Poly(ADP-Ribose) Polymerases in Plants and Their Human Counterparts: Parallels and Peculiarities
Abstract
1. Introduction
2. Poly(ADP-Ribosyl)ation and Poly(ADP-Ribose) Polymerases (PARPs) in Humans
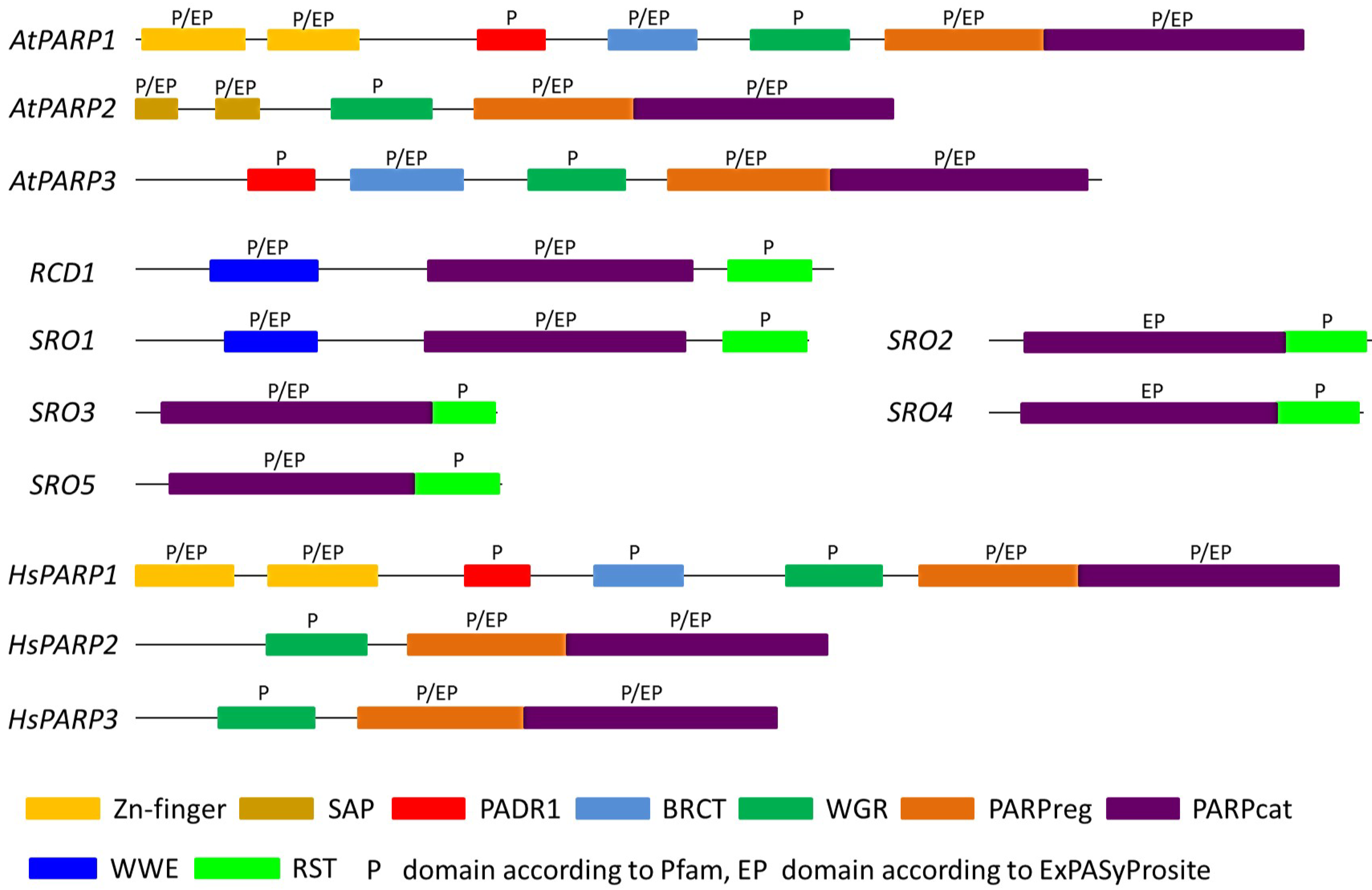
2.1. PARPs Constitute a Heterogeneous Protein Family in Humans
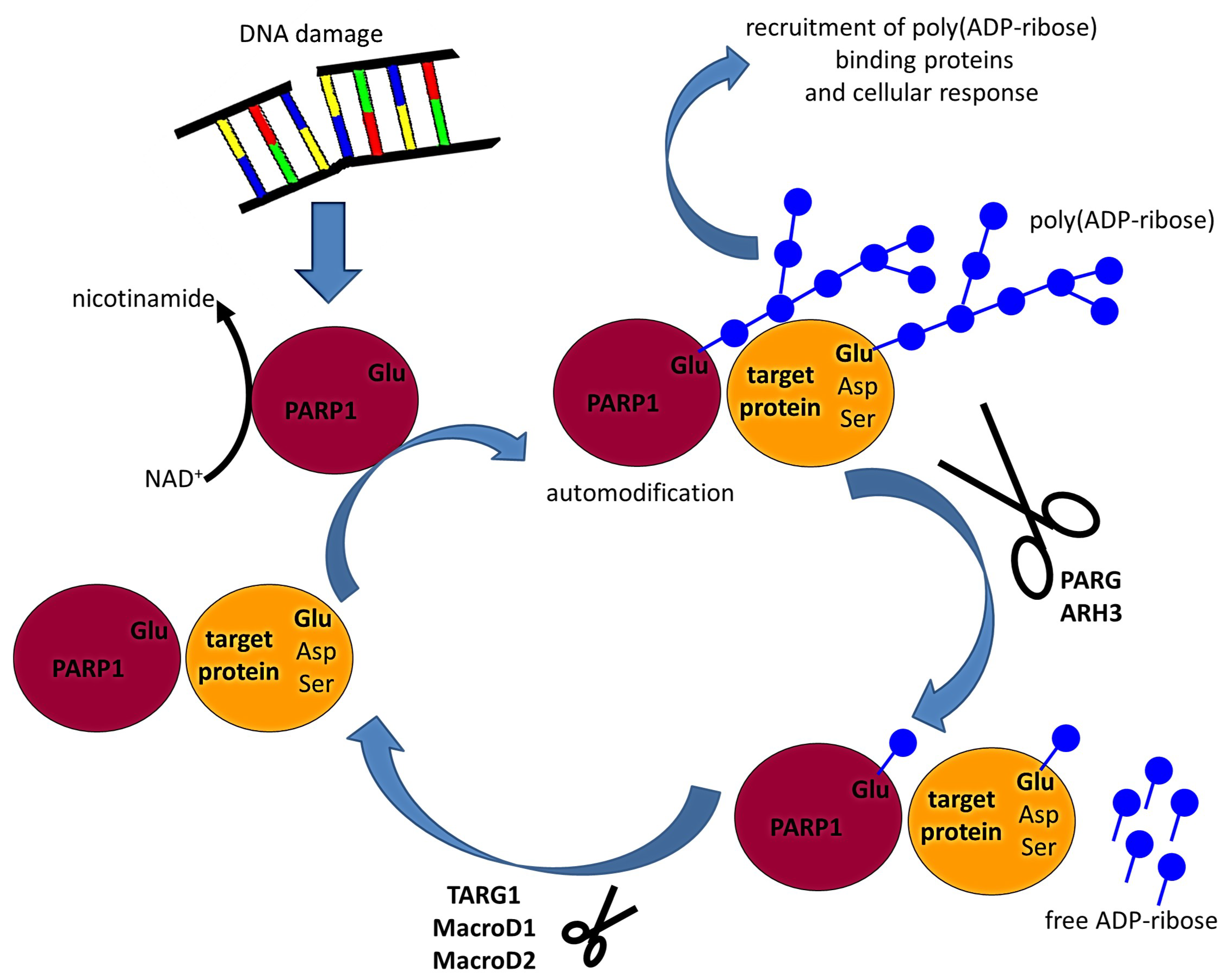
2.2. PARP1, PARP2, and PARP3 Are Activated upon DNA Damage
2.3. The Removal of Poly(ADP-Ribose) Is a Two-Step Process
3. Poly(ADP-Ribosyl)ation in Plants
3.1. Three Canonical PARP Proteins Have Been Identified in the Model Plant Arabidopsis thaliana
3.2. In Contrast to Mammals, Plants Possess More Than One Poly(ADP-ribose) Glycohydrolase (PARG) Gene
3.3. Plant PARPs Play a Role in DNA Damage Response and Genome Integrity
3.3.1. PARP1 and PARP2 Are Actors in Various DNA Damage Response Pathways in Plants
3.3.2. PARP3 Is a Core Component of Genome Integrity in Seeds
3.3.3. Like PARPs, Plant PARG Proteins Are Linked to DNA Damage Response
3.3.4. Similar to Their Human Counterparts, Plant PARPs Are Capable of Chromosome Modification
3.4. Plant PARPs Play Diverse Roles in Cell Death, Development, and Metabolism
3.5. Poly(ADP-Ribosyl)ation and Plant Responses to Abiotic Stress
3.6. Poly(ADP-Ribosyl)ation and Plant Responses to Biotic Attack
3.7. Non-Canonical PARP Domain Proteins Act in Plant Stress Responses
3.8. Potential Off-Target Effects of PARP Inhibitors
4. PARPs Under Stress—Concluding Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 3AB | 3-aminobenzamide |
| ADP-ribose | Adenosine diphosphate ribose |
| ALY | Ally of AML-1 and LEF-1 |
| 4-ANI | 4-amino-1,8-naphtalimde |
| AP2/ERF | APETALA2/ethylene response factor |
| APP | Arabidopsis thaliana homologue of PARP |
| ARH3 | ADP-ribose hydrolase 3 |
| ATM | Ataxia telangiectasia-mutated |
| ATP | Adenosine triphosphate |
| ATR | Ataxia telangiectasia and Rad3-related protein |
| BER | Base excision repair |
| BRCT | Breast cancer susceptibility gene 1 C-terminus |
| cADPR | Cyclic ADP-ribose |
| DIP | DNA-binding domain interacting protein |
| DREB | Dehydration-responsive element/C-repeat-binding proteins |
| DSB | DNA double strand break |
| elf18 | N-terminal 18 amino acids of EF-Tu |
| flg22 | N-terminally conserved 22 amino acids of flagellin |
| GUS | β-glucoronidase |
| HR | Homologous recombination |
| MacroD1 | Macrodomain-containing protein D |
| 3MB | 3-methoxybenzamide |
| MMEJ | Micro-homology-mediated end joining |
| MMS21 | Methyl methanesulfonate sensitivity gene 21 |
| MMS | Methyl methanesulfonate |
| MRE11 | Mitotic recombination 11 |
| MYB15 | Myeloblastosis transcription factor 15 |
| NAC | NAM, ATAF1 and −2, and CUC2 |
| NAD | Nicotinamide adenine dinucleotide |
| NAP | Non-classical poly(ADP-ribose)polymerase |
| NHEJ | Non-homologous end joining |
| NIC2 | Nicotinamidase2 |
| NTR | N-terminal region |
| PAMP | Pathogen-associated molecular pattern |
| PARG | Poly(ADP-ribose) glycohydrolase |
| PARP | Poly(ADP-ribose) polymerase |
| PCD | Programmed cell death |
| P5CDH | 1-pyrroline-5-carboxylate dehydrogenase |
| Pst | Pseudomonas syringae pv. tomato |
| RCD1 | Radical-induced cell death1 |
| REV7 | Protein putatively involved in translesion synthesis |
| RST domain | RCD1-SRO-TAF4 protein domain |
| SAP domain | Protein domain named after SAF-A/B, Acinus, and PIAS |
| SMC | Structural maintenance of chromosomes proteins |
| SOS1 | Salt overly sensitive 1 |
| SRO | Similar to Radical-induced cell death one |
| SUMO E3 | Small ubiquitin-like modifier E3 |
| SSB | DNA single strand break |
| SWI | Protein involved in involved in sister chromatid cohesion and chromosome organization |
| TARG1 | Terminal ADP-ribose glycohydrolase 1 |
| TEJ | Means “bright” in Sanskrit, name given to the PARG1 after its discovery as a regulator of circadian clock output |
| WGR domain | Protein domain named after the most conserved central motif of the domain |
| WWE domain | Protein domain named after three of its conserved residues |
| XRCC | X-ray repair cross-complementing protein |
| ZAP | Zinc-finger poly(ADP-ribose)polymerase |
References
- Hayashi, K.; Tanaka, M.; Shimada, T.; Miwa, M.; Sugimura, T. Size and shape of poly(ADP-ribose): Examination by gel filtration, gel electrophoresis and electron microscopy. Biochem. Biophys. Res. Commun. 1983, 112, 102–107. [Google Scholar] [CrossRef]
- Kiehlbauch, C.C.; Aboulela, N.; Jacobson, E.L.; Ringer, D.P.; Jacobson, M.K. High-resolution fractionation and characterization of ADP-ribose polymers. Anal. Biochem. 1993, 208, 26–34. [Google Scholar] [CrossRef] [PubMed]
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342, 249–268. [Google Scholar] [CrossRef]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef]
- Brady, P.N.; Goel, A.; Johnson, M.A. Poly(ADP-ribose) polymerases in host-pathogen interactions, inflammation, and immunity. Microbiol. Mol. Biol. Rev. 2019, 83, e00038-18. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Gonzalez, R.; Mendoza-Alvarez, H. Dissection of ADP-ribose polymer synthesis into individual steps of initiation, elongation, and branching. Biochimie 1995, 77, 403–407. [Google Scholar] [CrossRef]
- Gakière, B.; Hao, J.; de Bont, L.; Pétriacq, P.; Nunes-Nesi, A.; Fernie, A.R. NAD+ biosynthesis and signaling in plants. Crit. Rev. Plant Sci. 2018, 37, 259–307. [Google Scholar] [CrossRef]
- Gupte, R.; Liu, Z.Y.; Kraus, W.L. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017, 31, 101–126. [Google Scholar] [CrossRef]
- Schuhwerk, H.; Atteya, R.; Siniuk, K.; Wang, Z.Q. PARPing for balance in the homeostasis of poly(ADP-ribosyl)ation. Semin. Cell Dev. Biol. 2017, 63, 81–91. [Google Scholar] [CrossRef]
- Pleschke, J.M.; Kleczkowska, H.E.; Strohm, M.; Althaus, F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000, 275, 40974–40980. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Alvarez, H.; Alvarez-Gonzalez, R. Regulation of p53 sequence-specific DNA-binding by covalent poly(ADP-ribosyl)ation. J. Biol. Chem. 2001, 276, 36425–36430. [Google Scholar] [CrossRef]
