A Temporal Order in 5′- and 3′- Processing of Eukaryotic tRNAHis
Abstract
1. Introduction
2. Results
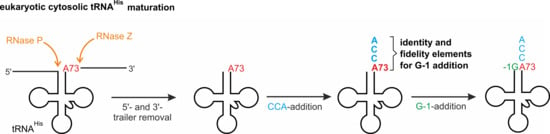
2.1. Eukaryotic Cytosolic tRNAHis Processing—A Temporal Order of 5′- and 3′- Nucleotide Incorporation
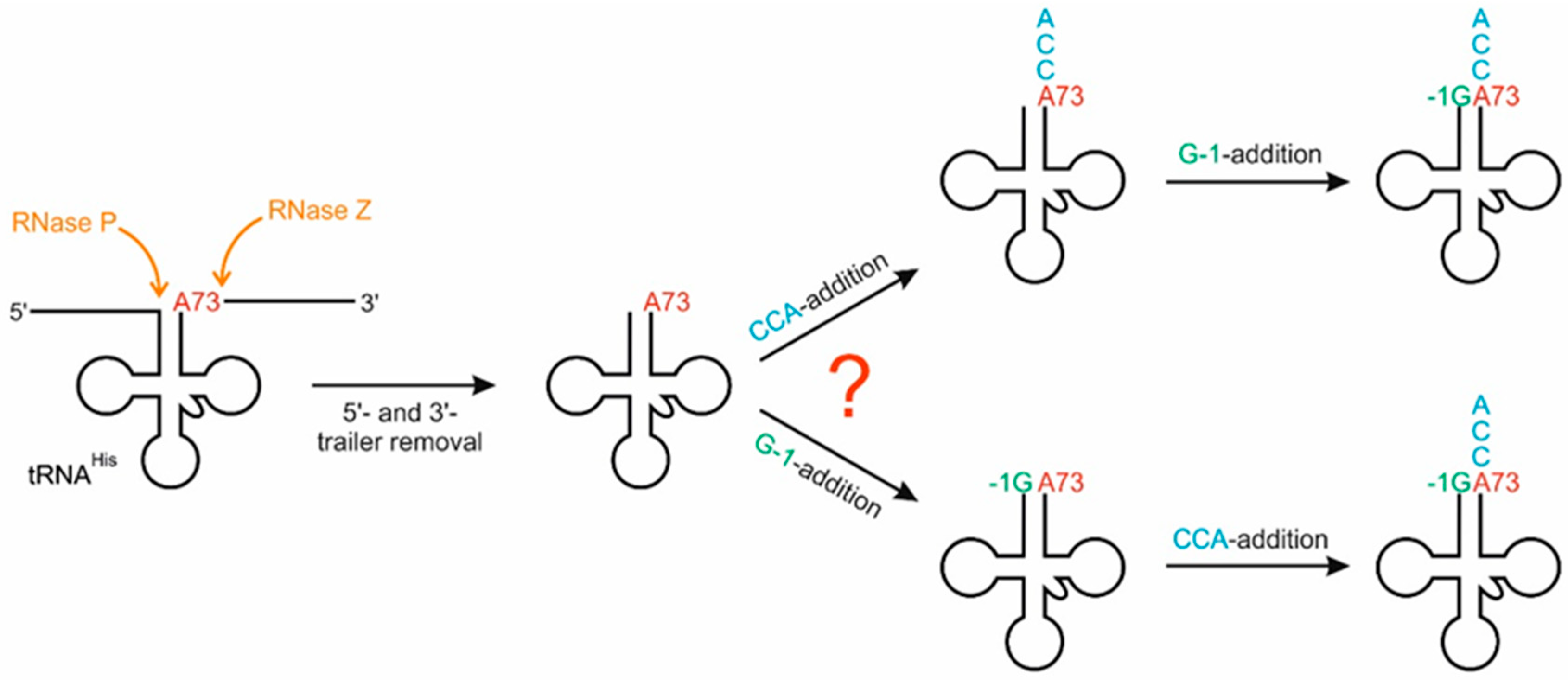
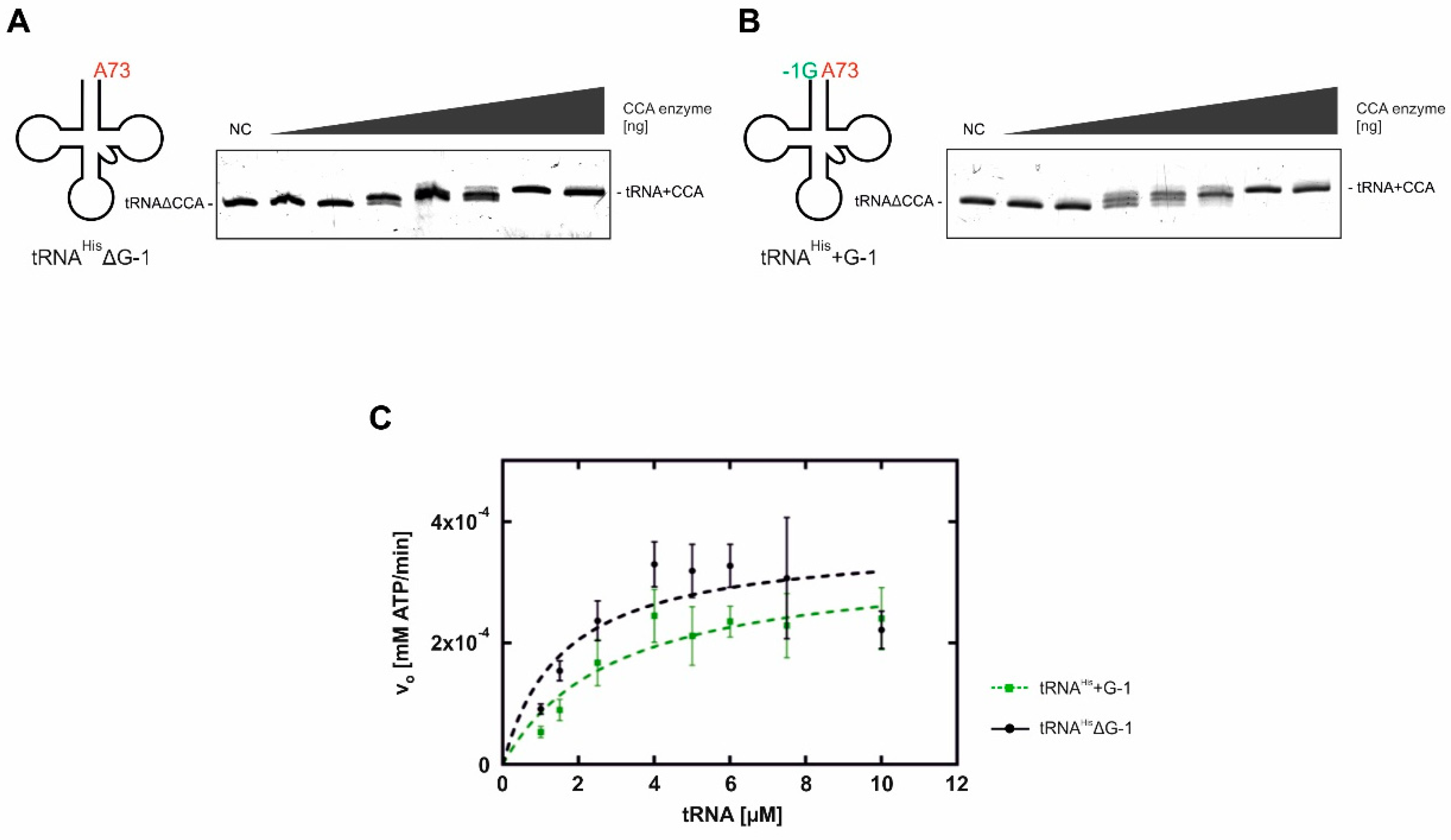
2.1.1. Addition of the 3′-terminal CCA Sequence
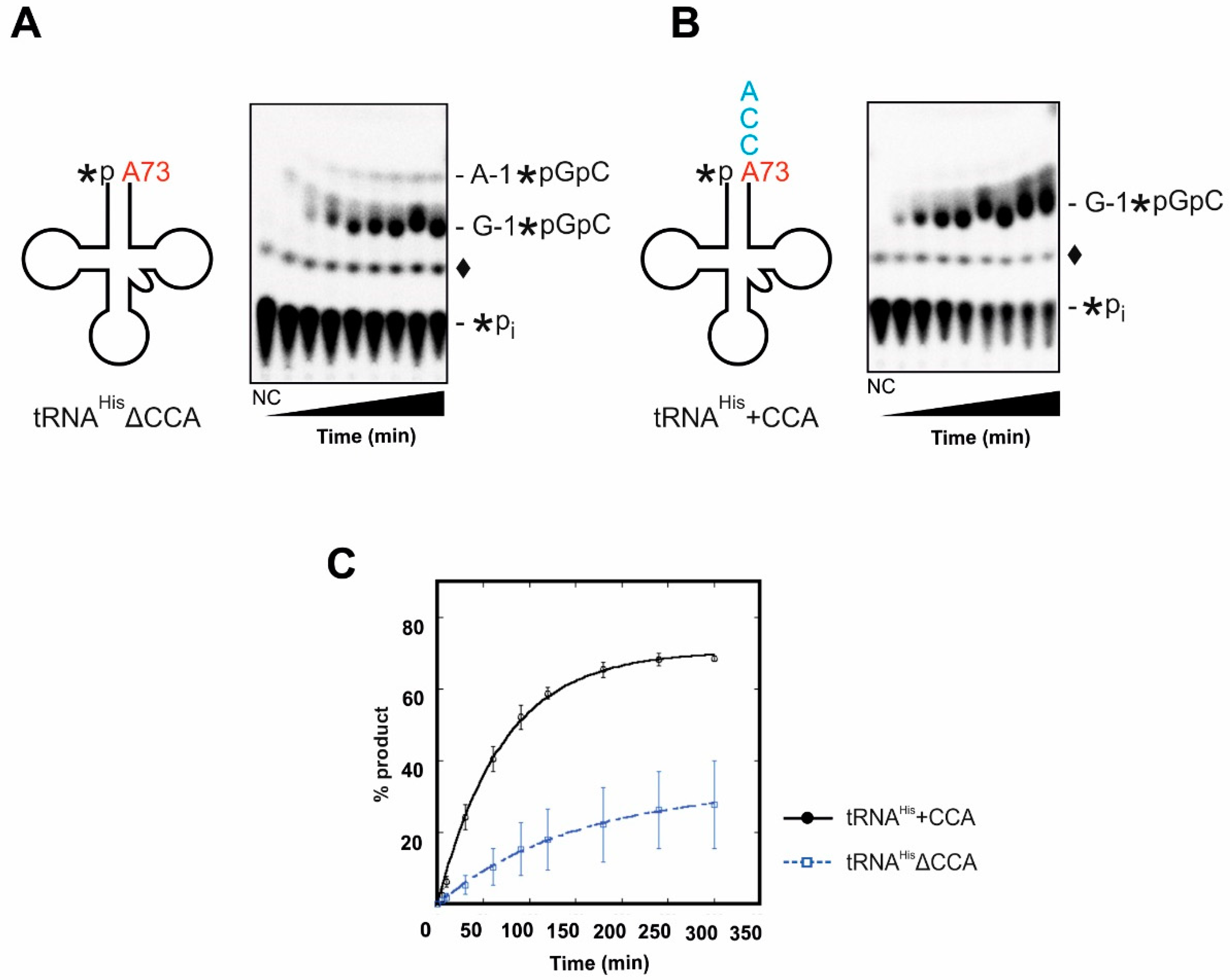
2.1.2. Incorporation of the G-1 Nucleotide
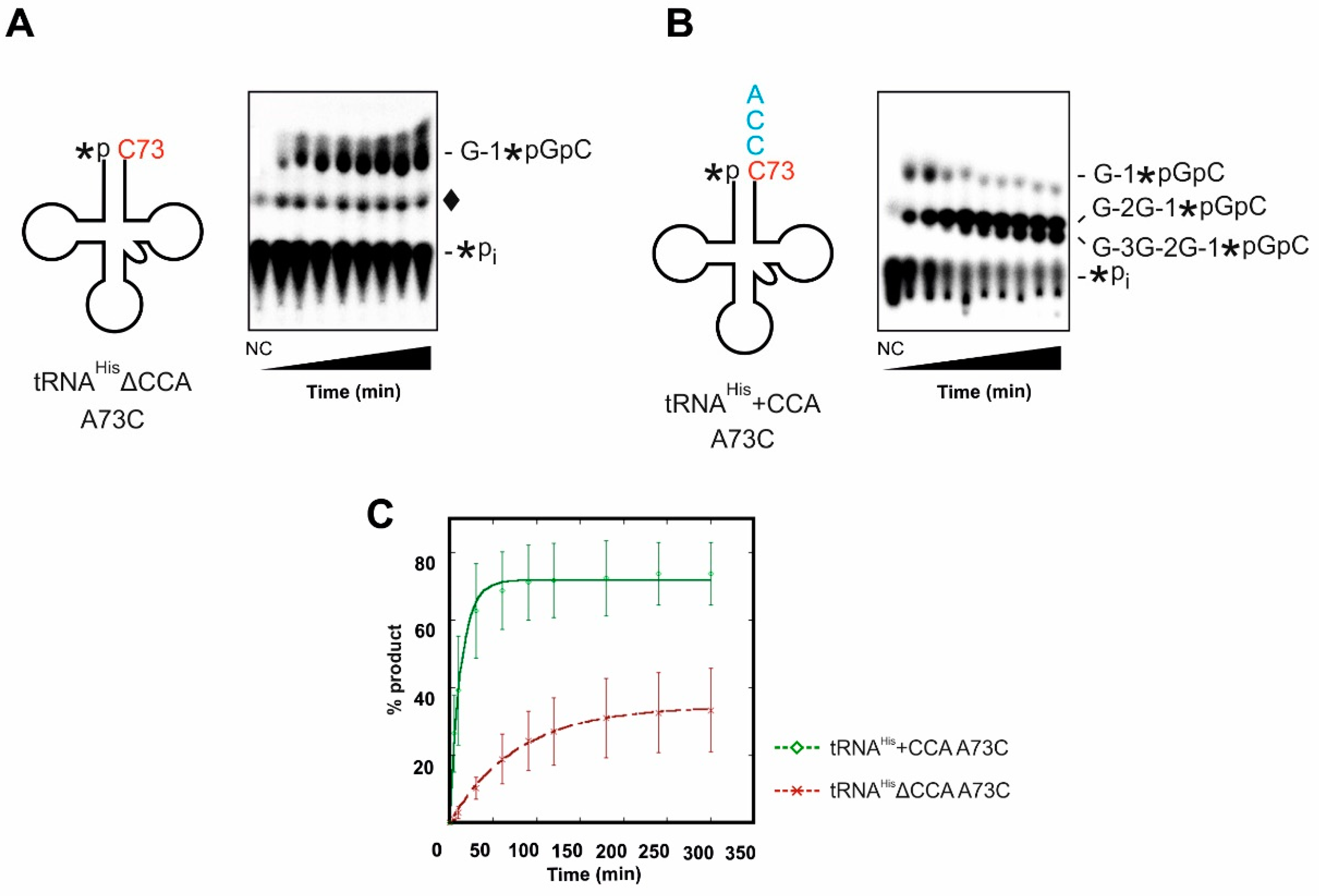
2.2. The 3′- A73CCA Sequence Serves as a Fidelity Determinant for Thg1
3. Discussion
4. Material and Methods
4.1. Preparation of Recombinat Enzymes
4.1.1. Yeast CCA-adding Enzyme
4.1.2. Yeast tRNAHis Guanylyltransferase (Thg1)
4.2. Preparation of tRNA Substrates
4.3. In Vitro CCA-Addition
4.4. In Vitro G-1 Addition: Phosphate Protection Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| aaRS | aminoacyl-tRNA synthetase |
| DTT | dithiothreitol |
| EDTA | 2,2′,2″,2‴-(Ethane-1,2-diyldinitrilo)tetraacetic acid |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HisRS | histidyl-tRNA synthetase |
| PEI | Polyethylenimine |
| Thg1 | tRNAHis guanylyltransferase |
| TLC | thin layer chromatography |
References
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed]
- Hopper, A.K. Transfer RNA Post-Transcriptional Processing, Turnover, and Subcellular Dynamics in the Yeast Saccharomyces cerevisiae. Genetics 2013, 194, 43–67. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.K.; Gößringer, M.; Späth, B.; Fischer, S.; Marchfelder, A. Chapter 8 The Making of tRNAs and More—RNase P and tRNase Z. In Molecular Biology of RNA Processing and Decay in Prokaryotes; Condon, C., Ed.; Elsevier: Amsterdam, The Netherlands, 2009; Volume 85, pp. 319–368. [Google Scholar]
