Cognizance of Molecular Methods for the Generation of Mutagenic Phage Display Antibody Libraries for Affinity Maturation
Abstract
1. Introduction
2. Phage Display Technology: Harnessing Novel Antibodies
3. Combinatorial Antibody Phage Library
3.1. Naïve Antibody Libraries
3.2. Immune Antibody Libraries
3.3. Semi-Synthetic and Synthetic Antibody Libraries
4. In Vitro Affinity Maturation Strategies
4.1. Random Mutagenesis
4.1.1. Error-Prone Polymerase Chain Reaction (PCR)
4.1.2. Chain Recombination
4.2. Site-Specific Mutagenesis
4.2.1. Enzyme-Based Mutagenesis
4.2.2. Chemical-Based Mutagenesis
4.3. Gene Synthesis Methods for Synthetic Antibody Gene Production
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| mAb | Monoclonal antibody |
| CDR | Complementarity determining regions |
| SHM | Somatic hypermutation |
| scFv | Single chain variable fragment |
| Fab | Antigen binding fragment |
| Fv | Variable fragment |
| GlySer | Glycine-serine linker |
| ssDNA | Single-stranded deoxyribonucleic acid |
| HC | Heavy chain |
| LC | Light chain |
| PCR | Polymerase chain reaction |
| RCA | Rolling circle amplification |
| dNTPs | Deoxyribonucleoside triphosphates |
| SM | Saturation mutagenesis |
| TAG | Amber stop codon |
| dsDNA | Double-stranded deoxyribonucleic acid |
| OE-PCR | Overlap extension polymerase chain reaction |
| sdAbs | Single domain antibodies |
| ISM | Iterative saturation mutagenesis |
| B-FIT | B-factor iterative test |
| TNF | Tumour necrosis factor |
| UV | Ultraviolet |
| HPLC | High performance liquid chromatography |
| PAGE | Polyacrylamide gel electrophoresis |
| NGS | Next generation sequencing |
References
- Frenzel, A.; Kügler, J.; Helmsing, S.; Meier, D.; Schirrmann, T.; Hust, M.; Dübel, S. Designing Human Antibodies by Phage Display. Transfus. Med. Hemother. 2017, 44, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Kaplon, H.; Reichert, J.M. Antibodies to watch in 2018. mAbs 2018, 10, 183–203. [Google Scholar] [CrossRef] [PubMed]
- Grilo, A.L.; Mantalaris, A. The Increasingly Human and Profitable Monoclonal Antibody Market. Trends Biotechnol. 2019, 37, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Ecker, D.M.; Jones, S.D.; Levine, H.L. The therapeutic monoclonal antibody market. mAbs 2014, 7, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Shaughnessy, A.F. Monoclonal antibodies: Magic bullets with a hefty price tag. BMJ 2012, 345, e8346. [Google Scholar] [CrossRef] [PubMed]
- Couzin-Frankel, J. Cancer Immunotherapy. Science 2013, 342, 1432–1433. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Rao, M.; Wallis, R.S.; Kaufmann, S.H.E.; Rustomjee, R.; Mwaba, P.; Vilaplana, C.; Yeboah-Manu, D.; Chakaya, J.; Ippolito, G.; et al. Host-directed therapies for infectious diseases: Current status, recent progress, and future prospects. Lancet Infect. Dis. 2016, 16, e47–e63. [Google Scholar] [CrossRef]
- Scott, A.M.; Wolchok, J.D.; Old, L.J. Antibody therapy of cancer. Nat. Rev. Cancer 2012, 12, 278. [Google Scholar] [CrossRef]
- Lagassé, H.; Alexaki, A.; Simhadri, V.; Katagiri, N.; Jankowski, W.; Sauna, Z.; Kimchi-Sarfaty, C. Recent advances in (therapeutic protein) drug development. F1000Research 2017, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.; Alt, F.W. Unraveling V(D)J Recombination: Insights into Gene Regulation. Cell 2004, 116, 299–311. [Google Scholar] [CrossRef]
- Little, A.J.; Matthews, A.; Oettinger, M.; Roth, D.B.; Schatz, D.G. The Mechanism of V(D)J Recombination. In Molecular Biology of B Cells, 2nd ed.; Alt, F.W., Honjo, T., Radbruch, A., Reth, M., Eds.; Academic Press: London, UK, 2015; pp. 13–34. [Google Scholar]
- Roth, D.B. V(D)J Recombination: Mechanism, Errors, and Fidelity. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Li, Z.; Woo, C.J.; Iglesias-Ussel, M.D.; Ronai, D.; Scharff, M.D. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004, 18, 1–11. [Google Scholar] [CrossRef]
- Papavasiliou, F.N.; Schatz, D.G. Somatic Hypermutation of Immunoglobulin Genes: Merging Mechanisms for Genetic Diversity. Cell 2002, 109, S35–S44. [Google Scholar] [CrossRef]
- Theam Soon, L.; Soo Khim, C. Immune Antibody Libraries: Manipulating The Diverse Immune Repertoire for Antibody Discovery. Curr. Pharm. Des. 2016, 22, 6480–6489. [Google Scholar] [CrossRef] [PubMed]
- Smith, G. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Aghebati-Maleki, L.; Bakhshinejad, B.; Baradaran, B.; Motallebnezhad, M.; Aghebati-Maleki, A.; Nickho, H.; Yousefi, M.; Majidi, J. Phage display as a promising approach for vaccine development. J. Biomed. Sci. 2016, 23, 66. [Google Scholar] [CrossRef]
- Zhao, A.; Tohidkia, M.R.; Siegel, D.L.; Coukos, G.; Omidi, Y. Phage antibody display libraries: A powerful antibody discovery platform for immunotherapy. Crit. Rev. Biotechnol. 2016, 36, 276–289. [Google Scholar] [CrossRef]
- Steinwand, M.; Droste, P.; Frenzel, A.; Hust, M.; Dübel, S.; Schirrmann, T. The influence of antibody fragment format on phage display based affinity maturation of IgG. mAbs 2014, 6, 204–218. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Douthwaite, J.A.; Chen, Y.; Kemp, B.; Kidd, S.; Percival-Alwyn, J.; Smith, A.; Goode, K.; Swerdlow, B.; Lowe, D.; et al. A high-throughput platform for population reformatting and mammalian expression of phage display libraries to enable functional screening as full-length IgG. mAbs 2017, 9, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Barbas, C.F.; Kang, A.S.; Lerner, R.A.; Benkovic, S.J. Assembly of combinatorial antibody libraries on phage surfaces: The gene III site. Proc. Natl. Acad. Sci. USA 1991, 88, 7978–7982. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. scFv antibody: Principles and clinical application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef]
- Huston, J.S.; Levinson, D.; Mudgett-Hunter, M.; Tai, M.S.; Novotný, J.; Margolies, M.N.; Ridge, R.J.; Bruccoleri, R.E.; Haber, E.; Crea, R. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 85, 5879–5883. [Google Scholar] [CrossRef] [PubMed]
- Beerli, R.R.; Rader, C. Mining human antibody repertoires. mAbs 2010, 2, 365–378. [Google Scholar] [CrossRef]
- FitzGerald, K. In vitro display technologies—New tools for drug discovery. Drug Discov. Today 2000, 5, 253–258. [Google Scholar] [CrossRef]
- Greenwood, J.; Willis, A.E.; Perham, R.N. Multiple display of foreign peptides on a filamentous bacteriophage: Peptides from Plasmodium falciparum circumsporozoite protein as antigens. J. Mol. Biol. 1991, 220, 821–827. [Google Scholar] [CrossRef]
- Scott, J.; Smith, G. Searching for peptide ligands with an epitope library. Science 1990, 249, 386–390. [Google Scholar] [CrossRef]
- Wilson, D.R.; Finlay, B.B. Phage display: Applications, innovations, and issues in phage and host biology. Can. J. Microbiol. 1998, 44, 313–329. [Google Scholar] [CrossRef]
- Qi, H.; Lu, H.; Qiu, H.-J.; Petrenko, V.; Liu, A. Phagemid Vectors for Phage Display: Properties, Characteristics and Construction. J. Mol. Biol. 2012, 417, 129–143. [Google Scholar] [CrossRef]
- Sidhu, S.S. Engineering M13 for phage display. Biomol. Eng. 2001, 18, 57–63. [Google Scholar] [CrossRef]
- Imai, S.; Mukai, Y.; Takeda, T.; Abe, Y.; Nagano, K.; Kamada, H.; Nakagawa, S.; Tsunoda, S.; Tsutsumi, Y. Effect of protein properties on display efficiency using the M13 phage display system. Die Pharmazie 2008, 63, 760–764. [Google Scholar] [CrossRef]
- Soltes, G.; Hust, M.; Ng, K.K.Y.; Bansal, A.; Field, J.; Stewart, D.I.H.; Dübel, S.; Cha, S.; Wiersma, E.J. On the influence of vector design on antibody phage display. J. Biotechnol. 2007, 127, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Tohidkia, M.R.; Barar, J.; Asadi, F.; Omidi, Y. Molecular considerations for development of phage antibody libraries. J. Drug Target. 2012, 20, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Carmen, S.; Jermutus, L. Concepts in antibody phage display. Brief. Funct. Genom. 2002, 1, 189–203. [Google Scholar] [CrossRef]
- Mai-Prochnow, A.; Hui, J.G.K.; McDougald, D.; Kjelleberg, S.; Rice, S.A.; Rakonjac, J. ‘Big things in small packages: The genetics of filamentous phage and effects on fitness of their host’. FEMS Microbiol. Rev. 2015, 39, 465–487. [Google Scholar] [CrossRef] [PubMed]
- Rakonjac, J.; Bennett, N.J.; Spagnuolo, J.; Gagic, D.; Russel, M. Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011, 13, 51. [Google Scholar]
- Loeb, T. Isolation of a Bacteriophage Specific for the F+ and Hfr Mating Types of Escherichia coli K-12. Science 1960, 131, 932–933. [Google Scholar] [CrossRef]
- Marvin, D.A.; Hoffmann-Berling, H. Physical and Chemical Properties of Two New Small Bacteriophages. Nature 1963, 197, 517–518. [Google Scholar] [CrossRef]
