S.K.; Giri, J. Identification of Purple Acid Phosphatases in Chickpea and Potential Roles of CaPAP7 in Seed Phytate Accumulation. Sci. Rep. 2017, 7, 11012. [Google Scholar] [CrossRef]
- Li, C.; Gui, S.; Yang, T.; Walk, T.; Wang, X.; Liao, H. Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann. Bot. 2012, 109, 275–285. [Google Scholar] [CrossRef]
- Li, D.; Zhu, H.; Liu, K.; Liu, X.; Leggewie, G.; Udvardi, M.; Wang, D. Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J. Biol. Chem. 2002, 277, 27772–27781. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Munoz, E.; Avendano-Vazquez, A.O.; Montes, R.A.; de Folter, S.; Andres-Hernandez, L.; Abreu-Goodger, C.; Sawers, R.J. The maize (Zea mays ssp. mays var. B73) genome encodes 33 members of the purple acid phosphatase family. Front. Plant Sci. 2015, 6, 341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, C.; Tian, J.; Li, K.; Shou, H. Identification of rice purple acid phosphatases related to phosphate starvation signalling. Plant. Biol. (Stuttg.) 2011, 13, 7–15. [Google Scholar] [CrossRef]
- Venkidasamy, B.; Selvaraj, D.; Ramalingam, S. Genome-wide analysis of purple acid phosphatase (PAP) family proteins Check for in Jatropha curcas L. Int. J. Biol. Macromol. 2019, 123, 648–656. [Google Scholar] [CrossRef]
- Tran, H.T.; Hurley, B.A.; Plaxton, W.C. Feeding hungry plants: The role of purple acid phosphatases in phosphate nutrition. Plant Sci. 2010, 179, 14–27. [Google Scholar] [CrossRef]
- Nuttleman, P.R.; Roberts, R.M. Transfer of iron from uteroferrin (purple acid phosphatase) to transferrin related to acid phosphatase activity. J. Biol. Chem. 1990, 265, 12192–12199. [Google Scholar]
- Kaija, H.; Alatalo, S.L.; Halleen, J.M.; Lindqvist, Y.; Schneider, G.; Vaananen, H.K.; Vihko, P. Phosphatase and oxygen radical-generating activities of mammalian purple acid phosphatase are functionally independent. Biochem. Biophys. Res. Commun. 2002, 292, 128–132. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Yu, L.; Yung, K.F.; Leung, D.Y.C.; Sun, F.; Lim, B.L. Over-expression of AtPAP2 in Camelina sativa leads to faster plant growth and higher seed yield. Biotechnol. Biofuels 2012, 5. [Google Scholar] [CrossRef] [PubMed]
- Kaida, R.; Serada, S.; Norioka, N.; Norioka, S.; Neumetzler, L.; Pauly, M.; Sampedro, J.; Zarra, I.; Hayashi, T.; Kaneko, T.S. Potential role for purple acid phosphatase in the dephosphorylation of wall proteins in tobacco cells. Plant Physiol. 2010, 153, 603–610. [Google Scholar] [CrossRef]
- Schultz, E.R.; Zupanska, A.K.; Sng, N.J.; Paul, A.L.; Ferl, R.J. Skewing in Arabidopsis roots involves disparate environmental signaling pathways. BMC Plant Biol. 2017, 17, 31. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Y.; Shao, G.; Lam, H.M. Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytol. 2008, 178, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Stone, S.L.; Benkel, B.; Zhang, J.; Berrue, F.; Prithiviraj, B. Optimal level of purple acid phosphatase5 is required for maintaining complete resistance to Pseudomonas syringae. Front. Plant Sci. 2015, 6, 568. [Google Scholar] [CrossRef]
- Lu, L.; Qiu, W.; Gao, W.; Tyerman, S.D.; Shou, H.; Wang, C. OsPAP10c, a novel secreted acid phosphatase in rice, plays an important role in the utilization of external organic phosphorus. Plant Cell Environ. 2016, 39, 2247–2259. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Lu, L.; Qiu, W.; Wang, C.; Shou, H. OsPAP26 Encodes a Major Purple Acid Phosphatase and Regulates Phosphate Remobilization in Rice. Plant Cell Physiol. 2017, 58, 885–892. [Google Scholar] [CrossRef]
- Mehra, P.; Pandey, B.K.; Giri, J. Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol. J. 2017, 15, 1054–1067. [Google Scholar] [CrossRef]