- Zhu, L.; Deutscher, M.P. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J. 1987, 6, 2473–2477. [Google Scholar] [CrossRef] [PubMed]
- Betat, H.; Rammelt, C.; Mörl, M. tRNA nucleotidyltransferases: Ancient catalysts with an unusual mechanism of polymerization. Cell. Mol. Life Sci. 2010, 67, 1447–1463. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Steitz, T.A. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature 2004, 430, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xiong, Y.; Wang, J.; Cho, H.D.; Tomita, K.; Weiner, A.M.; Steitz, T.A. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell 2002, 111, 815–824. [Google Scholar] [CrossRef]
- Wellner, K.; Betat, H.; Mörl, M. A tRNA’s fate is decided at its 3′ end: Collaborative actions of CCA-adding enzyme and RNases involved in tRNA processing and degradation. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Betat, H.; Mörl, M. The CCA-adding enzyme: A central scrutinizer in tRNA quality control. BioEssays 2015, 37, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Whipple, J.M.; Phizicky, E.M.; Sharp, P.A. tRNAs marked with CCACCA are targeted for degradation. Science 2011, 334, 817–821. [Google Scholar] [CrossRef]
- Hou, Y.-M. CCA Addition to tRNA: Implications for tRNA Quality Control. IUBMB Life 2010, 62, 251–260. [Google Scholar] [CrossRef]