- Tikunova, N.V.; Morozova, V.V. Phage display on the base of filamentous bacteriophages: Application for recombinant antibodies selection. Acta Naturae 2009, 1, 20–28. [Google Scholar]
- Ledsgaard, L.; Kilstrup, M.; Karatt-Vellatt, A.; McCafferty, J.; Laustsen, A.H. Basics of Antibody Phage Display Technology. Toxins 2018, 10, 236. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Geiser, M. Evaluation of antibodies fused to minor coat protein III and major coat protein VIII of bacteriophage M 13. Gene 1995, 155, 61–65. [Google Scholar] [CrossRef]
- Bazan, J.; Całkosiński, I.; Gamian, A. Phage display—A powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications. Hum. Vaccines Immunother. 2012, 8, 1817–1828. [Google Scholar] [CrossRef]
- Petrenko, V.A. Landscape Phage: Evolution from Phage Display to Nanobiotechnology. Viruses 2018, 10, 311. [Google Scholar] [CrossRef]
- Rondot, S.; Koch, J.; Breitling, F.; Dübel, S. A helper phage to improve single-chain antibody presentation in phage display. Nat. Biotechnol. 2001, 19, 75. [Google Scholar] [CrossRef] [PubMed]
- Chasteen, L.; Ayriss, J.; Pavlik, P.; Bradbury, A.R.M. Eliminating helper phage from phage display. Nucleic Acids Res. 2006, 34, e145. [Google Scholar] [CrossRef]
- Gao, C.; Mao, S.; Kaufmann, G.; Wirsching, P.; Lerner, R.A.; Janda, K.D. A method for the generation of combinatorial antibody libraries using pIX phage display. Proc. Natl. Acad. Sci. USA 2002, 99, 12612–12616. [Google Scholar] [CrossRef]
- Huovinen, T.; Syrjänpää, M.; Sanmark, H.; Seppä, T.; Akter, S.; Khan, L.M.F.; Lamminmäki, U. The selection performance of an antibody library displayed on filamentous phage coat proteins p9, p3 and truncated p3. BMC Res. Notes 2014, 7, 661. [Google Scholar] [CrossRef][Green Version]
- Porcar, M. Beyond directed evolution: Darwinian selection as a tool for synthetic biology. Syst. Synth. Biol. 2010, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ullman, C.G.; Frigotto, L.; Cooley, R.N. In vitro methods for peptide display and their applications. Brief. Funct. Genom. 2011, 10, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Barbas, C.F.; Burton, D.R.; Scott, J.K.; Silverman, G.J. Phage Display: A Laboratory Manual; CSHL Press: Cold Spring Harbor, NY, USA, 2004. [Google Scholar]
- AC’t Hoen, P.; Jirka, S.M.G.; ten Broeke, B.R.; Schultes, E.A.; Aguilera, B.; Pang, K.H.; Heemskerk, H.; Aartsma-Rus, A.; van Ommen, G.J.; den Dunnen, J.T. Phage display screening without repetitious selection rounds. Anal. Biochem. 2012, 421, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G.; Ippolito, G.C.; Beausang, J.; Busse, C.E.; Wardemann, H.; Quake, S.R. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 2014, 32, 158–168. [Google Scholar] [CrossRef]
- Lerner, R.A. Combinatorial antibody libraries: New advances, new immunological insights. Nat. Rev. Immunol. 2016, 16, 498. [Google Scholar] [CrossRef]
- Hammers, C.M.; Stanley, J.R. Antibody phage display: Technique and applications. J. Investig. Dermatol. 2014, 134, 1–5. [Google Scholar] [CrossRef]
- Griffiths, A.D.; Duncan, A.R. Strategies for selection of antibodies by phage display. Curr. Opin. Biotechnol. 1998, 9, 102–108. [Google Scholar] [CrossRef]
- Kügler, J.; Wilke, S.; Meier, D.; Tomszak, F.; Frenzel, A.; Schirrmann, T.; Dübel, S.; Garritsen, H.; Hock, B.; Toleikis, L.; et al. Generation and analysis of the improved human HAL9/10 antibody phage display libraries. BMC Biotechnol. 2015, 15, 10. [Google Scholar] [CrossRef]
- Könning, D.; Zielonka, S.; Grzeschik, J.; Empting, M.; Valldorf, B.; Krah, S.; Schröter, C.; Sellmann, C.; Hock, B.; Kolmar, H. Camelid and shark single domain antibodies: Structural features and therapeutic potential. Curr. Opin. Struct. Biol. 2017, 45, 10–16. [Google Scholar] [CrossRef]
- Noy-Porat, T.; Rosenfeld, R.; Ariel, N.; Epstein, E.; Alcalay, R.; Zvi, A.; Kronman, C.; Ordentlich, A.; Mazor, O. Isolation of Anti-Ricin Protective Antibodies Exhibiting High Affinity from Immunized Non-Human Primates. Toxins 2016, 8, 64. [Google Scholar] [CrossRef]
- Koti, M.; Saini, S.S.; Sachan, A.; Kaushik, A.K. Engineered Bovine Antibodies in the Development of Novel Therapeutics, Immunomodulators and Vaccines. Antibodies 2014, 3, 205–214. [Google Scholar] [CrossRef]
- Mathonet, P.; Ullman, C.G. The application of next generation sequencing to the understanding of antibody repertoires. Front. Immunol. 2013, 4, 265. [Google Scholar] [CrossRef]
- Schwimmer, L.J.; Huang, B.; Giang, H.; Cotter, R.L.; Chemla-Vogel, D.S.; Dy, F.V.; Tam, E.M.; Zhang, F.; Toy, P.; Bohmann, D.J.; et al. Discovery of diverse and functional antibodies from large human repertoire antibody libraries. J. Immunol. Methods 2013, 391, 60–71. [Google Scholar] [CrossRef]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef]
- Clackson, T.; Hoogenboom, H.R.; Griffiths, A.D.; Winter, G. Making antibody fragments using phage display libraries. Nature 1991, 352, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Glanville, J.; Zhai, W.; Berka, J.; Telman, D.; Huerta, G.; Mehta, G.R.; Ni, I.; Mei, L.; Sundar, P.D.; Day, G.M.R.; et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc. Natl. Acad. Sci. USA 2009, 106, 20216–20221. [Google Scholar] [CrossRef]
- Vaughan, T.J.; Williams, A.J.; Pritchard, K.; Osbourn, J.K.; Pope, A.R.; Earnshaw, J.C.; McCafferty, J.; Hodits, R.A.; Wilton, J.; Johnson, K.S. Human Antibodies with Sub-nanomolar Affinities Isolated from a Large Non-immunized Phage Display Library. Nat. Biotechnol. 1996, 14, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Marks, J.D.; Hoogenboom, H.R.; Bonnert, T.P.; McCafferty, J.; Griffiths, A.D.; Winter, G. By-passing immunization: Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991, 222, 581–597. [Google Scholar] [CrossRef]
- Barbas, C.F., 3rd; Bain, J.D.; Hoekstra, D.M.; Lerner, R.A. Semisynthetic combinatorial antibody libraries: A chemical solution to the diversity problem. Proc. Natl. Acad. Sci. USA 1992, 89, 4457–4461. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, A.D.; Williams, S.C.; Hartley, O.; Tomlinson, I.M.; Waterhouse, P.; Crosby, W.L.; Kontermann, R.E.; Jones, P.T.; Low, N.M.; Allison, T.J. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994, 13, 3245–3260. [Google Scholar] [CrossRef] [PubMed]
- Rothe, C.; Urlinger, S.; Löhning, C.; Prassler, J.; Stark, Y.; Jäger, U.; Hubner, B.; Bardroff, M.; Pradel, I.; Boss, M.; et al. The Human Combinatorial Antibody Library HuCAL GOLD Combines Diversification of All Six CDRs According to the Natural Immune System with a Novel Display Method for Efficient Selection of High-Affinity Antibodies. J. Mol. Biol. 2008, 376, 1182–1200. [Google Scholar] [CrossRef]
- Benner, R.; Hijmans, W.; Haaijman, J.J. The bone marrow: The major source of serum immunoglobulins, but still a neglected site of antibody formation. Clin. Exp. Immunol. 1981, 46, 1–8. [Google Scholar] [PubMed]
- Ponsel, D.; Neugebauer, J.; Ladetzki-Baehs, K.; Tissot, K. High affinity, developability and functional size: The holy grail of combinatorial antibody library generation. Molecules 2011, 16, 3675–3700. [Google Scholar] [CrossRef] [PubMed]
- Dantas-Barbosa, C.; de Macedo Brigido, M.; Maranhao, A.Q. Antibody phage display libraries: Contributions to oncology. Int. J. Mol. Sci. 2012, 13, 5420–5440. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.C.; Carter, P.J. Therapeutic antibodies for autoimmunity and inflammation. Nat. Rev. Immunol. 2010, 10, 301. [Google Scholar] [CrossRef]
- Osbourn, J.; Jermutus, L.; Duncan, A. Current methods for the generation of human antibodies for the treatment of autoimmune diseases. Drug Discov. Today 2003, 8, 845–851. [Google Scholar] [CrossRef]
- Frenzel, A.; Schirrmann, T.; Hust, M. Phage display-derived human antibodies in clinical development and therapy. mAbs 2016, 8, 1177–1194. [Google Scholar] [CrossRef] [PubMed]
- Shukra, A.M.; Sridevi, N.V.; Dev, C.; Kapil, M. Production of recombinant antibodies using bacteriophages. Eur. J. Microbiol. Immunol. 2014, 4, 91–98. [Google Scholar] [CrossRef]
- Hoogenboom, H.R. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 2005, 23, 1105. [Google Scholar] [CrossRef]
- Roovers, R.C.; van der Linden, E.; Zijlema, H.; de Bruïne, A.; Arends, J.-W.; Hoogenboom, H.R. Evidence for a bias toward intracellular antigens in the local humoral anti-tumor immune response of a colorectal cancer patient revealed by phage display. Int. J. Cancer 2001, 93, 832–840. [Google Scholar] [CrossRef] [PubMed]
- Rader, C.; Steinberger, P.; Barbas, C., III. Selection from Antibody Libraries; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001; pp. 10.12–10.20. [Google Scholar]