- Wang, X.R.; Wang, Y.X.; Tian, J.; Lim, B.L.; Yan, X.L.; Liao, H. Overexpressing AtPAP15 Enhances Phosphorus Efficiency in Soybean. Plant Physiol. 2009, 151, 233–240. [Google Scholar] [CrossRef]
- Mai, H.J.; Pateyron, S.; Bauer, P. Iron homeostasis in Arabidopsis thaliana: Transcriptomic analyses reveal novel FIT-regulated genes, iron deficiency marker genes and functional gene networks. BMC Plant Biol. 2016, 16, 211. [Google Scholar] [CrossRef]
- Zhu, H.; Qian, W.; Lu, X.; Li, D.; Liu, X.; Liu, K.; Wang, D. Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol. Biol. 2005, 59, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol. 2008, 146, 431–440. [Google Scholar] [CrossRef]
- Kuang, R.; Chan, K.H.; Yeung, E.; Lim, B.L. Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol. 2009, 151, 199–209. [Google Scholar] [CrossRef]
- Dionisio, G.; Madsen, C.K.; Holm, P.B.; Welinder, K.G.; Jorgensen, M.; Stoger, E.; Arcalis, E.; Brinch-Pedersen, H. Cloning and characterization of purple acid phosphatase phytases from wheat, barley, maize, and rice. Plant Physiol. 2011, 156, 1087–1100. [Google Scholar] [CrossRef]
- Holme, I.B.; Dionisio, G.; Madsen, C.K.; Brinch-Pedersen, H. Barley HvPAPhy_a as transgene provides high and stable phytase activities in mature barley straw and in grains. Plant Biotechnol. J. 2017, 15, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Reddy, C.S.; Kim, S.C.; Kaul, T. Genetically modified phytase crops role in sustainable plant and animal nutrition and ecological development: A review. 3 Biotech 2017, 7, 195. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.V.; Tetens, I.; Meyer, A.S. Potential of phytase-mediated iron release from cereal-based foods: A quantitative view. Nutrients 2013, 5, 3074–3098. [Google Scholar] [CrossRef] [PubMed]
- Kochian, L.V.; Hoekenga, O.A.; Pineros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef]
- Briat, J.F.; Dubos, C.; Gaymard, F. Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 2015, 20, 33–40. [Google Scholar] [CrossRef]
- Xie, L.; Shang, Q. Genome-wide analysis of purple acid phosphatase structure and expression in ten vegetable species. BMC Genom. 2018, 19, 646. [Google Scholar] [CrossRef] [PubMed]
- Olczak, M.; Watorek, W. Two subfamilies of plant purple acid phosphatases. Physiol. Plant. 2003, 118, 491–498. [Google Scholar] [CrossRef]
- Cashikar, A.G.; Rao, N.M. Role of the intersubunit disulfide bond in the unfolding pathway of dimeric red kidney bean purple acid phosphatase. Biochim. Biophys. Acta 1996, 1296, 76–84. [Google Scholar] [CrossRef]
- Antonyuk, S.V.; Olczak, M.; Olczak, T.; Ciuraszkiewicz, J.; Strange, R.W. The structure of a purple acid phosphatase involved in plant growth and pathogen defence exhibits a novel immunoglobulin-like fold. IUCrJ 2014, 1, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Cashikar, A.G.; Rao, M.N. Unique structural features of red kidney bean purple acid phosphatase. Indian J. Biochem. Biophys. 1995, 32, 130–136. [Google Scholar] [PubMed]
- Olczak, M.; Morawiecka, B.; Watorek, W. Plant purple acid phosphatases—Genes, structures and biological function. Acta Biochim. Pol. 2003, 50, 1245–1256. [Google Scholar]
- Madsen, C.K.; Dionisio, G.; Holme, I.B.; Holm, P.B.; Brinch-Pedersen, H. High mature grain phytase activity in the Triticeae has evolved by duplication followed by neofunctionalization of the purple acid phosphatase phytase (PAPhy) gene. J. Exp. Bot. 2013, 64, 3111–3123. [Google Scholar] [CrossRef]