- Chang, P.; Jacobson, M.K.; Mitchison, T.J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature 2004, 432, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Haince, J.F.; Kozlov, S.; Dawson, V.L.; Dawson, T.M.; Hendzel, M.J.; Lavin, M.F.; Poirier, G.G. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 2007, 282, 16441–16453. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Quesada, R.; Munoz-Gamez, J.A.; Martin-Oliva, D.; Peralta, A.; Valenzuela, M.T.; Matinez-Romero, R.; Quiles-Perez, R.; Murcia, J.M.D.; de Murcia, G.; de Almodovar, M.R.; et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 2007, 8, 29. [Google Scholar] [CrossRef]
- Kanai, M.; Hanashiro, K.; Kim, S.H.; Hanai, S.; Boulares, A.H.; Miwa, M.; Fukasawa, K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 2007, 9, 1175–1183. [Google Scholar] [CrossRef]
- Kedar, P.S.; Stefanick, D.F.; Horton, J.K.; Wilson, S.H. Interaction between PARP-1 and ATR in mouse fibroblasts is blocked by PARP inhibition. DNA Rep. 2008, 7, 1787–1798. [Google Scholar] [CrossRef]
- Ahel, I.; Ahel, D.; Matsusaka, T.; Clark, A.J.; Pines, J.; Boulton, S.J.; West, S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 2008, 451, 81–85. [Google Scholar] [CrossRef]
- Gagné, J.P.; Isabelle, M.; Lo, K.S.; Bourassa, S.; Hendzel, M.J.; Dawson, V.L.; Dawson, T.M.; Poirier, G.G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucl. Acid Res. 2008, 36, 6959–6976. [Google Scholar] [CrossRef]
- Ahel, D.; Horejsi, Z.; Wiechens, N.; Polo, S.E.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.C.; Jackson, S.P.; et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 2009, 325, 1240–1243. [Google Scholar] [CrossRef]
- Wang, Y.F.; Dawson, V.L.; Dawson, T.M. Poly(ADP-ribose) signals to mitochondrial AIF: A key event in parthanatos. Exp. Neurol. 2009, 218, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.C.; Lee, Y.I.; Shin, J.H.; Andrabi, S.A.; Chi, Z.K.; Gagne, J.P.; Lee, Y.J.; Ko, H.S.; Lee, B.D.; Poirier, G.G.; et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. USA 2011, 108, 14103–14108. [Google Scholar] [CrossRef]
- Min, W.; Bruhn, C.; Grigaravicius, P.; Zhou, Z.W.; Li, F.; Kruger, A.; Siddeek, B.; Greulich, K.O.; Popp, O.; Meisezahl, C.; et al. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat. Commun. 2013, 4, 2993. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, J.X.; Paudyal, S.C.; You, Z.S.; Yu, X.C. CHFR is important for the first wave of ubiquitination at DNA damage sites. Nucl. Acid Res. 2013, 41, 1698–1710. [Google Scholar] [CrossRef] [PubMed]
- Aredia, F.; Scovassi, A.I. Poly(ADP-ribose): A signaling molecule in different paradigms of cell death. Biochem. Pharmacol. 2014, 92, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Stadler, J.; Richly, H. Regulation of DNA repair mechanisms: How the chromatin environment regulates the DNA damage response. Int. J. Mol. Sci. 2017, 18, 1715. [Google Scholar] [CrossRef]
- Tao, Z.; Gao, P.; Liu, H.-W. Studies of the expression of human poly(ADP-ribose) polymerase-1 in Saccharomyces cerevisiae and identification of PARP-1 substrates by yeast proteome microarray screening. Biochemistry 2009, 48, 11745–11754. [Google Scholar] [CrossRef] [PubMed]
- Crawford, K.; Bonfiglio, J.J.; Mikoc, A.; Matic, I.; Ahel, I. Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 64–82. [Google Scholar] [CrossRef]
- Cohen, M.S.; Chang, P. Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat. Chem. Biol. 2018, 14, 236–243. [Google Scholar] [CrossRef]
- Bonfiglio, J.J.; Fontana, P.; Zhang, Q.; Colby, T.; Gibbs-Seymour, I.; Atanassov, I.; Bartlett, E.; Zaja, R.; Ahel, I.; Matic, I. Serine ADP-ribosylation depends on HPF1. Mol. Cell 2017, 65, 932–940. [Google Scholar] [CrossRef]
- Slade, D.; Dunstan, M.S.; Barkauskaite, E.; Weston, R.; Lafite, P.; Dixon, N.; Ahel, M.; Leys, D.; Ahel, I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 2011, 477, 616–620. [Google Scholar] [CrossRef]
- Perina, D.; Mikoc, A.; Ahel, J.; Cetkovic, H.T.; Zaja, R.; Ahel, I. Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Rep. 2014, 23, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Amé, J.C.; Spenlehauer, C.; de Murcia, G. The PARP superfamily. Bioessays 2004, 26, 882–893. [Google Scholar] [CrossRef]
- Otto, H.; Reche, P.A.; Bazan, F.; Dittmar, K.; Haag, F.; Koch-Nolte, F. In silico characterization of the family of PARP-like poly(ADP-ribosyl) transferases (pARTs). BMC Genom. 2005, 6, 139. [Google Scholar] [CrossRef] [PubMed]
- Hottiger, M.O.; Hassa, P.O.; Luscher, B.; Schuler, H.; Koch-Nolte, F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010, 35, 208–219. [Google Scholar] [CrossRef]
- Vyas, S.; Chesarone-Cataldo, M.; Todorova, T.; Huang, Y.H.; Chang, P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013, 4, 2240. [Google Scholar] [CrossRef]
- Rolli, V.; O’Farrell, M.; Menissier de Murcia, J.; de Murcia, G. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry 1997, 36, 12147–12154. [Google Scholar] [CrossRef]
- Vyas, S.; Matic, I.; Uchima, L.; Rood, J.; Zaja, R.; Hay, R.T.; Ahel, I.; Chang, P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014, 5, 4426. [Google Scholar] [CrossRef]
- Kraus, W.L.; Lis, J.T. PARP goes transcription. Cell 2003, 113, 677–683. [Google Scholar] [CrossRef]
- Rouleau, M.; Patel, A.; Hendzel, M.J.; Kaufmann, S.H.; Poirier, G.G. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 2010, 10, 293–301. [Google Scholar] [CrossRef]
- Ikejima, M.; Noguchi, S.; Yamashita, R.; Ogura, T.; Sugimura, T.; Gill, D.M.; Miwa, M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J. Biol. Chem. 1990, 265, 21907–21913. [Google Scholar] [PubMed]
- Gradwohl, G.; Demurcia, J.M.; Molinete, M.; Simonin, F.; Koken, M.; Hoeijmakers, J.H.J.; Demurcia, G. The 2nd zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc. Natl. Acad. Sci. USA 1990, 87, 2990–2994. [Google Scholar] [CrossRef]
- Langelier, M.F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef]
- Langelier, M.F.; Ruhl, D.D.; Planck, J.L.; Kraus, W.L.; Pascal, J.M. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 2010, 285, 18877–18887. [Google Scholar] [CrossRef] [PubMed]
- Dawicki-McKenna, J.M.; Langelier, M.F.; DeNizio, J.E.; Riccio, A.A.; Cao, C.D.; Karch, K.R.; McCauley, M.; Steffen, J.D.; Black, B.E.; Pascal, J.M. PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol. Cell 2015, 60, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Eustermann, S.; Wu, W.F.; Langelier, M.F.; Yang, J.C.; Easton, L.E.; Riccio, A.A.; Pascal, J.M.; Neuhaus, D. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell 2015, 60, 742–754. [Google Scholar] [CrossRef]