- Reddy, M.M.; Wilson, R.; Wilson, J.; Connell, S.; Gocke, A.; Hynan, L.; German, D.; Kodadek, T. Identification of candidate IgG biomarkers for Alzheimer’s disease via combinatorial library screening. Cell 2011, 144, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Rahumatullah, A.; Abdul Karim, I.Z.; Noordin, R.; Lim, T.S. Antibody-Based Protective Immunity against Helminth Infections: Antibody Phage Display Derived Antibodies against BmR1 Antigen. Int. J. Mol. Sci. 2017, 18, 2376. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.; Griffiths, A.D.; Hawkins, R.E.; Hoogenboom, H.R. Making Antibodies by Phage Display Technology. Ann. Rev. Immunol. 1994, 12, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Söderlind, E.; Strandberg, L.; Jirholt, P.; Kobayashi, N.; Alexeiva, V.; Åberg, A.-M.; Nilsson, A.; Jansson, B.; Ohlin, M.; Wingren, C.; et al. Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat. Biotechnol. 2000, 18, 852. [Google Scholar] [CrossRef] [PubMed]
- Hoet, R.M.; Cohen, E.H.; Kent, R.B.; Rookey, K.; Schoonbroodt, S.; Hogan, S.; Rem, L.; Frans, N.; Daukandt, M.; Pieters, H.; et al. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nat. Biotechnol. 2005, 23, 344. [Google Scholar] [CrossRef] [PubMed]
- Hoogenboom, H.R.; Winter, G. By-passing immunisation: Human antibodies from synthetic repertoires of germline VH gene segments rearranged in vitro. J. Mol. Biol. 1992, 227, 381–388. [Google Scholar] [CrossRef]
- Silacci, M.; Brack, S.; Schirru, G.; Mårlind, J.; Ettorre, A.; Merlo, A.; Viti, F.; Neri, D. Design, construction, and characterization of a large synthetic human antibody phage display library. Proteomics 2005, 5, 2340–2350. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Y.; Kang, K.J.; Chung, J.E.; Shim, H. Construction of a large synthetic human scFv library with six diversified CDRs and high functional diversity. Mol. Cells 2009, 27, 225–235. [Google Scholar] [CrossRef]
- Chen, W.; Zhu, Z.; Feng, Y.; Xiao, X.; Dimitrov, D.S. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J. Mol. Biol. 2008, 382, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Viti, F.; Nilsson, F.; Demartis, S.; Huber, A.; Neri, D. Design and use of phage display libraries for the selection of antibodies and enzymes. Methods Enzymol. 2000, 326, 480–505. [Google Scholar]
- Villa, A.; Lovato, V.; Bujak, E.; Wulhfard, S.; Pasche, N.; Neri, D. A novel synthetic naïve human antibody library allows the isolation of antibodies against a new epitope of oncofetal fibronectin. mAbs 2011, 3, 264–272. [Google Scholar] [CrossRef]
- Ge, X.; Mazor, Y.; Hunicke-Smith, S.P.; Ellington, A.D.; Georgiou, G. Rapid construction and characterization of synthetic antibody libraries without DNA amplification. Biotechnol. Bioeng. 2010, 106, 347–357. [Google Scholar] [CrossRef]
- Pini, A.; Viti, F.; Santucci, A.; Carnemolla, B.; Zardi, L.; Neri, P.; Neri, D. Design and Use of a Phage Display Library: Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J. Biol. Chem. 1998, 273, 21769–21776. [Google Scholar] [CrossRef]
- Rouet, R.; Lowe, D.; Christ, D. Stability engineering of the human antibody repertoire. FEBS Lett. 2014, 588, 269–277. [Google Scholar] [CrossRef]
- Tiller, T.; Schuster, I.; Deppe, D.; Siegers, K.; Strohner, R.; Herrmann, T.; Berenguer, M.; Poujol, D.; Stehle, J.; Stark, Y.; et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. mAbs 2013, 5, 445–470. [Google Scholar] [CrossRef]
- Nissim, A.; Hoogenboom, H.R.; Tomlinson, I.M.; Flynn, G.; Midgley, C.; Lane, D.; Winter, G. Antibody fragments from a ‘single pot’ phage display library as immunochemical reagents. EMBO J. 1994, 13, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.; Milstein, C. Man-made antibodies. Nature 1991, 349, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Clementi, N.; Mancini, N.; Solforosi, L.; Castelli, M.; Clementi, M.; Burioni, R. Phage display-based strategies for cloning and optimization of monoclonal antibodies directed against human pathogens. Int. J. Mol. Sci. 2012, 13, 8273–8292. [Google Scholar] [CrossRef]
- Sidhu, S.S.; Fellouse, F.A. Synthetic therapeutic antibodies. Nat. Chem. Biol. 2006, 2, 682. [Google Scholar] [CrossRef] [PubMed]
- Knappik, A.; Ge, L.; Honegger, A.; Pack, P.; Fischer, M.; Wellnhofer, G.; Hoess, A.; Wölle, J.; Plückthun, A.; Virnekäs, B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 2000, 296, 57–86. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, S.S.; Li, B.; Chen, Y.; Fellouse, F.A.; Eigenbrot, C.; Fuh, G. Phage-displayed Antibody Libraries of Synthetic Heavy Chain Complementarity Determining Regions. J. Mol. Biol. 2004, 338, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Nussinov, R.; Wu, W.-J.; Ma, B. In Silico Methods in Antibody Design. Antibodies 2018, 7, 22. [Google Scholar] [CrossRef]
- Lloyd, C.; Lowe, D.; Edwards, B.; Welsh, F.; Dilks, T.; Hardman, C.; Vaughan, T. Modelling the human immune response: Performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng. Des. Sel. 2008, 22, 159–168. [Google Scholar] [CrossRef]
- de Haard, H.J.; van Neer, N.; Reurs, A.; Hufton, S.E.; Roovers, R.C.; Henderikx, P.; de Bruїne, A.P.; Arends, J.-W.; Hoogenboom, H.R. A Large Non-immunized Human Fab Fragment Phage Library That Permits Rapid Isolation and Kinetic Analysis of High Affinity Antibodies. J. Biol. Chem. 1999, 274, 18218–18230. [Google Scholar] [CrossRef]
- Omar, N.; Hamidon, N.H.; Yunus, M.H.; Noordin, R.; Choong, Y.S.; Lim, T.S. Generation and selection of naïve Fab library for parasitic antigen: Anti-BmSXP antibodies for lymphatic filariasis. Biotechnol. Appl. Biochem. 2018, 65, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.N.; Chin, C.F.; Choong, Y.S.; Ismail, A.; Lim, T.S. Generation of a naïve human single chain variable fragment (scFv) library for the identification of monoclonal scFv against Salmonella Typhi Hemolysin E antigen. Toxicon 2016, 117, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zettlitz, K.A.; Lipianskaya, J.; Zhou, Y.; Marks, J.D.; Mallick, P.; Reiter, R.E.; Wu, A.M. A fully human scFv phage display library for rapid antibody fragment reformatting. Protein Eng. Des. Sel. 2015, 28, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, I.; Park, S.G.; Cho, S.; Kim, J.H.; Ipper, N.S.; Choi, S.S.; Lee, E.S.; Hong, H.J. Generation, Diversity Determination, and Application to Antibody Selection of a Human Naïve Fab Library. Mol. Cells 2017, 40, 655–666. [Google Scholar] [CrossRef]
- Huse, W.; Sastry, L.; Iverson, S.; Kang, A.; Alting-Mees, M.; Burton, D.; Benkovic, S.; Lerner, R. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science 1989, 246, 1275–1281. [Google Scholar] [CrossRef]
- Kramer, R.A.; Marissen, W.E.; Goudsmit, J.; Visser, T.J.; Clijsters-Van der Horst, M.; Bakker, A.Q.; de Jong, M.; Jongeneelen, M.; Thijsse, S.; Backus, H.H.J.; et al. The human antibody repertoire specific for rabies virus glycoprotein as selected from immune libraries. Eur. J. Immunol. 2005, 35, 2131–2145. [Google Scholar] [CrossRef] [PubMed]
- Burton, D.R.; Barbas, C.F., 3rd; Persson, M.A.; Koenig, S.; Chanock, R.M.; Lerner, R.A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 1991, 88, 10134–10137. [Google Scholar] [CrossRef]
- Hamidon, N.H.; Suraiya, S.; Sarmiento, M.E.; Acosta, A.; Norazmi, M.N.; Lim, T.S. Immune TB Antibody Phage Display Library as a Tool To Study B Cell Immunity in TB Infections. Appl. Biochem. Biotechnol. 2018, 184, 852–868. [Google Scholar] [CrossRef] [PubMed]
- Rahumatullah, A.; Ahmad, A.; Noordin, R.; Lim, T.S. Delineation of BmSXP antibody V-gene usage from a lymphatic filariasis based immune scFv antibody library. Mol. Immunol. 2015, 67, 512–523. [Google Scholar] [CrossRef]
- de Kruif, J.; Boel, E.; Logtenberg, T. Selection and Application of Human Single Chain Fv Antibody Fragments from a Semi-synthetic Phage Antibody Display Library with Designed CDR3 Regions. J. Mol. Biol. 1995, 248, 97–105. [Google Scholar] [CrossRef]
- Yan, J.P.; Ko, J.H.; Qi, Y.P. Generation and characterization of a novel single-chain antibody fragment specific against human fibrin clots from phage display antibody library. Thromb. Res. 2004, 114, 205–211. [Google Scholar] [CrossRef]
- Hairul Bahara, N.H.; Chin, S.T.; Choong, Y.S.; Lim, T.S. Construction of a Semisynthetic Human VH Single-Domain Antibody Library and Selection of Domain Antibodies against α-Crystalline of Mycobacterium tuberculosis. J. Biomol. Screen. 2016, 21, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.V.; Liang, W.-C.; Dennis, M.S.; Eigenbrot, C.; Sidhu, S.S.; Fuh, G. High-affinity Human Antibodies from Phage-displayed Synthetic Fab Libraries with a Single Framework Scaffold. J. Mol. Biol. 2004, 340, 1073–1093. [Google Scholar] [CrossRef]
- Rauchenberger, R.; Borges, E.; Thomassen-Wolf, E.; Rom, E.; Adar, R.; Yaniv, Y.; Malka, M.; Chumakov, I.; Kotzer, S.; Resnitzky, D.; et al. Human Combinatorial Fab Library Yielding Specific and Functional Antibodies against the Human Fibroblast Growth Factor Receptor 3. J. Biol. Chem. 2003, 278, 38194–38205. [Google Scholar] [CrossRef]