- Brinch-Pedersen, H.; Borg, S.; Tauris, B.; Holm, P.B. Molecular genetic approaches to increasing mineral availability and vitamin content of cereals. J. Cereal Sci. 2007, 46, 308–326. [Google Scholar] [CrossRef]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef]
- Kupper, H.; Andresen, E. Mechanisms of metal toxicity in plants. Metallomics 2016, 8, 269–285. [Google Scholar] [CrossRef]
- Li, G.J.; Xu, W.F.; Kronzucker, H.J.; Shi, W.M. Ethylene is critical to the maintenance of primary root growth and Fe homeostasis under Fe stress in Arabidopsis. J. Exp. Bot. 2015, 66, 2041–2054. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, D. Functions and regulation of phosphate starvation-induced secreted acid phosphatases in higher plants. Plant Sci. 2018, 271, 108–116. [Google Scholar] [CrossRef]
- Grillet, L.; Ouerdane, L.; Flis, P.; Hoang, M.T.T.; Isaure, M.P.; Lobinski, R.; Curie, C.; Mari, S. Ascorbate Efflux as a New Strategy for Iron Reduction and Transport in Plants. J. Biol. Chem. 2014, 289, 2515–2525. [Google Scholar] [CrossRef]
- Li, W.; Lin, W.D.; Ray, P.; Lan, P.; Schmidt, W. Genome-wide detection of condition-sensitive alternative splicing in Arabidopsis roots. Plant Physiol. 2013, 162, 1750–1763. [Google Scholar] [CrossRef] [PubMed]
- Ravet, K.; Touraine, B.; Kim, S.A.; Cellier, F.; Thomine, S.; Guerinot, M.L.; Briat, J.F.; Gaymard, F. Post-translational regulation of AtFER2 ferritin in response to intracellular iron trafficking during fruit development in Arabidopsis. Mol. Plant 2009, 2, 1095–1106. [Google Scholar] [CrossRef]
- Curie, C.; Mari, S. New routes for plant iron mining. New Phytol. 2017, 214, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.H.; Zhang, H.B.; Sheng, J.; Li, K.; Zhang, Q.J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The Tea Tree Genome Provides Insights into Tea Flavor and Independent Evolution of Caffeine Biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, S.; Cowley, A.; Lee, J.; Foix, A.; Lopez, R. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 2017, 45, W550–W553. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Clifton, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Nielsen, H. Predicting Secretory Proteins with SignalP. In Protein Function Prediction; Kihara, D., Ed.; Springer: Berlin, Germany, 2017; Volume 1611, pp. 59–73. [Google Scholar]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Ghanati, F.; Morita, A.; Yokota, H. Effects of aluminum on the growth of tea plant and activation of antioxidant system. Plant Soil 2005, 276, 133–141. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Lan, P. Re-analysis of RNA-seq transcriptome data reveals new aspects of gene activity in Arabidopsis root hairs. Front. Plant Sci. 2015, 6, 421. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.H.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [PubMed]
| Gene ID | Proposed Name | PAP Conserved Residues | Conserved Domain a | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GDXG | GDXXY | GNH(D/E) | VXXH | GHXH | |||||
| CSA004804 | CsPAP2 | GDISY | GNHE | VQGH | GHVH | MPP_PAPs | |||
| CSA005153 | CsPAP15b | GDLG | GDVTY | GNHD | ATWH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA006013 | CsPAP1 | GDLG | GDICY | GNHE | FLAH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA006536 | CsPAP29a | ADMH | GDNIF | GNHD | VYFH | GHDH | MPP_Dcr2 | ||
| CSA006537 | CsPAP29b | GDNIF | GNHD | VYFH | GHDH | MPP_Dcr2 | |||
| CSA006539 | CsPAP29c | ADMH | GDNIF | GNHD | VYFH | GHDH | MPP_Dcr2 | ||
| CSA008696 | CsPAP12 | GDLG | GDLSY | GNHE | VLMH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA011989 | CsPAP15a | GDLG | GDATY | GNHD | ASWH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA018804 | CsPAP27a | GDMG | GDLSY | GNHE | FAAH | GHVH | MPP_PAPs | Pur_ac_phosph_N | Metallophos_C |