- Steffen, J.D.; McCauley, M.M.; Pascal, J.M. Fluorescent sensors of PARP-1 structural dynamics and allosteric regulation in response to DNA damage. Nucl. Acid Res. 2016, 44, 9771–9783. [Google Scholar] [CrossRef] [PubMed]
- Rissel, D.; Heym, P.P.; Thor, K.; Brandt, W.; Wessjohann, L.A.; Peiter, E. No silver bullet - Canonical poly(ADP-ribose) polymerases (PARPs) are no universal factors of abiotic and biotic stress resistance of Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Masson, M.; Niedergang, C.; Schreiber, V.; Muller, S.; Menissier-de Murcia, J.; de Murcia, G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998, 18, 3563–3571. [Google Scholar] [CrossRef] [PubMed]
- Okano, S.; Lan, L.; Caldecott, K.W.; Mori, T.; Yasui, R. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003, 23, 3974–3981. [Google Scholar] [CrossRef]
- El-Khamisy, S.F.; Masutani, M.; Suzuki, H.; Caldecott, K.W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucl. Acid Res. 2003, 31, 5526–5533. [Google Scholar] [CrossRef]
- Haince, J.F.; McDonald, D.; Rodrigue, A.; Dery, U.; Masson, J.Y.; Hendzel, M.J.; Poirier, G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008, 283, 1197–1208. [Google Scholar] [CrossRef]
- Hochegger, H.; Dejsuphong, D.; Fukushima, T.; Morrison, C.; Sonoda, E.; Schreiber, V.; Zhao, G.Y.; Saberi, A.; Masutani, M.; Adachi, N.; et al. PARP-1 protects homologous recombination from interference by Ku and ligase IV in vertebrate cells. EMBO J. 2006, 25, 1305–1314. [Google Scholar] [CrossRef]
- Beck, C.; Robert, I.; Reina-San-Martin, B.; Schreiber, V.; Dantzer, F. Poly(ADP-ribose) polymerases in double-strand break repair: Focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 2014, 329, 18–25. [Google Scholar] [CrossRef]
- Galande, S.; Kohwi-Shigematso, T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 1999, 274, 20521–20528. [Google Scholar] [CrossRef]
- Wang, M.L.; Wu, W.Z.; Wu, W.Q.; Rosidi, B.; Zhang, L.H.; Wang, H.C.; Iliakis, G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucl. Acid Res. 2006, 34, 6170–6182. [Google Scholar] [CrossRef]
- Frizzell, K.M.; Gamble, M.J.; Berrocal, J.G.; Zhang, T.; Krishnakumar, R.; Cen, Y.; Sauve, A.A.; Kraus, W.L. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J. Biol. Chem. 2009, 284, 33926–33938. [Google Scholar] [CrossRef]
- Amé, J.C.; Rolli, V.; Schreiber, V.; Niedergang, C.; Apiou, F.; Decker, P.; Muller, S.; Hoger, T.; Murcia, J.M.D.; de Murcia, G. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999, 274, 17860–17868. [Google Scholar] [CrossRef] [PubMed]
- Kutuzov, M.M.; Khodyreva, S.N.; Schreiber, V.; Lavrik, O.I. Role of PARP2 in DNA repair. Mol. Biol. 2014, 48, 485–495. [Google Scholar] [CrossRef]
- Riccio, A.A.; Cingolani, G.; Pascal, J.M. PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucl. Acid Res. 2016, 44, 1691–1702. [Google Scholar] [CrossRef]
- Schreiber, V.; Ame, J.C.; Dolle, P.; Schultz, I.; Rinaldi, B.; Fraulob, V.; Menissier-de Murcia, J.; de Murcia, G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002, 277, 23028–23036. [Google Scholar] [CrossRef] [PubMed]
- Isabelle, M.; Moreel, X.; Gagne, J.P.; Rouleau, M.; Ethier, C.; Gagne, P.; Hendzel, M.J.; Poirier, G.G. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 2010, 8, 22. [Google Scholar] [CrossRef]
- Hanzlikova, H.; Gittens, W.; Krejcikova, K.; Zeng, Z.H.; Caldecott, K.W. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucl. Acid Res. 2017, 45, 2546–2557. [Google Scholar] [CrossRef]
- De Murcia, J.M.N.; Ricoul, M.; Tartier, L.; Niedergang, C.; Huber, A.; Dantzer, F.; Schreiber, V.; Ame, J.C.; Dierich, A.; LeMeur, M.; et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003, 22, 2255–2263. [Google Scholar] [CrossRef]
- Bryant, H.E.; Petermann, E.; Schultz, N.; Jemth, A.S.; Loseva, O.; Issaeva, N.; Johansson, F.; Fernandez, S.; McGlynn, P.; Helleday, T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009, 28, 2601–2615. [Google Scholar] [CrossRef]
- Fouquin, A.; Guirouilh-Barbat, J.; Lopez, B.; Hall, J.; Amor-Gueret, M.; Pennaneach, V. PARP2 controls double-strand break repair pathway choice by limiting 53BP1 accumulation at DNA damage sites and promoting end-resection. Nucl. Acid Res. 2017, 45, 12325–12339. [Google Scholar] [CrossRef] [PubMed]
- Boehler, C.; Dantzer, F. PARP-3, a DNA-dependent PARP with emerging roles in double-strand break repair and mitotic progression. Cell Cycle 2011, 10, 1023–1024. [Google Scholar] [CrossRef][Green Version]
- Rulten, S.L.; Fisher, A.E.O.; Robert, I.; Zuma, M.C.; Rouleau, M.; Ju, L.M.; Poirier, G.; Reina-San-Martin, B.; Caldecott, K.W. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell 2011, 41, 33–45. [Google Scholar] [CrossRef]
- Rouleau, M.; McDonald, D.; Gagné, P.; Ouellet, M.-E.; Droit, A.; Hunter, J.M.; Dutertre, S.; Prigent, C.; Hendzel, M.J.; Poirier, G.G. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J. Cell. Biochem. 2007, 100, 385–401. [Google Scholar] [CrossRef]
- Loseva, O.; Jemth, A.S.; Bryant, H.E.; Schuler, H.; Lehtio, L.; Karlberg, T.; Helleday, T. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 2010, 285, 8054–8060. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.S.; Ame, J.C.; AboulEla, N.; Jacobson, E.L.; Jacobson, M.K. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 1997, 272, 11895–11901. [Google Scholar] [CrossRef] [PubMed]
- Barkauskaite, E.; Brassington, A.; Tan, E.S.; Warwicker, J.; Dunstan, M.S.; Banos, B.; Lafite, P.; Ahel, M.; Mitchison, T.J.; Ahel, I.; et al. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat. Commun. 2013, 4, 2164. [Google Scholar] [CrossRef] [PubMed]
- Thomassin, H.; Menard, L.; Hengartner, C.; Kirkland, J.B.; Poirier, G.G. Poly(ADP-ribosyl)ation of chromatin in an in vitro poly(ADP-ribose)-turnover system. Biochim. Biophys. Act. 1992, 1137, 171–181. [Google Scholar] [CrossRef]
- Brochu, G.; Duchaine, C.; Thibeault, L.; Lagueux, J.; Shah, G.M.; Poirier, G.G. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim. Biophys. Act. 1994, 1219, 342–350. [Google Scholar] [CrossRef]
