Identification of miRNA Reference Genes in Extracellular Vesicles from Adipose Derived Mesenchymal Stem Cells for Studying Osteoarthritis
Abstract
1. Introduction
2. Results
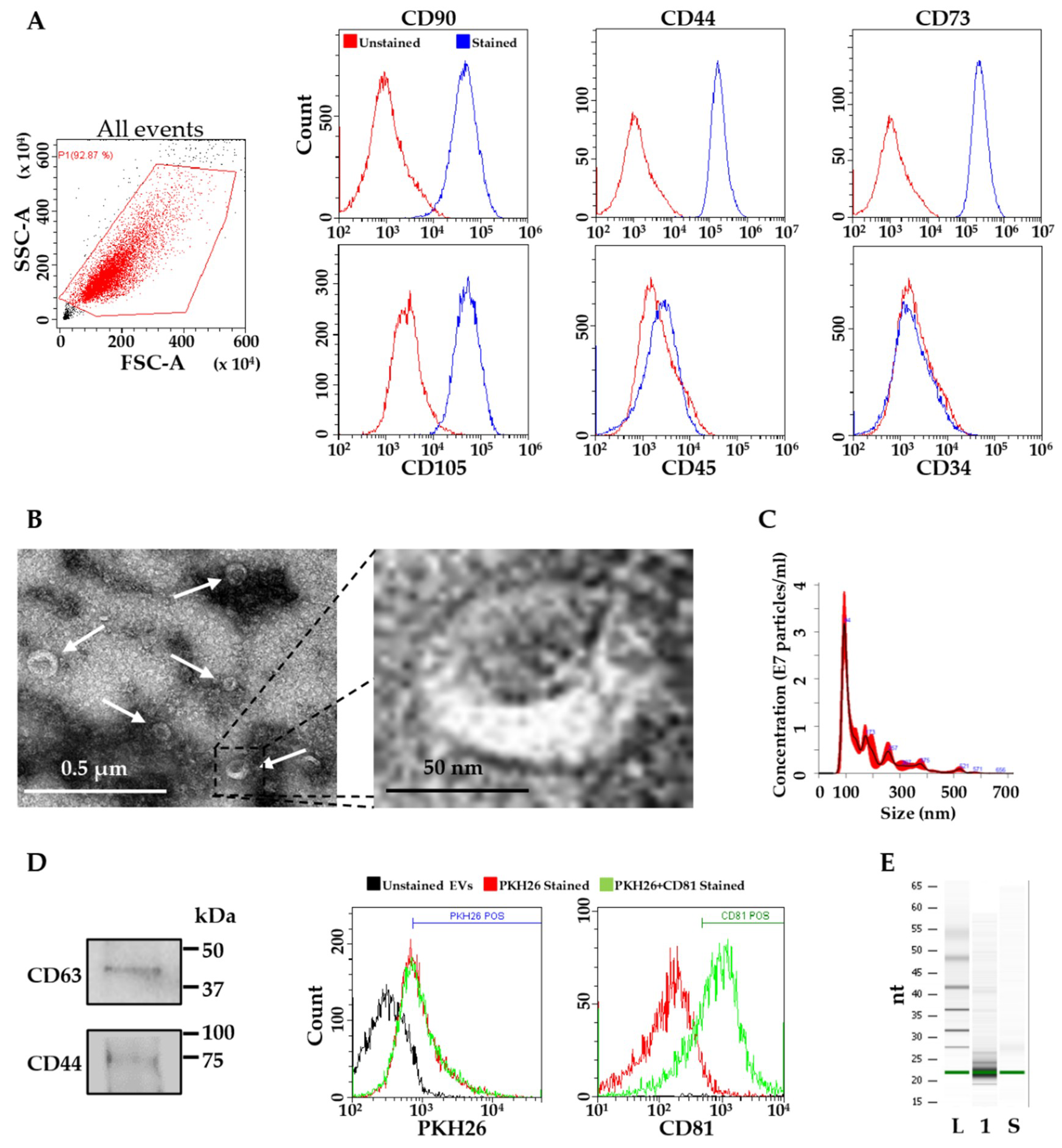
2.1. ASCs and EVs Characterization
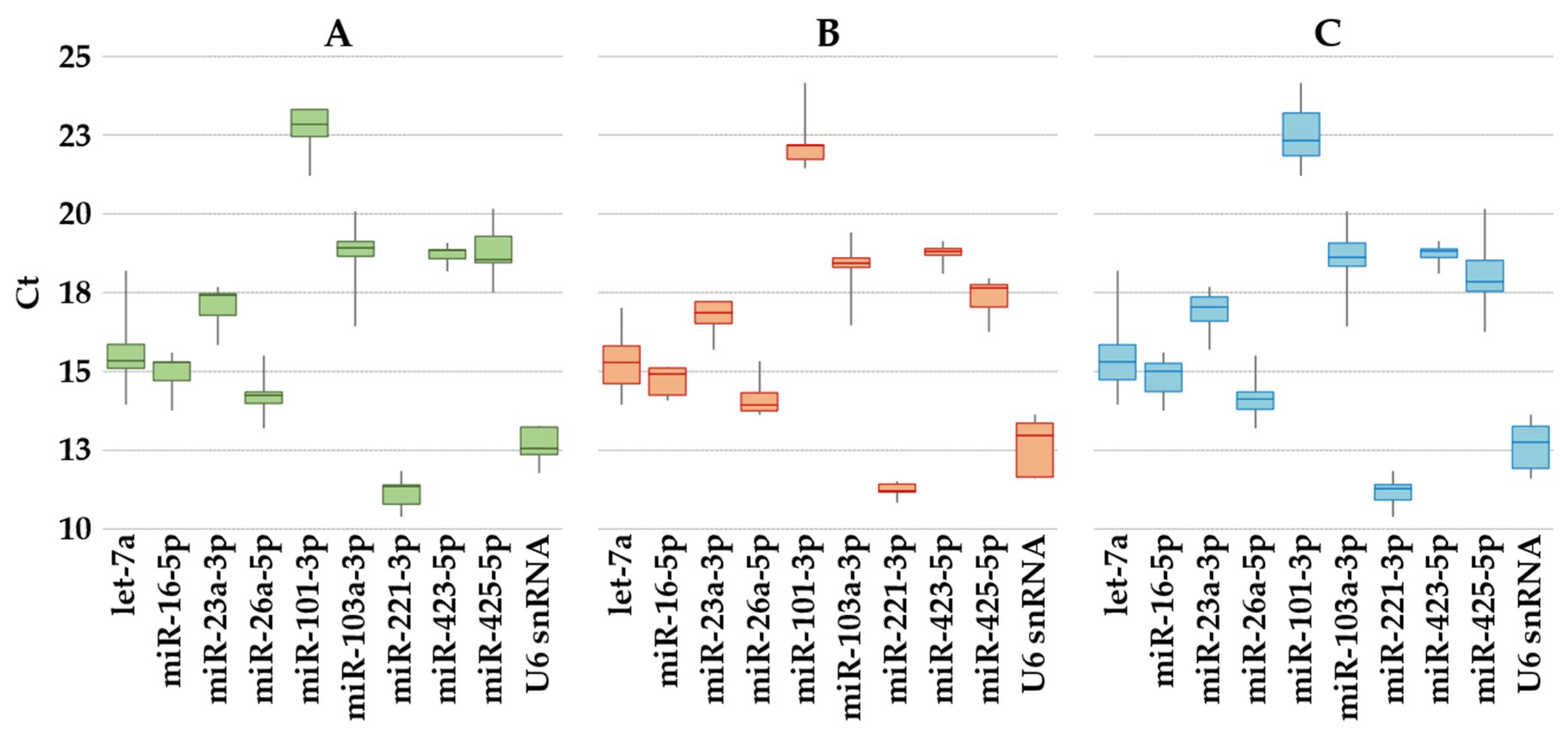
2.2. Expression of Candidate Reference Genes
2.3. Reference Genes Expression Stability Analysis
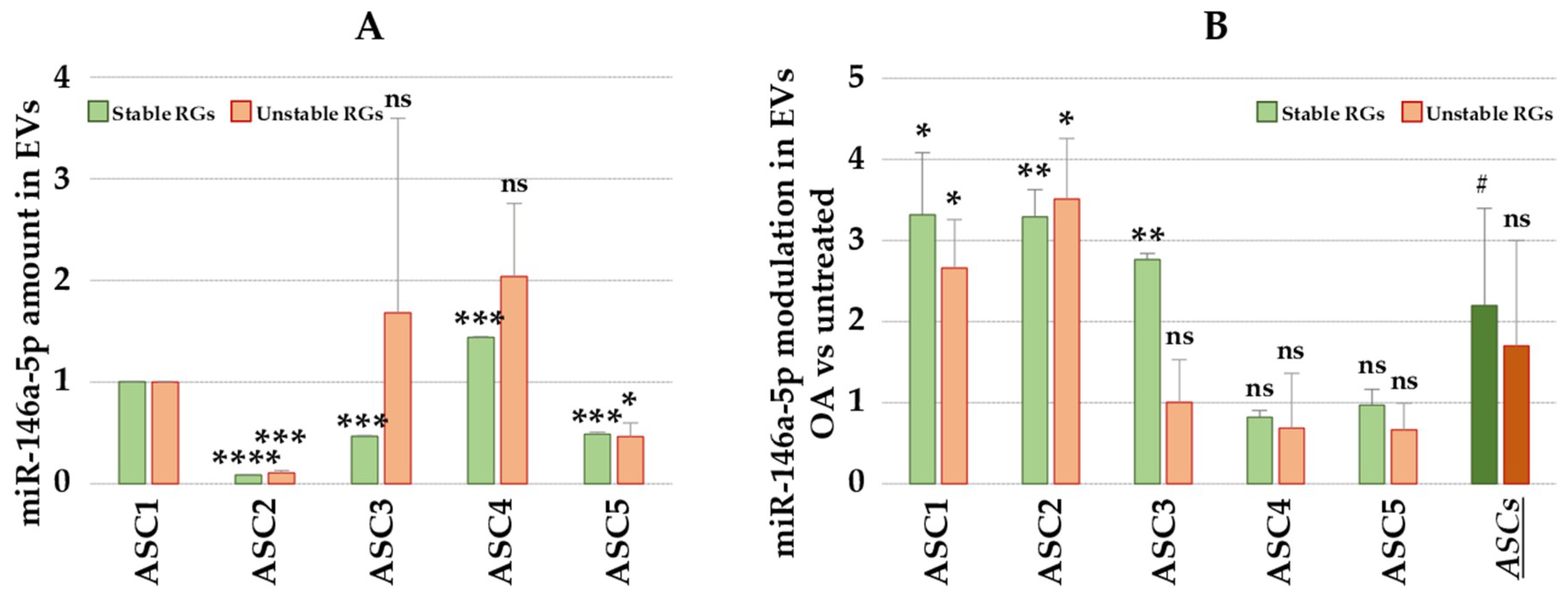
2.4. Impact of RGs Choice on the Expression Levels of Target Genes
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. ASCs Isolation and Expansion
4.3. ASC-EVs Isolation and Characterization
4.4. Selection of Candidate Reference Genes
4.5. Total RNA Isolation and miRNA Profiling
4.6. Data Analysis
4.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MSCs | Mesenchymal stem cells |
| ASCs | Adipose derived-MSCs |
| EVs | Extracellular vesicles |
| miRNA | microRNA |
| qRT-PCR | Quantitative-real time polymerase chain reaction |
| RGs | Reference genes |
| OA | Osteoarthritis |
References
- Cross, M.; Smith, E.; Hoy, D.; Nolte, S.; Ackerman, I.; Fransen, M.; Bridgett, L.; Williams, S.; Guillemin, F.; Hill, C.L.; et al. The global burden of hip and knee osteoarthritis: Estimates from the global burden of disease 2010 study. Ann. Rheum. Dis. 2014, 73, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Turkiewicz, A.; Petersson, I.F.; Björk, J.; Hawker, G.; Dahlberg, L.E.; Lohmander, L.S.; Englund, M. Current and future impact of osteoarthritis on health care: A population-based study with projections to year 2032. Osteoarthr. Cartil. 2014, 22, 1826–1832. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, C.; Nguyen, C.; Lefevre-Colau, M.M.; Rannou, F.; Poiraudeau, S. Risk factors and burden of osteoarthritis. Ann. Phys. Rehabil. Med. 2016, 59, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Wittenauer, R.; Smith, L.; Aden, K. Background Paper 6.12 Osteoarthritis; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Balmaceda, C.M. Evolving guidelines in the use of topical nonsteroidal anti-inflammatory drugs in the treatment of osteoarthritis. BMC Musculoskelet. Disord. 2014, 21, 27. [Google Scholar] [CrossRef] [PubMed]
- Sinusas, K. Osteoarthritis: Diagnosis and treatment. Am. Fam. Physician 2012, 85, 49–56. [Google Scholar]
- Barry, F.; Murphy, M. Mesenchymal stem cells in joint disease and repair. Nat. Rev. Rheumatol. 2013, 9, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Lopa, S.; Colombini, A.; Moretti, M.; de Girolamo, L. Injective mesenchymal stem cell-based treatments for knee osteoarthritis: From mechanisms of action to current clinical evidences. Knee Surg. Sports Traumatol. Arthrosc. 2018. [Google Scholar] [CrossRef] [PubMed]
- Caplan, A. New MSC: MSCs as pericytes are Sentinels and gatekeepers. J. Orthop. Res. 2017, 35, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Marmotti, A.; de Girolamo, L.; Bonasia, D.E.; Bruzzone, M.; Mattia, S.; Rossi, R.; Montaruli, A.; Dettoni, F.; Castoldi, F.; Peretti, G. Bone marrow derived stem cells in joint and bone diseases: A concise review. Int. Orthop. 2014, 38, 1787–1801. [Google Scholar] [CrossRef] [PubMed]
- Hass, R.; Kasper, C.; Böhm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 2011, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Iijima, H.; Isho, T.; Kuroki, H.; Takahashi, M.; Aoyama, T. Effectiveness of mesenchymal stem cells for treating patients with knee osteoarthritis: A meta-analysis toward the establishment of effective regenerative rehabilitation. NPJ Regen. Med. 2018, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Jevotovsky, D.S.; Alfonso, A.R.; Einhorn, T.A.; Chiu, E.S. Osteoarthritis and stem cell therapy in humans: A systematic review. Osteoarthr. Cartil. 