- Wang, C.; Sobral, B.W.; Williams, K.P. Loss of a universal tRNA feature. J. Bacteriol. 2007, 189, 1954–1962. [Google Scholar] [CrossRef]
- Rao, B.S.; Mohammad, F.; Gray, M.W.; Jackman, J.E. Absence of a universal element for tRNAHis identity in Acanthamoeba castellanii. Nucleic Acids Res. 2013, 41, 1885–1894. [Google Scholar] [CrossRef]
- Orellana, O.; Cooley, L.; Söll, D. The additional guanylate at the 5′ terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol. Cell. Biol. 1986, 6, 525–529. [Google Scholar] [CrossRef]
- Jackman, J.E.; Phizicky, E.M. tRNAHis guanylyltransferase adds G-1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA 2006, 12, 1007–1014. [Google Scholar] [CrossRef]
- Gu, W.; Jackman, J.E.; Lohan, A.J.; Gray, M.W.; Phizicky, E.M. tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev. 2003, 17, 2889–2901. [Google Scholar] [CrossRef]
- Ghaemmaghami, S.; Huh, W.-K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef]
- Tomita, K.; Ishitani, R.; Fukai, S.; Nureki, O. Complete crystallographic analysis of the dynamics of CCA sequence addition. Nature 2006, 443, 956–960. [Google Scholar] [CrossRef]
- Ernst, F.G.M.; Erber, L.; Sammler, J.; Jühling, F.; Betat, H.; Mörl, M. Cold adaptation of tRNA nucleotidyltransferases: A tradeoff in activity, stability and fidelity. RNA Biol. 2018, 15, 144–155. [Google Scholar] [CrossRef]
- Cho, H.D.; Verlinde, C.L.; Weiner, A.M. Archaeal CCA-adding enzymes: Central role of a highly conserved beta-turn motif in RNA polymerization without translocation. J. Biol. Chem. 2005, 280, 9555–9566. [Google Scholar] [CrossRef]