- Prassler, J.; Thiel, S.; Pracht, C.; Polzer, A.; Peters, S.; Bauer, M.; Nörenberg, S.; Stark, Y.; Kölln, J.; Popp, A.; et al. HuCAL PLATINUM, a Synthetic Fab Library Optimized for Sequence Diversity and Superior Performance in Mammalian Expression Systems. J. Mol. Biol. 2011, 413, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Bujak, E.; Putelli, A.; Villa, A.; Matasci, M.; Gualandi, L.; Hemmerle, T.; Wulhfard, S.; Neri, D. A Highly Functional Synthetic Phage Display Library Containing over 40 Billion Human Antibody Clones. PLoS ONE 2014, 9, e100000. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, A.R.M.; Sidhu, S.; Dübel, S.; McCafferty, J. Beyond natural antibodies: The power of in vitro display technologies. Nat. Biotechnol. 2011, 29, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Awwad, S.; Angkawinitwong, U. Overview of Antibody Drug Delivery. Pharmaceutics 2018, 10, 83. [Google Scholar] [CrossRef]
- Gaj, T.; Sirk, S.J.; Shui, S.-l.; Liu, J. Genome-Editing Technologies: Principles and Applications. Cold Spring Harbor Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef]
- Chen, C.; Roberts, V.A.; Rittenberg, M.B. Generation and analysis of random point mutations in an antibody CDR2 sequence: Many mutated antibodies lose their ability to bind antigen. J. Exp. Med. 1992, 176, 855–866. [Google Scholar] [CrossRef]
- Krykbaev, R.A.; Liu, W.R.; Jeffrey, P.D.; Margolies, M.N. Phage Display-selected Sequences of the Heavy-chain CDR3 Loop of the Anti-digoxin Antibody 26-10 Define a High Affinity Binding Site for Position 16-substituted Analogs of Digoxin. J. Biol. Chem. 2001, 276, 8149–8158. [Google Scholar] [CrossRef] [PubMed]
- Douthwaite, J.A.; Sridharan, S.; Huntington, C.; Hammersley, J.; Marwood, R.; Hakulinen, J.K.; Ek, M.; Sjögren, T.; Rider, D.; Privezentzev, C.; et al. Affinity maturation of a novel antagonistic human monoclonal antibody with a long VH CDR3 targeting the Class A GPCR formyl-peptide receptor 1. mAbs 2015, 7, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Stephens, D.E.; Singh, S.; Permaul, K. Error-prone PCR of a fungal xylanase for improvement of its alkaline and thermal stability. FEMS Microbiol. Lett. 2009, 293, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C.; Fischhaber, P.L.; Kisker, C. Error-Prone DNA Polymerases: Novel Structures and the Benefits of Infidelity. Cell 2001, 107, 9–12. [Google Scholar] [CrossRef]
- Biles, B.D.; Connolly, B.A. Low-fidelity Pyrococcus furiosus DNA polymerase mutants useful in error-prone PCR. Nucleic Acids Res. 2004, 32, e176. [Google Scholar] [CrossRef] [PubMed]
- Gram, H.; Marconi, L.A.; Barbas, C.F., 3rd; Collet, T.A.; Lerner, R.A.; Kang, A.S. In vitro selection and affinity maturation of antibodies from a naive combinatorial immunoglobulin library. Proc. Natl. Acad. Sci. USA 1992, 89, 3576–3580. [Google Scholar] [CrossRef] [PubMed]
- Martineau, P. Error-prone polymerase chain reaction for modification of scFvs. Methods Mol. Biol. 2002, 178, 287–294. [Google Scholar] [PubMed]
- Ye, J.; Wen, F.; Xu, Y.; Zhao, N.; Long, L.; Sun, H.; Yang, J.; Cooley, J.; Todd Pharr, G.; Webby, R.; et al. Error-prone pcr-based mutagenesis strategy for rapidly generating high-yield influenza vaccine candidates. Virology 2015, 482, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Cárcamo, E.; Roldán-Salgado, A.; Osuna, J.; Bello-Sanmartin, I.; Yáñez, J.A.; Saab-Rincón, G.; Viadiu, H.; Gaytán, P. Spiked Genes: A Method to Introduce Random Point Nucleotide Mutations Evenly throughout an Entire Gene Using a Complete Set of Spiked Oligonucleotides for the Assembly. ACS Omega 2017, 2, 3183–3191. [Google Scholar] [CrossRef] [PubMed]
- Cazaux, C.; Hoffmann, J.S.; Louat, T.; Servant, L.; Bouayadi, K.; Kharrat, H. Use of mutagenic DNA polymerase for producing random mutations. U.S. Patents US20040110294A1, 2 March 2010. [Google Scholar]
- Mondon, P.; Souyris, N.; Douchy, L.; Crozet, F.; Bouayadi, K.; Kharrat, H. Method for generation of human hyperdiversified antibody fragment library. Biotechnol. J. 2007, 2, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Huovinen, T.; Brockmann, E.-C.; Akter, S.; Perez-Gamarra, S.; Ylä-Pelto, J.; Liu, Y.; Lamminmäki, U. Primer Extension Mutagenesis Powered by Selective Rolling Circle Amplification. PLoS ONE 2012, 7, e31817. [Google Scholar] [CrossRef]
- Fujii, R.; Kitaoka, M.; Hayashi, K. Error-prone rolling circle amplification: The simplest random mutagenesis protocol. Nat. Protoc. 2006, 1, 2493. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, R.E.; Russell, S.J.; Winter, G. Selection of phage antibodies by binding affinity: Mimicking affinity maturation. J. Mol. Biol. 1992, 226, 889–896. [Google Scholar] [CrossRef]
- Boder, E.T.; Midelfort, K.S.; Wittrup, K.D. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl. Acad. Sci. USA 2000, 97, 10701–10705. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Gorman, K.T.; Vinci, C.R.; Dobrovetsky, E.; Gräslund, S.; Kay, B.K. Streamlining the Pipeline for Generation of Recombinant Affinity Reagents by Integrating the Affinity Maturation Step. Int. J. Mol. Sci. 2015, 16, 23587–23603. [Google Scholar] [CrossRef]
- McCullum, E.O.; Williams, B.A.; Zhang, J.; Chaput, J.C. Random mutagenesis by error-prone PCR. Methods Mol. Biol. 2010, 634, 103–109. [Google Scholar] [PubMed]
- Ducancel, F.; Muller, B.H. Molecular engineering of antibodies for therapeutic and diagnostic purposes. mAbs 2012, 4, 445–457. [Google Scholar] [CrossRef]
- Tee, K.L.; Wong, T.S. Polishing the craft of genetic diversity creation in directed evolution. Biotechnol. Adv. 2013, 31, 1707–1721. [Google Scholar] [CrossRef]
- Drummond, D.A.; Iverson, B.L.; Georgiou, G.; Arnold, F.H. Why High-error-rate Random Mutagenesis Libraries are Enriched in Functional and Improved Proteins. J. Mol. Biol. 2005, 350, 806–816. [Google Scholar] [CrossRef] [PubMed]
- van den Beucken, T.; Pieters, H.; Steukers, M.; van der Vaart, M.; Ladner, R.C.; Hoogenboom, H.R.; Hufton, S.E. Affinity maturation of Fab antibody fragments by fluorescent-activated cell sorting of yeast-displayed libraries. FEBS Lett. 2003, 546, 288–294. [Google Scholar] [CrossRef]
- Lewis, L.; Lloyd, C. Optimisation of antibody affinity by ribosome display using error-prone or site-directed mutagenesis. Methods Mol. Biol. 2012, 805, 139–161. [Google Scholar]
- Fukuda, I.; Kojoh, K.; Tabata, N.; Doi, N.; Takashima, H.; Miyamoto-Sato, E.; Yanagawa, H. In vitro evolution of single-chain antibodies using mRNA display. Nucleic Acids Res. 2006, 34, e127. [Google Scholar] [CrossRef] [PubMed]
- Marks, J.D. Antibody affinity maturation by chain shuffling. Methods Mol. Biol. 2004, 248, 327–343. [Google Scholar] [PubMed]
- Kang, A.S.; Jones, T.M.; Burton, D.R. Antibody redesign by chain shuffling from random combinatorial immunoglobulin libraries. Proc. Natl. Acad. Sci. USA 1991, 88, 11120–11123. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K. Evolutionary Affinity and Selectivity Optimization of a Pesticide-Selective Antibody Utilizing a Hapten-Selective Immunoglobulin Repertoire. Environ. Sci. Technol. 2002, 36, 4892–4898. [Google Scholar] [CrossRef] [PubMed]
- Marks, J.D.; Griffiths, A.D.; Malmqvist, M.; Clackson, T.P.; Bye, J.M.; Winter, G. By–Passing Immunization: Building High Affinity Human Antibodies by Chain Shuffling. Bio/Technology 1992, 10, 779–783. [Google Scholar] [CrossRef]
- Schier, R.; Bye, J.; Apell, G.; McCall, A.; Adams, G.P.; Malmqvist, M.; Weiner, L.M.; Marks, J.D. Isolation of High-affinity Monomeric Human Anti-c-erbB-2 Single chain Fv Using Affinity-driven Selection. J. Mol. Biol. 1996, 255, 28–43. [Google Scholar] [CrossRef]
- Christensen, P.A.; Danielczyk, A.; Ravn, P.; Larsen, M.; Stahn, R.; Karsten, U.; Goletz, S. Modifying Antibody Specificity by Chain Shuffling of VH / VL between Antibodies with Related Specificities. Scand. J. Immunol. 2009, 69, 1–10. [Google Scholar] [CrossRef]
- Lu, D.; Shen, J.; Vil, M.D.; Zhang, H.; Jimenez, X.; Bohlen, P.; Witte, L.; Zhu, Z. Tailoring in Vitro Selection for a Picomolar Affinity Human Antibody Directed against Vascular Endothelial Growth Factor Receptor 2 for Enhanced Neutralizing Activity. J. Biol. Chem. 2003, 278, 43496–43507. [Google Scholar] [CrossRef]
- Stemmer, W.P. DNA shuffling by random fragmentation and reassembly: In vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 10747–10751. [Google Scholar] [CrossRef] [PubMed]
- Alexandra, J.R. DNA Shuffling: Modifying the Hand That Nature Dealt. In Vitro Cell. Dev. Bioly. Plant 2000, 36, 331–337. [Google Scholar]