- Meyer-Ficca, M.L.; Meyer, R.G.; Coyle, D.L.; Jacobson, E.L.; Jacobson, M.K. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 2004, 297, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.G.; Meyer-Ficca, M.L.; Whatcott, C.J.; Jacobson, E.L.; Jacobson, M.K. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp. Cell Res. 2007, 313, 2920–2936. [Google Scholar] [CrossRef] [PubMed]
- Koh, D.W.; Lawler, A.M.; Poitras, M.F.; Sasaki, M.; Wattler, S.; Nehls, M.C.; Stoger, T.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA 2004, 101, 17699–17704. [Google Scholar] [CrossRef] [PubMed]
- Hanai, S.; Kanai, M.; Ohashi, S.; Okamoto, K.; Yamada, M.; Takahashi, H.; Miwa, M. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2004, 101, 82–86. [Google Scholar] [CrossRef]
- Oka, S.; Kato, J.; Moss, J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. FASEB J. 2006, 20, A45. [Google Scholar] [CrossRef]
- Niere, M.; Kernstock, S.; Koch-Nolte, F.; Ziegler, M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol. Cell. Biol. 2008, 28, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Jankevicius, G.; Hassler, M.; Golia, B.; Rybin, V.; Zacharias, M.; Timinszky, G.; Ladurner, A.G. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013, 20, 508–514. [Google Scholar] [CrossRef]
- Rosenthal, F.; Feijs, K.L.H.; Frugier, E.; Bonalli, M.; Forst, A.H.; Imhof, R.; Winkler, H.C.; Fischer, D.; Caflisch, A.; Hassa, P.O.; et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013, 20, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, R.; Morra, R.; Appel, C.D.; Tallis, M.; Chioza, B.; Jankevicius, G.; Simpson, M.A.; Matic, I.; Ozkan, E.; Golia, B.; et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013, 32, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.F.; Bal, A.K. Cytological detection of poly (ADP-ribose) polymerase. Exp. Cell Res. 1976, 99, 428–432. [Google Scholar] [CrossRef]
- Whitby, A.J.; Whish, W.J. Poly(adenoise diphosphate ribose) in wheat [proceedings]. Biochem. Soc. Transact. 1977, 5, 948–949. [Google Scholar] [CrossRef]
- Whitby, A.J.; Whish, W.J. Poly(adenosine diphosphate ribose) glycohydrolase in germinating wheat embryos [proceedings]. Biochem. Soc. Transact. 1978, 6, 619–620. [Google Scholar] [CrossRef]
- Whitby, A.J.; Stone, P.R.; Whish, W.J.D. Effect of polyamines and Mg++ on poly(ADP-ribose) synthesis and ADP-ribosylation of histones in wheat. Biochem. Biophys. Res. Commun. 1979, 90, 1295–1304. [Google Scholar] [CrossRef]
- Willmitzer, L. Demonstration of in vitro covalent modification of chromosomal-proteins by poly(ADP) ribosylation in plant nuclei. FEBS Lett. 1979, 108, 13–16. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Shall, S.; O’Farrell, M. Poly(ADP-ribose) polymerase in plant nuclei. Eur. J. Biochem. 1994, 224, 135–142. [Google Scholar] [CrossRef]
- Lepiniec, L.; Babiychuk, E.; Kushnir, S.; Van Montagu, M.; Inzé, D. Characterization of an Arabidopsis thaliana cDNA homologue to animal poly(ADP-ribose) polymerase. FEBS Lett. 1995, 364, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. SAP—A putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000, 25, 112–114. [Google Scholar] [CrossRef]
- Babiychuk, E.; Cottrill, P.B.; Storozhenko, S.; Fuangthong, M.; Chen, Y.; O’Farrell, M.K.; Van Montagu, M.; Inzé, D.; Kushnir, S. Higher plants possess two structurally different poly(ADP-ribose) polymerases. Plant J. 1998, 15, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Keppler, B.D.; Wise, R.R.; Bent, A.F. PARP2 is the predominant poly(ADP-ribose) polymerase in Arabidopsis DNA damage and immune responses. PLOS Genet. 2015, 11, e1005200. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.A.; Wahl, V.; Tohge, T.; de Souza, L.R.; Zhang, Y.; Do, P.T.; Olas, J.J.; Stitt, M.; Araújo, W.L.; Fernie, A.R. Analysis of knockout mutants reveals non-redundant functions of poly(ADP-ribose)polymerase isoforms in Arabidopsis. Plant Mol. Biol. 2015, 89, 319–338. [Google Scholar] [CrossRef]
- Chen, C.; De Masi, R.; Lintermann, R.; Wirthmueller, L. Nuclear import of Arabidopsis poly(ADP-ribose) polymerase 2 is mediated by importin-α and a nuclear localization sequence located between the predicted SAP domains. Front. Plant Sci. 2018, 9, 1581. [Google Scholar] [CrossRef] [PubMed]
- Doucet-Chabeaud, G.; Godon, C.; Brutesco, C.; de Murcia, G.; Kazmaier, M. Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol. Genet. Genom. 2001, 265, 954–963. [Google Scholar]
- Rissel, D.; Heym, P.P.; Peiter, E. A yeast growth assay to characterize plant poly(ADP-ribose) polymerase (PARP) proteins and inhibitors. Anal. Biochem. 2017, 527, 20–23. [Google Scholar] [CrossRef]
- Boltz, K.A.; Jasti, M.; Townley, J.M.; Shippen, D.E. Analysis of poly(ADP-ribose) polymerases in Arabidopsis telomere biology. PLoS ONE 2014, 9, e88872. [Google Scholar] [CrossRef][Green Version]
- Briggs, A.G.; Bent, A.F. Poly(ADP-ribosyl)ation in plants. Trends Plant Sci. 2011, 16, 372–380. [Google Scholar] [CrossRef]
- De Block, M.; Verduyn, C.; De Brouwer, D.; Cornelissen, M. Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 2005, 41, 95–106. [Google Scholar] [CrossRef]
- Jia, Q.; den Dulk-Ras, A.; Shen, H.; Hooykaas, P.J.J.; de Pater, S. Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana. Plant Mol. Biol. 2013, 82, 339–351. [Google Scholar] [CrossRef]
- Feng, B.; Liu, C.; de Oliveira, M.V.V.; Intorne, A.C.; Li, B.; Babilonia, K.; de Souza Filho, G.A.; Shan, L.; He, P. Protein poly(ADP-ribosyl)ation regulates Arabidopsis immune gene expression and defense responses. PLOS Genet. 2015, 11, e1004936. [Google Scholar] [CrossRef] [PubMed]
- Keppler, B.D.; Song, J.Q.; Nyman, J.; Voigt, C.A.; Bent, A.F. 3-aminobenzamide blocks MAMP-induced callose deposition independently of its poly(ADPribosyl)ation inhibiting activity. Front. Plant Sci. 2018, 9, 1907. [Google Scholar] [CrossRef]
- Lamb, R.S.; Citarelli, M.; Teotia, S. Functions of the poly(ADP-ribose) polymerase superfamily in plants. Cell. Mol. Life Sci. 2012, 69, 175–189. [Google Scholar] [CrossRef]
- Ogawa, T.; Ishikawa, K.; Harada, K.; Fukusaki, E.; Yoshimura, K.; Shigeoka, S. Overexpression of an ADP-ribose pyrophosphatase, AtNUDX2, confers enhanced tolerance to oxidative stress in Arabidopsis plants. Plant J. 2009, 57, 289–301. [Google Scholar] [CrossRef]