| CSA019400 | CsPAP3b | GDNFY | GNHD | VVGH | GHDH | MPP_ACP5 | |||
| CSA021776 | CsPAP3c | GDMG | GDISY | GNHE | FAGH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA026817 | CsPAP27c | GDMG | GDIVY | GNHE | FIAH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA027593 | CsPAP27b | GDMG | GDLPY | GNHE | FAAH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA028799 | CsPAP10b | GDLG | GDLSY | GNHE | VLMH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA030189 | CsPAP6a | GDLG | GDLSY | GNHE | VILH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA030190 | CsPAP6b | GDLG | GDLSY | GNHE | VILH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA031017 | CsPAP10a | GDLG | GDLSY | GNHE | VLLH | GHVH | MPP_PAPs | Pur_ac_phosph_N | |
| CSA034553 | CsPAP3a | GDWG | GDNFY | GNHD | VVGH | GHDH | MPP_ACP5 | ||
| CSA036115 | CsPAP23 | GDLG | GDLTY | GNHE | AAWH | GHVH | MPP_PAPs | Pur_ac_phosph_N | Metallophos_C |
| Gene | Scaffold | Location (Start-Stop) | Length (aa) | Predicted Size (kDa) | Signal Peptide Length | Subcellular Location # | Transmembrane Domain Site | Predicted Number of N-Glycosylation Site |
|---|---|---|---|---|---|---|---|---|
| CsPAP2 | Sc0000741 | 607594-608772 | 392 | 44.0 | No | _ | 350-371 | 2 * |
| CsPAP15b | Sc0002098 | 204497-207829 | 572 | 64.5 | 24 | S | 313-333 | 9 |
| CsPAP1d | xpSc0055840 | 6841-16105 | 700 | 78.8 | 22 | S | no | 6 |
| CsPAP29a | Sc0000663 | 233422-236333 | 387 | 42.1 | 29 | S | no | 1 |
| CsPAP29b | Sc0000663 | 239793-242521 | 304 | 33.4 | No | _ | no | 1 * |
| CsPAP29c | Sc0000663 | 250447-253464 | 387 | 42.2 | 29 | S | no | 2 |
| CsPAP12 | Sc0000340 | 248681-252365 | 469 | 53.8 | 24 | S | no | 4 |
| CsPAP15a | Sc0000525 | 77490- 80649 | 398 | 45.5 | No | _ | no | 4 * |
| CsPAP1a | Sc0001074 | 222761-234424 | 761 | 86.1 | No | _ | no | 7 * |
| CsPAP3b | Sc0001483 | 375068-377024 | 267 | 30.5 | No | _ | no | 0 * |
| CsPAP3c | Sc0000211 | 481949-485198 | 407 | 45.04 | No | _ | no | 1 * |
| CsPAP1c | Sc0001926 | 105169-108133 | 522 | 59.1 | No | M | no | 2 * |
| CsPAP1b | Sc0002467 | 157423-166705 | 631 | 71.1 | No | S | no | 7 * |
| CsPAP10b | Sc0002450 | 126798-130541 | 364 | 41.8 | 21 | S | no | 4 |
| CsPAP6a | xpSc0053256 | 290822-293152 | 402 | 46.5 | 22 | _ | no | 2 |
| CsPAP6b | xpSc0053256 | 310322-313230 | 472 | 54.6 | 22 | S | no | 4 |
| CsPAP10a | xpSc0053660 | 137781-142512 | 777 | 89.0 | No | _ | no | 5 * |
| CsPAP3a | Sc0004847 | 59846-70900 | 370 | 42.0 | No | C | no | 1 * |
| CsPAP23 | Sc0001836 | 97264-106048 | 683 | 76.0 | 23 | S | 656-677 | 5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, C.; Wang, F.; Fan, H.; Fang, Y.; Li, W. Identification of Tea Plant Purple Acid Phosphatase Genes and Their Expression Responses to Excess Iron. Int. J. Mol. Sci. 2019, 20, 1954. https://doi.org/10.3390/ijms20081954
Yin C, Wang F, Fan H, Fang Y, Li W. Identification of Tea Plant Purple Acid Phosphatase Genes and Their Expression Responses to Excess Iron. International Journal of Molecular Sciences. 2019; 20(8):1954. https://doi.org/10.3390/ijms20081954
Chicago/Turabian StyleYin, Chaoyan, Fei Wang, Huiqin Fan, Yanming Fang, and Wenfeng Li. 2019. "Identification of Tea Plant Purple Acid Phosphatase Genes and Their Expression Responses to Excess Iron" International Journal of Molecular Sciences 20, no. 8: 1954. https://doi.org/10.3390/ijms20081954
APA StyleYin, C., Wang, F., Fan, H., Fang, Y., & Li, W. (2019). Identification of Tea Plant Purple Acid Phosphatase Genes and Their Expression Responses to Excess Iron. International Journal of Molecular Sciences, 20(8), 1954. https://doi.org/10.3390/ijms20081954