- Meyer, A.J.; Ellefson, J.W.; Ellington, A.D. Library generation by gene shuffling. Curr. Protoc. Mol. Biol. 2014, 105, Unit-15.12. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, W.P.C. Rapid evolution of a protein in vitro by DNA shuffling. Nature 1994, 370, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, P.S.; Chen, G.; Iverson, B.L.; Georgiou, G. Quantitative analysis of the effect of the mutation frequency on the affinity maturation of single chain Fv antibodies. Proc. Natl. Acad. Sci. USA 2000, 97, 2029–2034. [Google Scholar] [CrossRef] [PubMed]
- Carter, P. Site-directed mutagenesis. Biochem. J. 1986, 237, 1–7. [Google Scholar] [CrossRef]
- Valetti, F.; Gilardi, G. Improvement of biocatalysts for industrial and environmental purposes by saturation mutagenesis. Biomolecules 2013, 3, 778–811. [Google Scholar] [CrossRef]
- Singh-Gasson, S.; Green, R.D.; Yue, Y.; Nelson, C.; Blattner, F.; Sussman, M.R.; Cerrina, F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999, 17, 974. [Google Scholar] [CrossRef] [PubMed]
- Nikolaos, E.L. Random Mutagenesis Methods for In Vitro Directed Enzyme Evolution. Curr. Protein Pept. Sci. 2010, 11, 91–100. [Google Scholar] [CrossRef]
- Ruff, A.J.; Dennig, A.; Schwaneberg, U. To get what we aim for—Progress in diversity generation methods. FEBS J. 2013, 280, 2961–2978. [Google Scholar] [CrossRef]
- Dubreuil, O.; Bossus, M.; Graille, M.; Bilous, M.; Savatier, A.; Jolivet, M.; Ménez, A.; Stura, E.; Ducancel, F. Fine Tuning of the Specificity of an Anti-progesterone Antibody by First and Second Sphere Residue Engineering. J. Biol. Chem. 2005, 280, 24880–24887. [Google Scholar] [CrossRef] [PubMed]
- Kusharyoto, W.; Pleiss, J.; Bachmann, T.T.; Schmid, R.D. Mapping of a hapten-binding site: Molecular modeling and site-directed mutagenesis study of an anti-atrazine antibody. Protein Eng. Des. Sel. 2002, 15, 233–241. [Google Scholar] [CrossRef]
- Casipit, C.L.; Tal, R.; Wittman, V.; Chavaillaz, P.A.; Arbuthnott, K.; Weidanz, J.A.; Jiao, J.A.; Wong, H.C. Improving the binding affinity of an antibody using molecular modeling and site-directed mutagenesis. Protein Sci. 1998, 7, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Chronopoulou, E.G.; Labrou, N.E. Site-saturation Mutagenesis: A Powerful Tool for Structure-Based Design of Combinatorial Mutation Libraries. Curr. Protoc. Protein Sci. 2011, 63, 26.6.1–26.6.10. [Google Scholar] [CrossRef]
- Steffens, D.L.; Williams, J.G.K. Efficient site-directed saturation mutagenesis using degenerate oligonucleotides. J. Biomol. Tech. 2007, 18, 147–149. [Google Scholar]
- Siloto, R.M.P.; Weselake, R.J. Site saturation mutagenesis: Methods and applications in protein engineering. Biocatal. Agric. Biotechnol. 2012, 1, 181–189. [Google Scholar] [CrossRef]
- Smith, M. Site-directed mutagenesis. Trends Biochem. Sci. 1982, 7, 440–442. [Google Scholar] [CrossRef]
- Zoller, M.J.; Smith, M. Oligonucleotide-Directed Mutagenesis: A Simple Method Using Two Oligonucleotide Primers and a Single-Stranded DNA Template. DNA 1984, 3, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Sauer, N.J.; Mozoruk, J.; Miller, R.B.; Warburg, Z.J.; Walker, K.A.; Beetham, P.R.; Schöpke, C.R.; Gocal, G.F.W. Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol. J. 2016, 14, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A.; Roberts, J.D.; Zakour, R.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987, 154, 367–382. [Google Scholar]
- Tonikian, R.; Zhang, Y.; Boone, C.; Sidhu, S.S. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc. 2007, 2, 1368. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Fang, P.; Kay, B.K. Improvements to the Kunkel mutagenesis protocol for constructing primary and secondary phage-display libraries. Methods 2012, 58, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Fellouse, F.A.; Esaki, K.; Birtalan, S.; Raptis, D.; Cancasci, V.J.; Koide, A.; Jhurani, P.; Vasser, M.; Wiesmann, C.; Kossiakoff, A.A.; et al. High-throughput Generation of Synthetic Antibodies from Highly Functional Minimalist Phage-displayed Libraries. J. Mol. Biol. 2007, 373, 924–940. [Google Scholar] [CrossRef] [PubMed]
- Michael, D.S.; John, W.K.; Brian, K.K. Efficient Construction of a Large Collection of Phage-Displayed Combinatorial Peptide Libraries. Comb. Chem. High Throughput Screen. 2005, 8, 545–551. [Google Scholar] [CrossRef]
- Vandeyar, M.A.; Weiner, M.P.; Hutton, C.J.; Batt, C.A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene 1988, 65, 129–133. [Google Scholar] [CrossRef]
- Sugimoto, M.; Esaki, N.; Tanaka, H.; Soda, K. A simple and efficient method for the oligonucleotide-directed mutagenesis using plasmid DNA template and phosphorothioate-modified nucleotide. Anal. Biochem. 1989, 179, 309–311. [Google Scholar] [CrossRef]
- Hsieh, P.-C.; Vaisvila, R. Protein Engineering: Single or Multiple Site-Directed Mutagenesis. In Enzyme Engineering: Methods and Protocols; Samuelson, J.C., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 173–186. [Google Scholar]
- Carrigan, P.E.; Ballar, P.; Tuzmen, S. Site-Directed Mutagenesis. In Disease Gene Identification: Methods and Protocols; DiStefano, J.K., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 107–124. [Google Scholar]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef]
- Heckman, K.L.; Pease, L.R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007, 2, 924. [Google Scholar] [CrossRef]
- Hussain, H.; Chong, N.F.-M. Combined Overlap Extension PCR Method for Improved Site Directed Mutagenesis. BioMed Res. Int. 2016, 2016, 7. [Google Scholar] [CrossRef]
- Kitzman, J.O.; Starita, L.M.; Lo, R.S.; Fields, S.; Shendure, J. Massively parallel single-amino-acid mutagenesis. Nat. Methods 2015, 12, 203. [Google Scholar] [CrossRef]
- Lim, B.N.; Choong, Y.S.; Ismail, A.; Glökler, J.; Konthur, Z.; Lim, T.S. Directed evolution of nucleotide-based libraries using lambda exonuclease. BioTechniques 2012, 53, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Muniyappa, K.; Radding, C.M. The homologous recombination system of phage lambda. Pairing activities of beta protein. J. Biol. Chem. 1986, 261, 7472–7478. [Google Scholar] [PubMed]
- Poteete, A.R. What makes the bacteriophage λ Red system useful for genetic engineering: Molecular mechanism and biological function. FEMS Microbiol. Lett. 2001, 201, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Sriprakash, K.S.; Lundh, N.; Huh, M.M.-O.; Radding, C.M. The specificity of lambda exonuclease. Interactions with single-stranded DNA. J. Biol. Chem. 1975, 250, 5438–5445. [Google Scholar]
- Lai, J.Y.; Loh, Q.; Choong, Y.S.; Lim, T.S. Cassette hybridization for vector assembly application in antibody chain shuffling. BioTechniques 2018, 65, 269–274. [Google Scholar] [CrossRef]
- Halemano, K.; Guo, K.; Heilman, K.J.; Barrett, B.S.; Smith, D.S.; Hasenkrug, K.J.; Santiago, M.L. Immunoglobulin somatic hypermutation by APOBEC3/Rfv3 during retroviral infection. Proc. Natl. Acad. Sci. USA 2014, 111, 7759–7764. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Chahwan, R.; Wang, S.; Wang, X.; Pham, P.T.; Goodman, M.F.; Bergman, A.; Scharff, M.D.; MacCarthy, T. Overlapping hotspots in CDRs are critical sites for V region diversification. Proc. Natl. Acad. Sci. USA 2015, 112, E728–E737. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Rada, C.; Neuberger, M.S. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J. Exp. Med. 2010, 207, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Yau, K.Y.; Dubuc, G.; Li, S.; Hirama, T.; MacKenzie, C.R.; Jermutus, L.; Hall, J.C.; Tanha, J. Affinity maturation of a VHH by mutational hotspot randomization. J. Immunol. Methods 2005, 297, 213–224. [Google Scholar] [CrossRef]
- Chaudhuri, J.; Tian, M.; Khuong, C.; Chua, K.; Pinaud, E.; Alt, F.W. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 2003, 422, 726. [Google Scholar] [CrossRef]
- Keim, C.; Kazadi, D.; Rothschild, G.; Basu, U. Regulation of AID, the B-cell genome mutator. Genes Dev. 2013, 27, 1–17. [Google Scholar] [CrossRef]
- McConnell, A.D.; Do, M.; Neben, T.Y.; Spasojevic, V.; MacLaren, J.; Chen, A.P.; Altobell, L., III; Macomber, J.L.; Berkebile, A.D.; Horlick, R.A.; et al. High Affinity Humanized Antibodies without Making Hybridomas; Immunization Paired with Mammalian Cell Display and In Vitro Somatic Hypermutation. PLoS ONE 2012, 7, e49458. [Google Scholar] [CrossRef]
- Ho, M.; Pastan, I. In vitro antibody affinity maturation targeting germline hotspots. Methods Mol. Biol. 2009, 525, 293–308. [Google Scholar] [CrossRef]
- Bowers, P.M.; Verdino, P.; Wang, Z.; da Silva Correia, J.; Chhoa, M.; Macondray, G.; Do, M.; Neben, T.Y.; Horlick, R.A.; Stanfield, R.L.; et al. Nucleotide insertions and deletions complement point mutations to massively expand the diversity created by somatic hypermutation of antibodies. J. Biol. Chem. 2014, 289, 33557–33567. [Google Scholar] [CrossRef]