- Pellny, T.K.; Locato, V.; Vivancos, P.D.; Markovic, J.; De Gara, L.; Pallardo, F.V.; Foyer, C.H. Pyridine nucleotide cycling and control of intracellular redox state in relation to poly (ADP-ribose) polymerase activity and nuclear localization of glutathione during exponential growth of Arabidopsis cells in culture. Mol. Plant 2009, 2, 442–456. [Google Scholar] [CrossRef]
- Schulz, P.; Neukermans, J.; Van der Kelen, K.; Mühlenbock, P.; Van Breusegem, F.; Noctor, G.; Teige, M.; Metzlaff, M.; Hannah, M.A. Chemical PARP inhibition enhances growth of Arabidopsis and reduces anthocyanin accumulation and the activation of stress protective mechanisms. PLoS ONE 2012, 7, e37284. [Google Scholar] [CrossRef]
- Vanderauwera, S.; De Block, M.; Van de Steene, N.; Van de Cotte, B.; Metzlaff, M.; Van Breusegem, F. Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc. Natl. Acad. Sci. USA 2007, 104, 15150–15155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gu, Z.; Wu, Q.; Yang, L.; Liu, C.; Ma, H.; Xia, Y.; Ge, X. Arabidopsis PARG1 is the key factor promoting cell survival among the enzymes regulating post-translational poly(ADP-ribosyl)ation. Sci. Rep. 2015, 5, 15892. [Google Scholar] [CrossRef] [PubMed]
- Hunt, L.; Lerner, F.; Ziegler, M. NAD—New roles in signalling and gene regulation in plants. New Phytol. 2004, 163, 31–44. [Google Scholar] [CrossRef]
- Rissel, D.; Losch, J.; Peiter, E. The nuclear protein Poly(ADP-ribose) polymerase 3 (AtPARP3) is required for seed storability in Arabidopsis thaliana. Plant Biol. 2014, 16, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Perkins, E.; Sun, D.; Nguyen, A.; Tulac, S.; Francesco, M.; Tavana, H.; Nguyen, H.; Tugendreich, S.; Barthmaier, P.; Couto, J.; et al. Novel inhibitors of poly(ADP-ribose) polymerase/PARP1 and PARP2 identified using a cell-based screen in yeast. Cancer Res. 2001, 61, 4175–4183. [Google Scholar] [PubMed]
- Panda, S.; Poirier, G.G.; Kay, S.A. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator. Dev. Cell 2002, 3, 51–61. [Google Scholar] [CrossRef]
- Yuan, D.; Lai, J.; Xu, P.; Zhang, S.; Zhang, J.; Li, C.; Wang, Y.; Du, J.; Liu, Y.; Yang, C. AtMMS21 regulates DNA damage response and homologous recombination repair in Arabidopsis. DNA Rep. 2014, 21, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Stolarek, M.; Gruszka, D.; Braszewska-Zalewska, A.; Maluszynski, M. Alleles of newly identified barley gene HvPARP3 exhibit changes in efficiency of DNA repair. DNA Rep. 2015, 28, 116–130. [Google Scholar] [CrossRef] [PubMed]
- Arena, C.; De Micco, V.; De Maio, A. Growth alteration and leaf biochemical responses in Phaseolus vulgaris exposed to different doses of ionising radiation. Plant Biol. 2014, 16, 194–202. [Google Scholar] [CrossRef]
- Liu, C.; Wu, Q.; Liu, W.; Gu, Z.; Wang, W.; Xu, P.; Ma, H.; Ge, X. Poly(ADP-ribose) polymerases regulate cell division and development in Arabidopsis roots. J. Integr. Plant Biol. 2017, 59, 459–474. [Google Scholar] [CrossRef]
- Klemm, T.; Mannuss, A.; Kobbe, D.; Knoll, A.; Trapp, O.; Dorn, A.; Puchta, H. The DNA translocase RAD5A acts independently of the other main DNA repair pathways, and requires both its ATPase and RING domain for activity in Arabidopsis thaliana. Plant J. 2017, 91, 725–740. [Google Scholar] [CrossRef]
- Song, J.Q.; Bent, A.F. Microbial pathogens trigger host DNA double-strand breaks whose abundance is reduced by plant defense responses. PLOS Path. 2014, 10, e1004030. [Google Scholar] [CrossRef]
- De Schutter, K.; Joubes, J.; Cools, T.; Verkest, A.; Corellou, F.; Babiychuk, E.; Van Der Schueren, E.; Beeckman, T.; Kushnir, S.; Inze, D.; et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 2007, 19, 211–225. [Google Scholar] [CrossRef]
- Tuteja, N.; Ahmad, P.; Panda, B.B.; Tuteja, R. Genotoxic stress in plants: Shedding light on DNA damage, repair and DNA repair helicases. Mut. Res. Rev. Mut. Res. 2009, 681, 134–149. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H.; Swoboda, P.; Hohn, B. Induction of intrachromosomal homologous recombination in whole plants. Plant J. 1995, 7, 203–210. [Google Scholar] [CrossRef]
- Ishikawa, K.; Ogawa, T.; Hirosue, E.; Nakayama, Y.; Harada, K.; Fukusaki, E.; Yoshimura, K.; Shigeoka, S. Modulation of the poly(ADP-ribosyl)ation reaction via the Arabidopsis ADP-Ribose/NADH pyrophosphohydrolase, AtNUDX7, is involved in the response to oxidative stress. Plant Physiol. 2009, 151, 741–754. [Google Scholar] [CrossRef]
- Shen, H.; Strunks, G.D.; Klemann, B.J.P.M.; Hooykaas, P.J.J.; de Pater, S. CRISPR/Cas9-induced double-strand break repair in Arabidopsis nonhomologous end-joining mutants. G3 Gen. Genom. Genet. 2017, 7, 193–202. [Google Scholar] [CrossRef]
- Spampinato, C.P. Protecting DNA from errors and damage: An overview of DNA repair mechanisms in plants compared to mammals. Cell. Mol. Life Sci. 2017, 74, 1693–1709. [Google Scholar] [CrossRef]
- Howell, K.A.; Narsai, R.; Carroll, A.; Ivanova, A.; Lohse, M.; Usadel, B.; Millar, A.H.; Whelan, J. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol. 2009, 149, 961–980. [Google Scholar] [CrossRef]
- Hunt, L.; Holdsworth, M.J.; Gray, J.E. Nicotinamidase activity is important for germination. Plant J. 2007, 51, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Hunt, L.; Gray, J.E. The relationship between pyridine nucleotides and seed dormancy. New Phytol. 2009, 181, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Rajjou, L.; North, H.M.; Debeaujon, I.; Marion-Poll, A.; Seo, M. Staying alive: Molecular aspects of seed longevity. Plant Cell Physiol. 2016, 57, 660–674. [Google Scholar] [CrossRef]
- Li, W.; Zhang, F.; Chang, Y.; Zhao, T.; Schranz, M.E.; Wang, G. Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell 2015, 27, 1907–1924. [Google Scholar] [CrossRef] [PubMed]
- Adams-Phillips, L.; Briggs, A.G.; Bent, A.F. Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol. 2010, 152, 267–280. [Google Scholar] [CrossRef]
- Briggs, A.G.; Adams-Phillips, L.C.; Keppler, B.D.; Zebell, S.G.; Arend, K.C.; Apfelbaum, A.A.; Smith, J.A.; Bent, A.F. A transcriptomics approach uncovers novel roles for poly(ADP-ribosyl) ation in the basal defense response in Arabidopsis thaliana. PLoS ONE 2017, 12, e0190268. [Google Scholar] [CrossRef] [PubMed]