- Betat, H.; Rammelt, C.; Martin, G.; Mörl, M. Exchange of regions between bacterial poly(A) polymerase and the CCA-adding enzyme generates altered specificities. Mol. Cell 2004, 15, 389–398. [Google Scholar] [CrossRef]
- Jahn, D.; Pande, S. Histidine tRNA guanylyltransferase from Saccharomyces cerevisiae. II. Catalytic mechanism. J. Biol. Chem. 1991, 266, 22832–22836. [Google Scholar]
- Abad, M.G.; Rao, B.S.; Jackman, J.E. Template-dependent 3′-5′ nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc. Natl. Acad. Sci. USA 2010, 107, 674–679. [Google Scholar] [CrossRef]
- Jackman, J.E.; Gott, J.M.; Gray, M.W. Doing it in reverse: 3′-to-5′ polymerization by the Thg1 superfamily. RNA 2012, 18, 886–899. [Google Scholar] [CrossRef]
- Jackman, J.E.; Phizicky, E.M. tRNAHis guanylyltransferase catalyzes a 3′-5′ polymerization reaction that is distinct from G-1 addition. Proc. Natl. Acad. Sci. USA 2006, 103, 8640–8645. [Google Scholar] [CrossRef]
- Chatterjee, K.; Nostramo, R.T.; Wan, Y.; Hopper, A.K. tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 373–386. [Google Scholar] [CrossRef]
- Wende, S.; Bonin, S.; Götze, O.; Betat, H.; Mörl, M. The identity of the discriminator base has an impact on CCA addition. Nucleic Acids Res. 2015, 43, 5617–5629. [Google Scholar] [CrossRef]
- Wolf, J.; Obermaier-Kusser, B.; Jacobs, M.; Milles, C.; Mörl, M.; von Pein, H.D.; Grau, A.J.; Bauer, M.F. A new mitochondrial point mutation in the transfer RNA(Lys) gene associated with progressive external ophthalmoplegia with impaired respiratory regulation. J. Neurol. Sci. 2012, 316, 108–111. [Google Scholar] [CrossRef]