- Li, B.; Zhao, L.; Wang, C.; Guo, H.; Wu, L.; Zhang, X.; Qian, W.; Wang, H.; Guo, Y. The protein-protein interface evolution acts in a similar way to antibody affinity maturation. J. Biol. Chem. 2010, 285, 3865–3871. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.A.; Watanabe, C.K.; Zhong, A.; Goddard, A.; Sidhu, S.S. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl. Acad. Sci. USA 2000, 97, 8950–8954. [Google Scholar] [CrossRef]
- Tiller, K.E.; Chowdhury, R.; Li, T.; Ludwig, S.D.; Sen, S.; Maranas, C.D.; Tessier, P.M. Facile Affinity Maturation of Antibody Variable Domains Using Natural Diversity Mutagenesis. Front. Immunol. 2017, 8, 986. [Google Scholar] [CrossRef]
- Morrison, K.L.; Weiss, G.A. Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 2001, 5, 302–307. [Google Scholar] [CrossRef]
- Chatellier, J.; Mazza, A.; Brousseau, R.; Vernet, T. Codon-Based Combinatorial Alanine Scanning Site-Directed Mutagenesis: Design, Implementation, and Polymerase Chain Reaction Screening. Anal. Biochem. 1995, 229, 282–290. [Google Scholar] [CrossRef]
- Vajdos, F.; Adams, C.W.; Breece, T.; Presta, L.G.; de Vos, A.M.; Sidhu, S.S. Comprehensive Functional Maps of the Antigen-binding Site of an Anti-ErbB2 Antibody Obtained with Shotgun Scanning Mutagenesis. J. Mol. Biol. 2002, 320, 415–428. [Google Scholar] [CrossRef]
- Robin, G.; Sato, Y.; Desplancq, D.; Rochel, N.; Weiss, E.; Martineau, P. Restricted Diversity of Antigen Binding Residues of Antibodies Revealed by Computational Alanine Scanning of 227 Antibody–Antigen Complexes. J. Mol. Biol. 2014, 426, 3729–3743. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre, F.; Rémy, M.-H.; Masson, J.-M. Alanine-stretch scanning mutagenesis: A simple and efficient method to probe protein structure and function. Nucleic Acids Res. 1997, 25, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T.; Carballeira, J.D. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat. Protoc. 2007, 2, 891. [Google Scholar] [CrossRef] [PubMed]
- Jeliazkov, J.R.; Sljoka, A.; Kuroda, D.; Tsuchimura, N.; Katoh, N.; Tsumoto, K.; Gray, J.J. Repertoire Analysis of Antibody CDR-H3 Loops Suggests Affinity Maturation Does Not Typically Result in Rigidification. Front. Immunol. 2018, 9, 413. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.A.; Vasser, M.; Powers, D.B. Cassette mutagenesis: An efficient method for generation of multiple mutations at defined sites. Gene 1985, 34, 315–323. [Google Scholar] [CrossRef]
- Kegler-Ebo, D.M.; Docktor, C.M.; DiMaio, D. Codon cassette mutagenesis: A general method to insert or replace individual codons by using universal mutagenic cassettes. Nucleic Acids Res. 1994, 22, 1593–1599. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hidalgo, A.; Schließmann, A.; Molina, R.; Hermoso, J.; Bornscheuer, U.T. A one-pot, simple methodology for cassette randomisation and recombination for focused directed evolution. Protein Eng. Des. Sel. 2008, 21, 567–576. [Google Scholar] [CrossRef]
- Lai, R.; Bekessy, A.; Chen, C.C.; Walsh, T.; Barnard, R. Megaprimer Mutagenesis Using Very Long Primers. BioTechniques 2003, 34, 52–56. [Google Scholar] [CrossRef]
- Hermes, J.D.; Parekh, S.M.; Blacklow, S.C.; Koster, H.; Knowles, J.R. A reliable method for random mutagenesis: The generation of mutant libraries using spiked oligodeoxyribonucleotide primers. Gene 1989, 84, 143–151. [Google Scholar] [CrossRef]
- Firnberg, E.; Ostermeier, M. PFunkel: Efficient, Expansive, User-Defined Mutagenesis. PLoS ONE 2012, 7, e52031. [Google Scholar] [CrossRef]
- Kowalsky, C.A.; Faber, M.S.; Nath, A.; Dann, H.E.; Kelly, V.W.; Liu, L.; Shanker, P.; Wagner, E.K.; Maynard, J.A.; Chan, C.; et al. Rapid Fine Conformational Epitope Mapping Using Comprehensive Mutagenesis and Deep Sequencing. J. Biol. Chem. 2015, 290, 26457–26470. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Ostermaier, M.K.; Heydenreich, F.M.; Mayer, D.; Jaussi, R.; Standfuss, J.; Veprintsev, D.B. AAscan, PCRdesign and MutantChecker: A suite of programs for primer design and sequence analysis for high-throughput scanning mutagenesis. PLoS ONE 2013, 8, e78878. [Google Scholar] [CrossRef]
- Acevedo-Rocha, C.G.; Reetz, M.T.; Nov, Y. Economical analysis of saturation mutagenesis experiments. Sci. Rep. 2015, 5, 10654. [Google Scholar] [CrossRef] [PubMed]
- Nov, Y. When second best is good enough: Another probabilistic look at saturation mutagenesis. Appl. Environ. Microbiol. 2012, 78, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Gao, H.; Zhu, X.; Wang, X.; Zhou, M.; Jiang, R. Construction of “small-intelligent” focused mutagenesis libraries using well-designed combinatorial degenerate primers. BioTechniques 2012, 52, 149–158. [Google Scholar] [CrossRef]
- Kille, S.; Acevedo-Rocha, C.G.; Parra, L.P.; Zhang, Z.-G.; Opperman, D.J.; Reetz, M.T.; Acevedo, J.P. Reducing Codon Redundancy and Screening Effort of Combinatorial Protein Libraries Created by Saturation Mutagenesis. ACS Synth. Biol. 2013, 2, 83–92. [Google Scholar] [CrossRef]
- Hughes, M.D.; Nagel, D.A.; Santos, A.F.; Sutherland, A.J.; Hine, A.V. Removing the Redundancy From Randomised Gene Libraries. J. Mol. Biol. 2003, 331, 973–979. [Google Scholar] [CrossRef]
- Ashraf, M.; Frigotto, L.; Smith, M.E.; Patel, S.; Hughes, M.D.; Poole, A.J.; Hebaishi, H.R.M.; Ullman, C.G.; Hine, A.V. ProxiMAX randomization: A new technology for non-degenerate saturation mutagenesis of contiguous codons. Biochem. Soc. Trans. 2013, 41, 1189–1194. [Google Scholar] [CrossRef]
- Frigotto, L.; Smith, M.E.; Brankin, C.; Sedani, A.; Cooper, S.E.; Kanwar, N.; Evans, D.; Svobodova, S.; Baar, C.; Glanville, J.; et al. Codon-Precise, Synthetic, Antibody Fragment Libraries Built Using Automated Hexamer Codon Additions and Validated through Next Generation Sequencing. Antibodies 2015, 4, 88–102. [Google Scholar] [CrossRef]
- Balint, R.F.; Larrick, J.W. Antibody engineering by parsimonious mutagenesis. Gene 1993, 137, 109–118. [Google Scholar] [CrossRef]
- Mena, M.A.; Daugherty, P.S. Automated design of degenerate codon libraries. Protein Eng. Des. Sel. 2005, 18, 559–561. [Google Scholar] [CrossRef]
- Patrick, W.M.; Firth, A.E. Strategies and computational tools for improving randomized protein libraries. Biomol. Eng. 2005, 22, 105–112. [Google Scholar] [CrossRef]
- Gupta, K.; Varadarajan, R. Insights into protein structure, stability and function from saturation mutagenesis. Curr. Opin. Struct. Biol. 2018, 50, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Nair, S.; Sen, N.; Soni, N.; Madhusudhan, M.S. In silico methods for design of biological therapeutics. Methods 2017, 131, 33–65. [Google Scholar] [CrossRef]
- Steinberg, D.; Glaser, F.; Nimrod, G.; Ben-Tal, N.; Pupko, T. In silico identification of functional regions in proteins. Bioinformatics 2005, 21, i328–i337. [Google Scholar] [CrossRef]
- Nimrod, G.; Schushan, M.; Steinberg, D.M.; Ben-Tal, N. Detection of Functionally Important Regions in “Hypothetical Proteins” of Known Structure. Structure 2008, 16, 1755–1763. [Google Scholar] [CrossRef]
- Shazman, S.; Celniker, G.; Haber, O.; Glaser, F.; Mandel-Gutfreund, Y. Patch Finder Plus (PFplus): A web server for extracting and displaying positive electrostatic patches on protein surfaces. Nucleic Acids Res. 2007, 35, W526–W530. [Google Scholar] [CrossRef] [PubMed]
- Graur, D.; Mayrose, I.; Ben-Tal, N.; Pupko, T. Comparison of Site-Specific Rate-Inference Methods for Protein Sequences: Empirical Bayesian Methods Are Superior. Mol. Biol. Evol. 2004, 21, 1781–1791. [Google Scholar] [CrossRef]
- Burks, E.A.; Chen, G.; Georgiou, G.; Iverson, B.L. In vitro scanning saturation mutagenesis of an antibody binding pocket. Proc. Natl. Acad. Sci. USA 1997, 94, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Dubrawsky, I.; Mendez, P.; Georgiou, G.; Iverson, B.L. In vitro scanning saturation mutagenesis of all the specificity determining residues in an antibody binding site. Protein Eng. Des. Sel. 1999, 12, 349–356. [Google Scholar] [CrossRef]
- Firth, A.E.; Patrick, W.M. GLUE-IT and PEDEL-AA: New programmes for analyzing protein diversity in randomized libraries. Nucleic Acids Res. 2008, 36, W281–W285. [Google Scholar] [CrossRef]
- Tang, L.; Wang, X.; Ru, B.; Sun, H.; Huang, J.; Gao, H. MDC-Analyzer: A novel degenerate primer design tool for the construction of intelligent mutagenesis libraries with contiguous sites. BioTechniques 2014, 56, 301–310. [Google Scholar] [CrossRef]
- Kosuri, S.; Church, G.M. Large-scale de novo DNA synthesis: Technologies and applications. Nat. Methods 2014, 11, 499. [Google Scholar] [CrossRef]
- Hughes, R.A.; Ellington, A.D. Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology. Cold Spring Harbor Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef]