- Babiychuk, E.; Van Montagu, M.; Kushnir, S. N-terminal domains of plant poly(ADP-ribose) polymerases define their association with mitotic chromosomes. Plant J. 2001, 28, 245–255. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Storozhenko, S.; Inzé, D.; Van Montagu, M.; Kushnir, S. Arabidopsis coactivator ALY-like proteins, DIP1 and DIP2, interact physically with the DNA-binding domain of the Zn-finger poly(ADP-ribose) polymerase. J. Exp. Bot. 2001, 52, 1375–1380. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cifuentes-Rojas, C.; Nelson, A.D.L.; Boltz, K.A.; Kannan, K.; She, X.T.; Shippen, D.E. An alternative telomerase RNA in Arabidopsis modulates enzyme activity in response to DNA damage. Genes Dev. 2012, 26, 2512–2523. [Google Scholar] [CrossRef] [PubMed]
- Amor, Y.; Babiychuk, E.; Inzé, D.; Levine, A. The involvement of poly(ADP-ribose) polymerase in the oxidative stress responses in plants. FEBS Lett. 1998, 440, 1–7. [Google Scholar] [CrossRef]
- Tian, R.-H.; Zhang, G.-Y.; Yan, C.-H.; Dai, Y.-R. Involvement of poly(ADP-ribose) polymerase and activation of caspase-3-like protease in heat shock-induced apoptosis in tobacco suspension cells. FEBS Lett. 2000, 474, 11–15. [Google Scholar] [CrossRef]
- Vainonen, J.P.; Shapiguzov, A.; Vaattovaara, A.; Kangasjarvi, J. Plant PARPs, PARGs and PARP-like proteins. Curr. Prot. Pept. Sci. 2016, 17, 713–723. [Google Scholar] [CrossRef]
- Phillips, R.; Hawkins, S.W. Characteristics of the inhibition of induced tracheary element differentiation by 3-aminobenzamide and related compounds. J. Exp. Bot. 1985, 36, 119–128. [Google Scholar] [CrossRef]
- Schulz, P.; Jansseune, K.; Degenkolbe, T.; Méret, M.; Claeys, H.; Skirycz, A.; Teige, M.; Willmitzer, L.; Hannah, M.A. Poly(ADP-ribose)polymerase activity controls plant growth by promoting leaf cell number. PLoS ONE 2014, 9, e90322. [Google Scholar] [CrossRef]
- Adams-Phillips, L.; Wan, J.; Tan, X.; Dunning, F.M.; Meyers, B.C.; Michelmore, R.W.; Bent, A.F. Discovery of ADP-ribosylation and other plant defense pathway elements through expression profiling of four different Arabidopsis-Pseudomonas R-avr interactions. Mol. Plant Micr. Interact. 2008, 21, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- Perera, I.Y.; Hung, C.-Y.; Moore, C.D.; Stevenson-Paulik, J.; Boss, W.F. Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 2008, 20, 2876–2893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, W.; Li, Z.; Deng, X.W.; Wu, W.; Xue, Y. F-Box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis. Plant Physiol. 2008, 148, 2121–2133. [Google Scholar] [CrossRef]
- Mizoguchi, M.; Umezawa, T.; Nakashima, K.; Kidokoro, S.; Takasaki, H.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 2010, 51, 842–847. [Google Scholar] [CrossRef]
- Chan, Z.; Grumet, R.; Loescher, W. Global gene expression analysis of transgenic, mannitol-producing, and salt-tolerant Arabidopsis thaliana indicates widespread changes in abiotic and biotic stress-related genes. J. Exp. Bot. 2011, 62, 4787–4803. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, G.B.; Nguyen, T.T.; Verslues, P.E. Unique drought resistance functions of the Highly ABA-Induced clade A protein phosphatase 2Cs. Plant Physiol. 2012, 160, 379–395. [Google Scholar] [CrossRef]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef]
- Berger, F.; Ramirez-Hernandez, M.H.; Ziegler, M. The new life of a centenarian: Signalling functions of NAD(P). Trends Biochem. Sci. 2004, 29, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.-P.; Duque, P.; Chua, N.-H. ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J. 2004, 38, 381–395. [Google Scholar] [CrossRef]
- Dodd, A.N.; Gardner, M.J.; Hotta, C.T.; Hubbard, K.E.; Dalchau, N.; Love, J.; Assie, J.-M.; Robertson, F.C.; Jakobsen, M.K.; Goncalves, J.; et al. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 2007, 318, 1789–1792. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.J.; Muir, S.R.; Sanders, D. Release of Ca2+ from individual plant vacuoles by both InsP3 and cyclic ADP-ribose. Science 1995, 268, 735–737. [Google Scholar] [CrossRef]
- Wu, Y.; Kuzma, J.; Maréchal, E.; Graeff, R.; Lee, H.C.; Foster, R.; Chua, N.-H. Abscisic acid signaling through cyclic ADP-ribose in plants. Science 1997, 278, 2126–2130. [Google Scholar] [CrossRef]
- Abdul-Awal, S.M.; Hotta, C.T.; Davey, M.P.; Dodd, A.N.; Smith, A.G.; Webb, A.A.R. NO-mediated [Ca2+]cyt increases depend on ADP-ribosyl cyclase activity in Arabidopsis. Plant Physiol. 2016, 171, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Leckie, C.P.; McAinsh, M.R.; Allen, G.J.; Sanders, D.; Hetherington, A.M. Abscisic acid-induced stomatal closure mediated by cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 15837–15842. [Google Scholar] [CrossRef] [PubMed]
- Peiter, E. The plant vacuole: Emitter and receiver of calcium signals. Cell Calcium 2011, 50, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Bouche, N.; Bouchez, D. Arabidopsis gene knockout: Phenotypes wanted. Curr. Opin. Plant Biol. 2001, 4, 111–117. [Google Scholar] [CrossRef]
- Lloyd, J.; Meinke, D. A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol. 2012, 158, 1115–1129. [Google Scholar] [CrossRef]
- Hauser, M.T.; Morikami, A.; Benfey, P.N. Conditional root expansion mutants of Arabidopsis. Development 1995, 121, 1237–1252. [Google Scholar]
- Kurata, T.; Yamamoto, K.T. petit1, a conditional growth mutant of Arabidopsis defective in sucrose-dependent elongation growth. Plant Physiol. 1998, 118, 793–801. [Google Scholar] [CrossRef]
- Hicks, K.A.; Millar, A.J.; Carre, I.A.; Somers, D.E.; Straume, M.; MeeksWagner, D.R.; Kay, S.A. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 1996, 274, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Lorrain, S.; Lina, B.Q.; Auriac, M.C.; Kroj, T.; Saindrenan, P.; Nicole, M.; Balague, C.; Roby, D. Vascular associated death1, a novel gram domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 2004, 16, 2217–2232. [Google Scholar] [CrossRef]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are MicroRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef]
- Geissler, T.; Wessjohann, L.A. A whole-plant microtiter plate assay for drought stress tolerance-inducing effects. J. Plant Growth Regul. 2011, 30, 504–511. [Google Scholar] [CrossRef]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 2004, 16, 3496–3507. [Google Scholar] [CrossRef] [PubMed]