- Williams, K.R.; Schofield, P. Kinetic mechanism of tRNA nucleotidyltransferase from Escherichia coli. J. Biol. Chem. 1977, 252, 5589–5597. [Google Scholar]
- Giegé, R.; Jühling, F.; Pütz, J.; Stadler, P.; Sauter, C.; Florentz, C. Structure of transfer RNAs: Similarity and variability. Wiley Interdiscip. Rev. RNA 2012, 3, 37–61. [Google Scholar] [CrossRef]
- Lizano, E.; Scheibe, M.; Rammelt, C.; Betat, H.; Mörl, M. A comparative analysis of CCA-adding enzymes from human and E. coli: Differences in CCA addition and tRNA 3′-end repair. Biochimie 2008, 90, 762–772. [Google Scholar] [CrossRef]
- Li, Z.; Sun, Y.; Thurlow, D.L. RNA minihelices as model substrates for ATP/CTP:tRNA nucleotidyltransferase. Biochem. J. 1997, 327, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Nameki, N.; Asahara, H.; Shimizu, M.; Okada, N.; Himeno, H. Identity elements of Saccharomyces cerevisiae tRNA(His). Nucleic Acids Res. 1995, 23, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Rosen, A.E.; Brooks, B.S.; Guth, E.; Francklyn, C.S.; Musier-Forsyth, K. Evolutionary conservation of a functionally important backbone phosphate group critical for aminoacylation of histidine tRNAs. RNA 2006, 12, 1315–1322. [Google Scholar] [CrossRef]
- Shigematsu, M.; Kirino, Y. 5′-Terminal nucleotide variations in human cytoplasmic tRNAHisGUG and its 5′-halves. RNA 2017, 23, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Traut, T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Preston, M.A.; Phizicky, E.M. The requirement for the highly conserved G-1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase. RNA 2010, 16, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Ohmichi, T.; Nakano, S.