- Beaucage, S.L.; Caruthers, M.H. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. [Google Scholar] [CrossRef]
- Lindner, T.; Kolmar, H.; Haberkorn, U.; Mier, W. DNA libraries for the construction of phage libraries: Statistical and structural requirements and synthetic methods. Molecules 2011, 16, 1625–1641. [Google Scholar] [CrossRef]
- Fodor, S.; Read, J.; Pirrung, M.; Stryer, L.; Lu, A.; Solas, D. Light-directed, spatially addressable parallel chemical synthesis. Science 1991, 251, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, A.P.; Kaiser, R.J.; Hood, L.E. High-density oligonucleotide arrays. Biosens. Bioelectron. 1996, 11, 687–690. [Google Scholar] [CrossRef]
- Gao, X.; LeProust, E.; Zhang, H.; Srivannavit, O.; Gulari, E.; Yu, P.; Nishiguchi, C.; Xiang, Q.; Zhou, X. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 2001, 29, 4744–4750. [Google Scholar] [CrossRef] [PubMed]
- Forsström, B.; Axnäs, B.B.; Stengele, K.-P.; Bühler, J.; Albert, T.J.; Richmond, T.A.; Hu, F.J.; Nilsson, P.; Hudson, E.P.; Rockberg, J.; et al. Proteome-wide epitope mapping of antibodies using ultra-dense peptide arrays. Mol. Cell. Proteomics 2014, 13, 1585–1597. [Google Scholar] [CrossRef]
- Ghindilis, A.L.; Smith, M.W.; Schwarzkopf, K.R.; Roth, K.M.; Peyvan, K.; Munro, S.B.; Lodes, M.J.; Stöver, A.G.; Bernards, K.; Dill, K.; et al. CombiMatrix oligonucleotide arrays: Genotyping and gene expression assays employing electrochemical detection. Biosens. Bioelectron. 2007, 22, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.; Yazvenko, N.; Peyvan, K.; Maurer, K.; Taitt, C.R.; Lyon, W.; Danley, D.L. Targeted deposition of antibodies on a multiplex CMOS microarray and optimization of a sensitive immunoassay using electrochemical detection. PLoS ONE 2010, 5, e9781. [Google Scholar] [CrossRef]
- Dill, K.; Montgomery, D.D.; Wang, W.; Tsai, J.C. Antigen detection using microelectrode array microchips. Anal. Chim. Acta 2001, 444, 69–78. [Google Scholar] [CrossRef]
- Yazdi, S.M.H.T.; Kiah, H.M.; Garcia-Ruiz, E.; Ma, J.; Zhao, H.; Milenkovic, O. DNA-Based Storage: Trends and Methods. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2015, 1, 230–248. [Google Scholar] [CrossRef]
- Tian, J.; Gong, H.; Sheng, N.; Zhou, X.; Gulari, E.; Gao, X.; Church, G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 2004, 432, 1050. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, S.; Ding, B.; Fei, C.; Wan, W.; Hu, D.; Du, R.; Zhou, X.; Hong, J.; Liu, H.; et al. Design and construction of small perturbation mutagenesis libraries for antibody affinity maturation using massive microchip-synthesized oligonucleotides. J. Biotechnol. 2015, 194, 27–36. [Google Scholar] [CrossRef]
- Kosuri, S.; Eroshenko, N.; LeProust, E.M.; Super, M.; Way, J.; Li, J.B.; Church, G.M. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. 2010, 28, 1295. [Google Scholar] [CrossRef]
- Hu, D.; Hu, S.; Wan, W.; Xu, M.; Du, R.; Zhao, W.; Gao, X.; Liu, J.; Liu, H.; Hong, J. Effective Optimization of Antibody Affinity by Phage Display Integrated with High-Throughput DNA Synthesis and Sequencing Technologies. PLoS ONE 2015, 10, e0129125. [Google Scholar] [CrossRef]
- Gunderson, K.L.; Kruglyak, S.; Graige, M.S.; Garcia, F.; Kermani, B.G.; Zhao, C.; Che, D.; Dickinson, T.; Wickham, E.; Bierle, J.; et al. Decoding randomly ordered DNA arrays. Genome Res. 2004, 14, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Nesterov-Mueller, A.; Maerkle, F.; Hahn, L.; Fortsch, T.; Schillo, S.; Bykovskaya, V.; Sedlmayr, M.; Weber, L.; Ridder, B.; Soehindrijo, M.; et al. Particle-Based Microarrays of Oligonucleotides and Oligopeptides. Microarrays 2014, 3, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Brulle, J.V.d.; Fischer, M.; Langmann, T.; Horn, G.; Waldmann, T.; Arnold, S.; Fuhrmann, M.; Schatz, O.; O’Connell, T.; O’Connell, D.; et al. A novel solid phase technology for high-throughput gene synthesis. BioTechniques 2008, 45, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, H.; Härd, T.; Löfblom, J.; Ståhl, S. A truncated and dimeric format of an Affibody library on bacteria enables FACS-mediated isolation of amyloid-beta aggregation inhibitors with subnanomolar affinity. Biotechnol. J. 2015, 10, 1707–1718. [Google Scholar] [CrossRef]
- Cortina-Ceballos, B.; Godoy-Lozano, E.E.; Téllez-Sosa, J.; Ovilla-Muñoz, M.; Sámano-Sánchez, H.; Aguilar-Salgado, A.; Gómez-Barreto, R.E.; Valdovinos-Torres, H.; López-Martínez, I.; Aparicio-Antonio, R.; et al. Longitudinal analysis of the peripheral B cell repertoire reveals unique effects of immunization with a new influenza virus strain. Genome Med. 2015, 7, 124. [Google Scholar] [CrossRef]
- Adler, A.S.; Bedinger, D.; Adams, M.S.; Asensio, M.A.; Edgar, R.C.; Leong, R.; Leong, J.; Mizrahi, R.A.; Spindler, M.J.; Bandi, S.R.; et al. A natively paired antibody library yields drug leads with higher sensitivity and specificity than a randomly paired antibody library. mAbs 2018, 10, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Larman, H.B.; Xu, G.J.; Pavlova, N.N.; Elledge, S.J. Construction of a rationally designed antibody platform for sequencing-assisted selection. Proc. Natl. Acad. Sci. USA 2012, 109, 18523–18528. [Google Scholar] [CrossRef]
- Kim, H.; Han, H.; Ahn, J.; Lee, J.; Cho, N.; Jang, H.; Kim, H.; Kwon, S.; Bang, D. ‘Shotgun DNA synthesis’ for the high-throughput construction of large DNA molecules. Nucleic Acids Res. 2012, 40, e140. [Google Scholar] [CrossRef] [PubMed]
- Matzas, M.; Stähler, P.F.; Kefer, N.; Siebelt, N.; Boisguérin, V.; Leonard, J.T.; Keller, A.; Stähler, C.F.; Häberle, P.; Gharizadeh, B.; et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing. Nat. Biotechnol. 2010, 28, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.B.; Naciri, J.; Liu, J.L.; Anderson, G.P.; Goldman, E.R.; Zabetakis, D. Next-Generation Sequencing of a Single Domain Antibody Repertoire Reveals Quality of Phage Display Selected Candidates. PLoS ONE 2016, 11, e0149393. [Google Scholar] [CrossRef]
- Yang, W.; Yoon, A.; Lee, S.; Kim, S.; Han, J.; Chung, J. Next-generation sequencing enables the discovery of more diverse positive clones from a phage-displayed antibody library. Exp. Mol. Med. 2017, 49, e308. [Google Scholar] [CrossRef] [PubMed]
- Rouet, R.; Jackson, K.J.L.; Langley, D.B.; Christ, D. Next-Generation Sequencing of Antibody Display Repertoires. Front. Immunol. 2018, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Kono, N.; Sun, L.; Toh, H.; Shimizu, T.; Xue, H.; Numata, O.; Ato, M.; Ohnishi, K.; Itamura, S. Deciphering antigen-responding antibody repertoires by using next-generation sequencing and confirming them through antibody-gene synthesis. Biochem. Biophys. Res. Commun. 2017, 487, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Paciello, G.; Acquaviva, A.; Pighi, C.; Ferrarini, A.; Macii, E.; Zamo’, A.; Ficarra, E. VDJSeq-Solver: In Silico V(D)J Recombination Detection Tool. PLoS ONE 2015, 10, e0118192. [Google Scholar] [CrossRef] [PubMed]
- Christley, S.; Scarborough, W.; Salinas, E.; Rounds, W.H.; Toby, I.T.; Fonner, J.M.; Levin, M.K.; Kim, M.; Mock, S.A.; Jordan, C.; et al. VDJServer: A Cloud-Based Analysis Portal and Data Commons for Immune Repertoire Sequences and Rearrangements. Front. Immunol. 2018, 9, 976. [Google Scholar] [CrossRef]
- Pavoni, E.; Flego, M.; Dupuis, M.L.; Barca, S.; Petronzelli, F.; Anastasi, A.M.; D’Alessio, V.; Pelliccia, A.; Vaccaro, P.; Monteriù, G.; et al. Selection, affinity maturation, and characterization of a human scFv antibody against CEA protein. BMC Cancer 2006, 6, 41. [Google Scholar] [CrossRef]
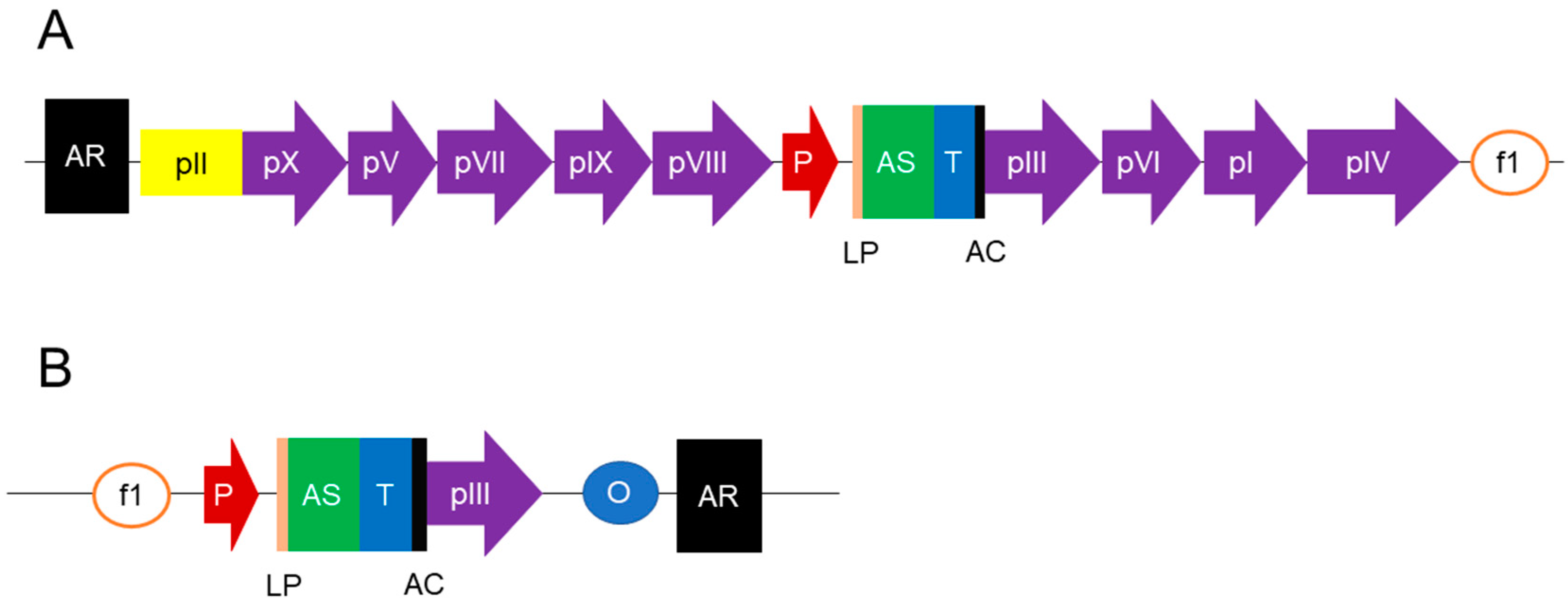
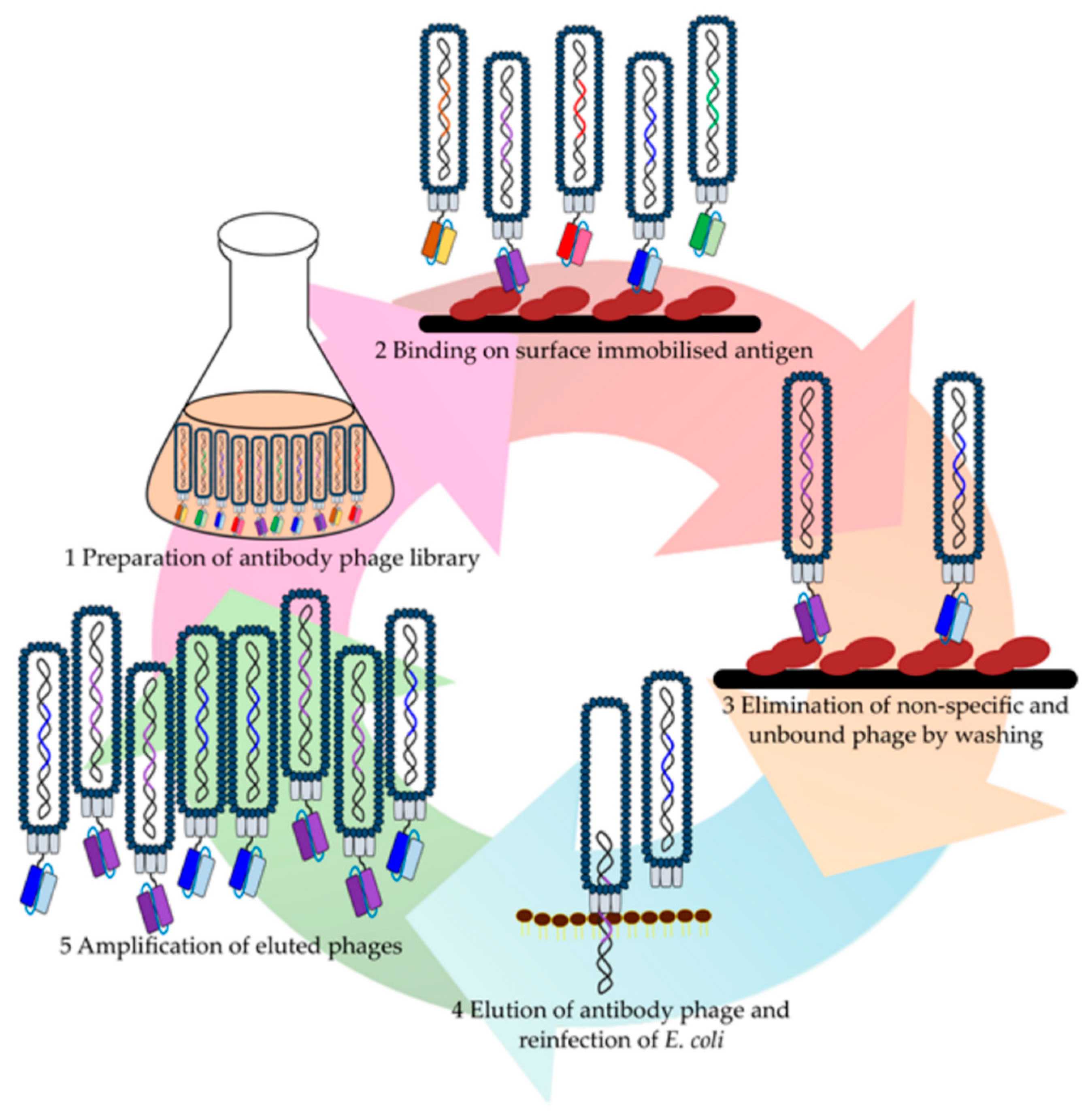
| Library | Library Name/Author | Format | Source/Diversity | Library Size | Targets | Affinities | Reference |
|---|---|---|---|---|---|---|---|
| Naïve | Marks et al. | scFv | human | 1.9 × 108 | Lysozyme, Haptens | nM | [66] |
| CAT 1.0 MedImmune | scFv | human | 1.4 × 1010 | Fluroscein, Hapten, hormones | nM | [65] | |
| CAT 2.0 MedImmune | scFv | human | 1.29 × 1011 | Peptides, Receptors, Chemokines, Cytokines, Growth factors, Protease inhibitor, IgE, gp41 | nM–pM | [102] | |
| de Haard et al. | Fab | human | 3.7 × 1010 | TTX, phOx, MUC1, human glycoprotein hormones | nM | [103] | |
| Omar et al. | Fab | human | 2.99 × 109 | LF recombinant BmSXP antigen | ND | [104] | |
| Lim et al. | scFv | human | 2 × 109 | HlyE | ND | [105] | |
| Li et al. | scFv | human | 9 × 109 | Human N-cadherin | nM | [106] | |
| Kim et al. | Fab | human | 3 × 1010 | Human recombinant proteins, peptides | nM | [107] | |
| Immune | Huse et al. | Fab | murine | 2.5 × 107 | NPN | nM | [108] |