- Thor, K.; Peiter, E. Cytosolic calcium signals elicited by the pathogen-associated molecular pattern flg22 in stomatal guard cells are of an oscillatory nature. New Phytol. 2014, 204, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Berglund, T.; Kalbin, G.; Strid, A.; Rydstrom, J.; Ohlsson, A.B. UV-B- and oxidative stress-induced increase in nicotinamide and trigonelline and inhibition of defensive metabolism induction by poly(ADP-ribose)polymerase inhibitor in plant tissue. FEBS Lett. 1996, 380, 188–193. [Google Scholar] [CrossRef]
- Feng, B.M.; Ma, S.S.; Chen, S.X.; Zhu, N.; Zhang, S.X.; Yu, B.; Yu, Y.; Le, B.; Chen, X.M.; Dinesh-Kumar, S.P.; et al. PARylation of the forkhead-associated domain protein DAWDLE regulates plant immunity. EMBO Rep. 2016, 17, 1799–1813. [Google Scholar] [CrossRef] [PubMed]
- Ahlfors, R.; Lang, S.; Overmyer, K.; Jaspers, P.; Brosché, M.; Taurianinen, A.; Kollist, H.; Tuominen, H.; Belles-Boix, E.; Piippo, M.; et al. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 2004, 16, 1925–1937. [Google Scholar] [CrossRef]
- Jaspers, P.; Overmyer, K.; Wrzaczek, M.; Vainonen, J.P.; Blomster, T.; Salojärvi, J.; Reddy, R.A.; Kangasjärvi, J. The RST and PARP-like domain containing SRO protein family: Analysis of protein structure, function and conservation in land plants. BMC Genom. 2010, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Zong, W.; Du, H.; Hu, H.; Xiong, L. A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses through interaction with various transcription factors. Plant Mol. Biol. 2014, 84, 693–705. [Google Scholar] [CrossRef]
- Siddiqua, B.; Qamarunnisa, S.; Azhar, A. RCD1 homologues and their constituent WWE domain in plants: Analysis of conservation through phylogeny methods. Biologia 2016, 71, 642–650. [Google Scholar] [CrossRef]
- Belles-Boix, E.; Babiychuk, E.; Van Montagu, M.; Inzé, D.; Kushnir, S. CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 2000, 482, 19–24. [Google Scholar] [CrossRef]
- Jaspers, P.; Blomster, T.; Brosché, M.; Salojärvi, J.; Ahlfors, R.; Vainonen, J.P.; Reddy, R.A.; Immink, R.; Angenent, G.; Turck, F.; et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 2009, 60, 268–279. [Google Scholar] [CrossRef]
- Liu, S.; Liu, S.; Wang, M.; Wei, T.; Meng, C.; Wang, M.; Xia, G. A wheat SIMILAR TO RCD-ONE gene enhances seedling growth and abiotic stress resistance by modulating redox homeostasis and maintaining genomic integrity. Plant Cell 2014, 26, 164–180. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.; Tuominen, H.; Kettunen, R.; Betz, C.; Langebartels, C.; Sandermann, H.; Kangasjärvi, J. Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 2000, 12, 1849–1862. [Google Scholar] [CrossRef]
- Fujibe, T.; Saji, H.; Arakawa, K.; Yabe, N.; Takeuchi, Y.; Yamamoto, K.T. A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 2004, 134, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Teotia, S.; Lamb, R.S. The paralogous genes RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 have partially redundant functions during Arabidopsis development. Plant Physiol. 2009, 151, 180–198. [Google Scholar] [CrossRef]
- Fujibe, T.; Saji, H.; Watahiki, M.K.; Yamamoto, K.T. Overexpression of the RADICAL-INDUCED CELL DEATH1 (RCD1) gene of Arabidopsis causes weak rcd1 phenotype with compromised oxidative-stress responses. Biosci. Biotechnol. Biochem. 2006, 70, 1827–1831. [Google Scholar] [CrossRef][Green Version]
- Katiyar-Agarwal, S.; Zhu, J.; Kim, K.; Agarwal, M.; Fu, X.; Huang, A.; Zhu, J.-K. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 18816–18821. [Google Scholar] [CrossRef]
- Overmyer, K.; Brosché, M.; Pellinen, R.; Kuittinen, T.; Tuominen, H.; Ahlfors, R.; Keinänen, M.; Saarma, M.; Scheel, D.; Kangasjärvi, J. Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death1 mutant. Plant Physiol. 2005, 137, 1092–1104. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, Y.; Bjorn, L.O.; Li, S. Arabidopsis radical-induced cell death1 is involved in UV-B signaling. Photochem. Photobiol. Sci. 2009, 8, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Morales, L.O.; Brosche, M.; Vainonen, J.P.; Sipari, N.; Lindfors, A.V.; Strid, A.; Aphalo, P.J. Are solar UV-B- and UV-A-dependent gene expression and metabolite accumulation in Arabidopsis mediated by the stress response regulator RADICAL-INDUCED CELL DEATH1? Plant Cell Environ. 2015, 38, 878–891. [Google Scholar] [CrossRef]
- Wirthmueller, L.; Asai, S.; Rallapalli, G.; Sklenar, J.; Fabro, G.; Kim, D.S.; Lintermann, R.; Jaspers, P.; Wrzaczek, M.; Kangasjärvi, J.; et al. Arabidopsis downy mildew effector HaRxL106 suppresses plant immunity by binding to RADICAL-INDUCED CELL DEATH1. New Phytol. 2018, 220, 232–248. [Google Scholar] [CrossRef]
- He, R.; Drury, G.E.; Rotari, V.I.; Gordon, A.; Willer, M.; Farzaneh, T.; Woltering, E.J.; Gallois, P. Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J. Biol. Chem. 2008, 283, 774–783. [Google Scholar] [CrossRef]
- Brosché, M.; Blomster, T.; Salojärvi, J.; Cui, F.; Sipari, N.; Leppälä, J.; Lamminmäki, A.; Tomai, G.; Narayanasamy, S.; Reddy, R.A.; et al. Transcriptomics and functional genomics of ROS-induced cell death regulation by RADICAL-INDUCED CELL DEATH1. PLoS Genet. 2014, 10, e1004112. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Du, B.; Qian, J.; Zou, B.; Hua, J. Disease resistance gene-induced growth inhibition is enhanced by rcd1 independent of defense activation in Arabidopsis. Plant Physiol. 2013, 161, 2005–2013. [Google Scholar] [CrossRef] [PubMed]
- Pelagio-Flores, R.; Ruiz-Herrera, L.F.; López-Bucio, J. Serotonin modulates Arabidopsis root growth via changes in reactive oxygen species and jasmonic acid-ethylene signaling. Physiol. Plant 2016, 158, 92–105. [Google Scholar] [CrossRef]
- Irani, S.; Todd, C.D. Exogenous allantoin increases Arabidopsis seedlings tolerance to NaCl stress and regulates expression of oxidative stress response genes. J. Plant Physiol. 2018, 221, 43–50. [Google Scholar] [CrossRef]
- Hiltscher, H.; Rudnik, R.; Shaikhali, J.; Heiber, I.; Mellenthin, M.; Duarte, I.M.; Schuster, G.; Kahmann, U.; Baier, M. The radical induced cell death protein 1 (RCD1) supports transcriptional activation of genes for chloroplast antioxidant enzymes. Front. Plant Sci. 2014, 5, 475. [Google Scholar] [CrossRef] [PubMed]