-I.; Miyoshi, D.; Sugimoto, N. Long RNA Dangling End Has Large Energetic Contribution to Duplex Stability. J. Am. Chem. Soc. 2002, 124, 10367–10372. [Google Scholar] [CrossRef]
- Betat, H.; Long, Y.; Jackman, J.E.; Mörl, M. From end to end: TRNA editing at 5′- and 3′-terminal positions. Int. J. Mol. Sci. 2014, 15, 23975–23998. [Google Scholar] [CrossRef]
- Sibler, A.P.; Martin, R.P.; Dirheimer, G. The nucleotide sequence of yeast mitochondrial histidine-tRNA. FEBS Lett. 1979, 107, 182–186. [Google Scholar] [CrossRef]
- De Wijn, R.; Hennig, O.; Ernst, F.G.M.; Lorber, B.; Betat, H.; Mörl, M.; Sauter, C. Combining crystallogenesis methods to produce diffraction-quality crystals of a psychrophilic tRNA-maturation enzyme. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018, 74, 747–753. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mörl, M.; Lizano, E.; Willkomm, D.K.; Hartmann, R.K. Production of RNAs with Homogeneous 5′- and 3′-Ends. In Handbook of RNA Biochemistry; Hartmann, R.K., Bindereif, A., Schön, A., Westhof, E., Eds.; Wiley-VCH: Weinheim, Germany, 2005; pp. 22–35. [Google Scholar]
- Schürer, H.; Lang, K.; Schuster, J.; Mörl, M. A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res. 2002, 30, e56. [Google Scholar] [CrossRef]
- Tomita, K.; Fukai, S.; Ishitani, R.; Ueda, T.; Takeuchi, N.; Vassylyev, D.G.; Nureki, O. Structural basis for template-independent RNA polymerization. Nature 2004, 430, 700–704. [Google Scholar] [CrossRef]