| Kramer et al. | scFv | human | 9.3 × 106, 1.2 × 107 | Rabies virus (RV) glycoproteins (gp) | ND | [109] | |
| Burton et al. | Fab | human | 107 | HIV-I surface glycoprotein gp120 | nM | [110] | |
| Hamidon et al. | scFv | human | 109 | Recombinant MTb α-crystalline | ND | [111] | |
| Rahumatullah et al. | scFv | human | 108 | LF recombinant BmSXP antigen | ND | [112] | |
| Semi-synthetic | Hoogenboom and Winter | scFv | H, 49 VH/1 VL | 2.2 × 107 | Haptens, TNF | ND | [85] |
| de Kruif et al. | scFv | H, 49 VH/7 VL | 3.6 × 108 | DNP, TTX, GBS, SpA, HMG, IgG, Tg, VWF, A2, ICAM-1, δEGP-2, BLT1, PBX1a | μM–nM | [113] | |
| Tomlinson I+J | scFv | H, 1 VH/1 Vκ | 1.47 × 108* | Human fibrin clots | ND | [114] | |
| Nissim et al. | scFv | H, 50 VH/1 VL | 1 × 108 | FITC, NIP, phOX, KLH, maltose BP, TCR, BiP, EF-1α, SRY, anti-erythrocyte rhesus D antibody, p53 | ND | [95] | |
| Pini et al. | scFv | H, 1 VH/1 Vκ | 3 × 108 | Recombinant fibronectin fragments | nM–pM | [92] | |
| Hairul et al. | sdAb | H, 1 VH | 6.6 × 109 | Recombinant MTb α-crystalline | ND | [115] | |
| n-CoDeR® | scFv | H, 1 VH/1 VL | 2 × 109 | Haptens, peptides, carbohydrates, proteins | nM | [83] | |
| Dyax | Fab | H, 1 VH/1 VL | 4.5 × 1010 | TIE-1, DESC1, MSPL, hK1 | nM | [84] | |
| Chen et al. | sdAb | H, 1 VH | 2.5 × 1010 | Vaccinia protein B5R | nM | [88] | |
| Lee et al. | Fab | 1 VH/1 VL | 4 × 1010 | mVEGF | nM | [116] | |
| Griffiths et al. | Fab | 49 VH/26 Vκ/21 Vλ | 6.5 × 1010 | NIP, FTIC | μM–nM | [68] | |
| Synthetic | Ylanthia | Fab | 36 VH/VL pairing | 1.3 × 1011 | rhTNF-α, M-CSF, rhErbB4, rhFZD-4, eGFP | nM–pM | [94] |
| HuCAL® | scFv | 49 VH/VL pairing | 2 × 109 | ICAM-1, Insulin, CD11b, hEGFR, Mac1p, Hagp, NFκBp | nM–pM | [99] | |
| HuCAL® | Fab | 49 VH/VL pairing | 2.1 × 1010 | rFGFR3 | nM | [117] | |
| HuCAL GOLD® | Fab | 49 VH/VL pairing | 1.6 × 1010 | IL18R-Fc, β-Gal, Est-BSA | pM | [69] | |
| HuCAL PLATINUM® | Fab | 49 VH/VL pairing | 4.5 × 1010 | Receptor, interleukin, virus, growth factor, peptide, cytokine, IgG1(1), IgG1(2) | nM–pM | [118] | |
| ETH-2-Gold | scFv | 2 VH/VL pairing | 3 × 109 | BSA, TNC, TTX, haptoglobin, hemoglobin, HCV envelope proteins, fibronectins | nM | [86] | |
| PHILO | scFv | 2 VH/VL pairing | 3.1 × 109 | Fibronectin domains, murine tenascin-C domain, fibrin | nM | [90] | |
| PHILODiamond | scFv | 2 VH/VL pairing | ND | Fibronectin, TNC, fibrinogen, GST, MMP1, MMP3, mycolactone, collagen I, follistatin-like protein I, PSMD6, serpin, TIMP, UBOL1, TOM | nM | [119] |
| Methods | Description | Mutational Rate * | Library Size/Diversity | Affinities | Affinity Selection | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| High | Moderate | Low | ||||||
| Error-prone PCR | Randomization of an scFv (digoxin/digoxigenin) | + | + | + | 105–106 | nM | FACS and SPR against 100 nM digoxin | [158] |
| Randomization of VH/VL of 3 Fab (progesterone) | + | + | ND | µM–nM | ND | [129] | ||
| Randomization of several monobodies (fibronectin type 3) | + | 107–109 | nM | Lowering antigen (MAP2K5 and SF3A1) concentrations sequentially from 300 nM to 10 nM | [139] | |||
| Randomization of CDR1 and CDR2 of an scFv (CEA) | + | + | 107 | nM | ND | [269] | ||
| Randomization of a Fab (streptavidin) | + | + | 106 | nM | Lowering antigen concentrations sequentially from 5% to 0.5% (initial is 6 nM) | [144] | ||
| Generation of a hemagglutinin mutant library | ND | ND | ND | Selected vaccine candidates were evaluated on mice protection study | [131] | |||
| Using spiked genes for random mutations | + | ND | ND | ND | [132] | |||
| Generation of hyperdiversified human antibody fragment mutant libraries using MutaGen™ | + | + | 106–107 | ND | ND | [134] | ||
| Generation of scFv gene mutant library using RCA | + | + | 107 | ND | ND | [135] | ||
| Randomization of an scFv (NP) | + | ND | nM | Gradually decreasing antigen concentration from 8 nM to 1 nM and repeat two rounds with 1 nM | [137] | |||
| Randomization of an scFv (fluorescein) | ND | 105–107 | fM | Competitive panning against fluorescein competitor and FACS | [138] | |||
| Chain recombination | Light chains shuffling (Vĸ and Vλ) of an scFv (phOx-15) | + | + | 106 | nM | Two rounds of panning against 10 µg/mL antigen | [150] | |
| VH chain shuffling of an scFv (phOx-15) | + | 105 | nM | Four rounds of panning against 1 µg/mL antigen | ||||
| VH/VL shuffling of several Fab (NPN) | ND | 106 | ND | ND | [148] | |||
| VH/VL shuffling of several scFv (s-triazine) #coupled with random point mutations | ND | 106–107 | nM | Three repetitive cycles using immunoaffinity chromatography | [149] | |||
| VH/VL shuffling of an scFv (c-erbB-2) | ND | 106 | nM | Gradually lowering antigen concentration from 100 nM to 1 nM vs. 40 nM to 0.01 nM | [151] | |||
| VH/VL shuffling of chimeric antibodies (Lewis Y) | ND | ND | ND | SPR | [152] | |||
| VL shuffling of a Fab (KDR) | ND | 108 | nM | Gradually decreasing phage input and time for binding | [153] | |||
| Site-specific mutagenesis | Saturation mutagenesis of an scFv (progesterone) #coupled with random mutagenesis | ND | 106 | nM | Five rounds of competitive selection against 5 nM antigen and 5 µM competitor | [164] | ||
| Kunkel mutagenesis and asymmetric PCR on FN3 monobodies | + | 108 | 2 to 4-fold higher | Gradually decreasing antigen concentration from 25 nM to 500 pM with increasing washes | [175] | |||
| Defined positions in the CDR to construct four Fab (VEFG) | ND | ND | nM | Competitive phage ELISA with as low as 100 nM antigen | [176] | |||
| Overlap-extension mutagenesis and microarray-based DNA synthesis of p53 and Gal4 | + | + | ND | NC | NC | [185] | ||
| DNA shuffling using ssDNA and lambda exonuclease #coupled with CDR3 mutagenesis using NNK nucleotides | ND | 107–108 | NC | NC | [186] | |||
| Chain recombination via specific DNA hybridization on an scFv | NC | ND | NC | NC | [190] | |||
| Mutational hotspot mutagenesis on CDR2/3 of a peptide (VHH) | ND | 1011 | nM | Three rounds of panning against 100 nM antigen | [194] | |||
| In vitro somatic hypermutation with AID of humanized antibodies | ND | ND | pM | SPR analysis | [197] | |||
| Germline hotspot mutagenesis of an antibody, RFB4 | NC | ND | ND | Subtractive biopanning with increasing washes | [198] | |||
| Single and multiple mutations on CDR2/3 of a nanobody (α-synuclein) | ND | 106 | nM | FACS against decreasing peptide concentration (from 50 nM to 5 nM) | [202] | |||
| Randomization of CDR3 of a VH/ VHH domain with controlled codon ratios using ProxiMAX strategy | ND | ND | ND | NC | [224] | |||
| Selected VH/VL framework pairs were randomly combined for constructing mutant Fab Ylanthia libraries | ND | 1011 | nM–pM | Few human recombinant antigens were used. | [94] | |||
| Single-site saturation mutagenesis was performed using PFunkel mutagenesis for TNF, pertussis toxin, and TROP2 mutant libraries | ND | ND | ND | Selection against 32 nM scFv (TNF), 5 nM anti-pertussis, 22 nM Fab (TROP2) using FACS | [216] | |||
| Affibody (Aβ) mutant libraries construction using SlonoMax® | ND | 107–108 | nM–pM | Gradually decreasing antigen concentration from 50 nM to 10 nM using FACS | [257] | |||
| Gene synthesis | Five scFv (erbB2) gene libraries were constructed on an array | ND | 106 | рM | Gradually decreasing antigen concentration from 0.1 nM to 0.001 nM | [253] | ||
| Sequence-defined oligonucleotide libraries were assembled on a microarray and combined into an scFv mutant library | ND | ND | ND | Four rounds of selection on PVRL4 | [260] | |||
| Random VH and VL paired libraries generation using OE-RT-PCR in microemulsion | ND | ND | nM | 9 mM of Human IgG1-Fc and 9 mM of IL-21 proteins were used in FACS | [259] | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, C.C.; Choong, Y.S.; Lim, T.S. Cognizance of Molecular Methods for the Generation of Mutagenic Phage Display Antibody Libraries for Affinity Maturation. Int. J. Mol. Sci. 2019, 20, 1861. https://doi.org/10.3390/ijms20081861
Lim CC, Choong YS, Lim TS. Cognizance of Molecular Methods for the Generation of Mutagenic Phage Display Antibody Libraries for Affinity Maturation. International Journal of Molecular Sciences. 2019; 20(8):1861. https://doi.org/10.3390/ijms20081861
Chicago/Turabian StyleLim, Chia Chiu, Yee Siew Choong, and Theam Soon Lim. 2019. "Cognizance of Molecular Methods for the Generation of Mutagenic Phage Display Antibody Libraries for Affinity Maturation" International Journal of Molecular Sciences 20, no. 8: 1861. https://doi.org/10.3390/ijms20081861
APA StyleLim, C. C., Choong, Y. S., & Lim, T. S. (2019). Cognizance of Molecular Methods for the Generation of Mutagenic Phage Display Antibody Libraries for Affinity Maturation. International Journal of Molecular Sciences, 20(8), 1861. https://doi.org/10.3390/ijms20081861