- Kjaersgaard, T.; Jensen, M.K.; Christiansen, M.W.; Gregersen, P.; Kragelund, B.B.; Skriver, K. Senescence-associated barley NAC (NAM, ATAF1,2, CUC) transcription factor interacts with Radical-induced Cell Death 1 through a disordered regulatory domain. J. Biol. Chem. 2011, 286, 35418–35429. [Google Scholar] [CrossRef] [PubMed]
- Vainonen, J.P.; Jaspers, P.; Wrzaczek, M.; Lamminmäki, A.; Reddy, R.A.; Vaahtera, L.; Brosche, M.; Kangasjärvi, J. RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 2012, 442, 573–581. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, P.; Brosché, M.; Overmyer, K.; Kangasjärvi, J. The transcription factor interacting protein RCD1 contains a novel conserved domain. Plant Signal. Behav. 2010, 5, 78–80. [Google Scholar] [CrossRef]
- Tossavainen, H.; Hellman, M.; Vainonen, J.P.; Kangasjärvi, J.; Permi, P. 1H, 13C and 15N NMR chemical shift assignments of A. thaliana RCD1 RST. Biomol. NMR Assign. 2017, 11, 207–210. [Google Scholar] [CrossRef]
- O’Shea, C.; Staby, L.; Bendsen, S.K.; Tidemand, F.G.; Redsted, A.; Willemoes, M.; Kragelund, B.B.; Skriver, K. Structures and short linear motif of disordered transcription factor regions provide clues to the interactome of the cellular hub protein Radical-induced Cell Death1. J. Biol. Chem. 2017, 292, 512–527. [Google Scholar] [CrossRef] [PubMed]
- Kragelund, B.B.; Jensen, M.K.; Skriver, K. Order by disorder in plant signaling. Trends Plant Sci. 2012, 17, 625–632. [Google Scholar] [CrossRef]
- Teotia, S.; Lamb, R.S. RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 1271–1284. [Google Scholar] [CrossRef]
- Babajani, G.; Effendy, J.; Plant, A.L. Sl-SROl1 increases salt tolerance and is a member of the radical-induced cell death 1-similar to RCD1 gene family of tomato. Plant Sci. 2009, 176, 214–222. [Google Scholar] [CrossRef]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.-K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef]
- You, J.; Zong, W.; Li, X.; Ning, J.; Hu, H.; Li, X.; Xiao, J.; Xiong, L. The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 2013, 64, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. OsSRO1a interacts with RNA binding domain-containing protein (OsRBD1) and functions in abiotic stress tolerance in yeast. Front. Plant Sci. 2016, 7, 62. [Google Scholar] [CrossRef]
- Li, X.; Xu, Y.; Liu, F.; Zhao, M.; Sun, Y.; Ma, Q. Maize similar to RCD1 gene induced by salt enhances Arabidopsis thaliana abiotic stress resistance. Biochem. Biophys. Res. Commun. 2018, 503, 2625–2632. [Google Scholar] [CrossRef]
- Jiang, H.; Xiao, Y.; Zhu, S. Genome-wide identification, systematic analysis and characterization of SRO family genes in maize (Zea mays L.). Acta Physiol. Plant 2018, 40, 176. [Google Scholar] [CrossRef]
- Wu, Z.; Liang, J.; Zhang, S.; Zhang, B.; Zhao, Q.; Li, G.; Yang, X.; Wang, C.; He, J.; Yi, M. A canonical DREB2-type transcription factor in lily is post-translationally regulated and mediates heat stress response. Front. Plant Sci. 2018, 9, 243. [Google Scholar] [CrossRef]
- Wahlberg, E.; Karlberg, T.; Kouznetsova, E.; Markova, N.; Macchiarulo, A.; Thorsell, A.-G.; Pol, E.; Frostell, A.; Ekblad, T.; Öncü, D.; et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 2012, 30, 283–288. [Google Scholar] [CrossRef]
- Ramegowda, V.; Senthil-Kumar, M. The interactive effects of simultaneous biotic and abiotic stresses on plants: Mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 2015, 176, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Zhang, H.N.; Sonnewald, U. Differences and commonalities of plant responses to single and combined stresses. Plant J. 2017, 90, 839–855. [Google Scholar] [CrossRef] [PubMed]
| Reference | At2g31320 | At4g02390 |
|---|---|---|
| Suggested Nomenclature | AtPARP1 | AtPARP2 |
| Boltz et al. (2014), PLoS One 9: e88872 [97] | AtPARP2 | AtPARP1 |
| Briggs and Bent (2011), Trends Plant Sci. 16: 372–380 [98] | AtPARP2 | AtPARP1 |
| Chen et al. (2018), Front. Plant Sci. 9: 1581 [94] | AtPARP1 | AtPARP2 |
| De Block et al. (2005), Plant J. 41: 95–106 [99] | AtPARP2 | AtPARP1 |
| Doucet-Chabeaud et al. (2001), Mol. Genet. Genom. 265: 954–963 [95] | AtPARP1 | AtPARP2 |
| Jia et al. (2013), Plant Mol. Biol. 82: 339–351 [100] | AtPARP1 | AtPARP2 |
| Feng et al. (2015), PLoS Genet. 11: e1004936 [101] | AtPARP1 | AtPARP2 |
| Keppler et al. (2018), Front Plant Sci. 9: 1907 [102] | AtPARP1 | AtPARP2 |
| Lamb et al. (2012), Cell Mol. Life Sci. 69: 175–189 [103] | AtPARP2 | AtPARP1 |
| Ogawa et al. (2009), Plant J. 57: 289–301 [104] | AtPARP1 | AtPARP2 |
| Pellny et al. (2009), Mol. Plant 2: 442–456 [105] | AtPARP1 | AtPARP2 |
| Pham et al. (2015), Plant Mol. Biol. 89: 319–338 [93] | AtPARP2 | AtPARP1 |
| Rissel et al. (2017), Front. Plant Sci. 8: 59 [47] | AtPARP1 | AtPARP2 |
| Rissel et al. (2017), Anal. Biochem. 527: 20–23 [96] | AtPARP1 | AtPARP2 |
| Schulz et al. (2012), PLoS One 7: e37287 [106] | AtPARP2 | AtPARP1 |
| Song et al. (2015), PLoS Genet. 11: e1005200 [92] | AtPARP1 | AtPARP2 |
| Vanderauwera et al. (2007), PNAS 104: 15150–15155 [107] | AtPARP2 | AtPARP1 |
| Zhang et al. (2015), Sci. Rep. 5:15892 [108] | AtPARP1 | AtPARP2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rissel, D.; Peiter, E. Poly(ADP-Ribose) Polymerases in Plants and Their Human Counterparts: Parallels and Peculiarities. Int. J. Mol. Sci. 2019, 20, 1638. https://doi.org/10.3390/ijms20071638
Rissel D, Peiter E. Poly(ADP-Ribose) Polymerases in Plants and Their Human Counterparts: Parallels and Peculiarities. International Journal of Molecular Sciences. 2019; 20(7):1638. https://doi.org/10.3390/ijms20071638
Chicago/Turabian StyleRissel, Dagmar, and Edgar Peiter. 2019. "Poly(ADP-Ribose) Polymerases in Plants and Their Human Counterparts: Parallels and Peculiarities" International Journal of Molecular Sciences 20, no. 7: 1638. https://doi.org/10.3390/ijms20071638
APA StyleRissel, D., & Peiter, E. (2019). Poly(ADP-Ribose) Polymerases in Plants and Their Human Counterparts: Parallels and Peculiarities. International Journal of Molecular Sciences, 20(7), 1638. https://doi.org/10.3390/ijms20071638