- Smith, B.A.; Jackman, J.E. Kinetic analysis of 3′-5′ nucleotide addition catalyzed by eukaryotic tRNA(His) guanylyltransferase. Biochemistry 2012, 51, 453–465. [Google Scholar] [CrossRef]
| Substrate | KM [µM] | kcat [min−1] |
|---|---|---|
| tRNAHis∆G-1 | 1.6 ± 0.6 | 27.5 ± 3.3 |
| tRNAHis+G-1 | 2.9 ± 1.0 | 25.1 ± 3.3 |
| tRNAHis+CCA | tRNAHis∆CCA | tRNAHis+CCA A73C | tRNAHis∆CCA A73C | |
|---|---|---|---|---|
| kobs (min−1) | 0.014 ± 0.001 | 0.0070 ± 0.0004 | 0.097 ± 0.008 | 0.013 ± 0.001 |
| KD,app,tRNA (µM) | 5.8 ± 4.3 | >30 * | 3.5 ± 1.7 | >30 * |
| kmax/KD,app (µM−1min−1) | 385 ± 45 | ND | 110 ± 7.7 | ND |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pöhler, M.-T.; Roach, T.M.; Betat, H.; Jackman, J.E.; Mörl, M. A Temporal Order in 5′- and 3′- Processing of Eukaryotic tRNAHis. Int. J. Mol. Sci. 2019, 20, 1384. https://doi.org/10.3390/ijms20061384
Pöhler M-T, Roach TM, Betat H, Jackman JE, Mörl M. A Temporal Order in 5′- and 3′- Processing of Eukaryotic tRNAHis. International Journal of Molecular Sciences. 2019; 20(6):1384. https://doi.org/10.3390/ijms20061384
Chicago/Turabian StylePöhler, Marie-Theres, Tracy M. Roach, Heike Betat, Jane E. Jackman, and Mario Mörl. 2019. "A Temporal Order in 5′- and 3′- Processing of Eukaryotic tRNAHis" International Journal of Molecular Sciences 20, no. 6: 1384. https://doi.org/10.3390/ijms20061384
APA StylePöhler, M.-T., Roach, T. M., Betat, H., Jackman, J. E., & Mörl, M. (2019). A Temporal Order in 5′- and 3′- Processing of Eukaryotic tRNAHis. International Journal of Molecular Sciences, 20(6), 1384. https://doi.org/10.3390/ijms20061384