2018, 26, 711–729. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.R.; Teixeira, G.Q.; Santos, S.G.; Barbosa, M.A.; Almeida-Porada, G.; Gonçalves, R.M. Mesenchymal Stromal Cell Secretome: Influencing Therapeutic Potential by Cellular Pre-conditioning. Front. Immunol. 2018, 9, 2837. [Google Scholar] [CrossRef] [PubMed]
- de Girolamo, L.; Kon, E.; Filardo, G.; Marmotti, A.G.; Soler, F.; Peretti, G.M.; Vannini, F.; Madry, H.; Chubinskaya, S. Regenerative approaches for the treatment of early OA. Knee Surg. Sports Traumatol. Arthrosc. 2016, 24, 1826–1835. [Google Scholar] [CrossRef] [PubMed]
- Phinney, D.G.; Pittenger, M.F. Concise Review: MSC-Derived Exosomes for Cell-Free Therapy. Stem Cells 2017, 35, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, S.; Ruiz, M.; Toupet, K.; Jorgensen, C.; Noël, D. Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Sci. Rep. 2017, 7, 16214. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Y.; Zhao, B.; Niu, X.; Hu, B.; Li, Q.; Zhang, J.; Ding, J.; Chen, Y.; Wang, Y. Comparison of exosomes secreted by induced pluripotent stem cell-derived mesenchymal stem cells and synovial membrane-derived mesenchymal stem cells for the treatment of osteoarthritis. Stem Cell Res. Ther. 2017, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chu, W.C.; Lai, R.C.; Lim, S.K.; Hui, J.H.; Toh, W.S. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthr. Cartil. 2016, 24, 2135–2140. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, D.; Liu, Z.; Zhou, F.; Dai, J.; Wu, B.; Zhou, J.; Heng, B.C.; Zou, X.H.; Ouyang, H.; et al. Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res. Ther. 2017, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.C.; Tao, S.C.; Yin, W.J.; Qi, X.; Sheng, J.G.; Zhang, C.Q. Exosomes from Human Synovial-Derived Mesenchymal Stem Cells Prevent Glucocorticoid-Induced Osteonecrosis of the Femoral Head in the Rat. Int. J. Biol. Sci. 2016, 12, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Casado, J.G.; Blázquez, R.; Vela, F.J.; Álvarez, V.; Tarazona, R.; Sánchez-Margallo, F.M. Mesenchymal Stem Cell-Derived Exosomes: Immunomodulatory Evaluation in an Antigen-Induced Synovitis Porcine Model. Front. Vet. Sci. 2017, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Viganò, M.; Rebulla, P.; Giordano, R.; Lazzari, L. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: How to choose the most reliable housekeeping genes. J. Cell. Mol. Med. 2013, 17, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Banfi, F.; Colombini, A.; Perucca Orfei, C.; Parazzi, V.; Ragni, E. Validation of reference and identity-defining genes in human mesenchymal stem cells cultured under unrelated fetal bovine serum batches for basic science and clinical application. Stem Cell Rev. 2018, 14, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Viganò, M.; Perucca Orfei, C.; de Girolamo, L.; Pearson, J.R.; Ragni, E.; De Luca, P.; Colombini, A. Housekeeping Gene Stability in Human Mesenchymal Stem and Tendon Cells Exposed to Tenogenic Factors. Tissue Eng. Part C Methods 2018, 24, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, L.; Liu, F.; Xiang, G.; Jiang, D.; Pu, X. Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Dis. Markers 2015, 2015, 893594. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiang, G.M.; Liu, L.L.; Liu, C.; Liu, F.; Jiang, D.N.; Pu, X.Y. Assessment of endogenous reference gene suitability for serum exosomal microRNA expression analysis in liver carcinoma resection studies. Mol. Med. Rep. 2015, 12, 4683–4691. [Google Scholar] [CrossRef] [PubMed]
- Cazzoli, R.; Buttitta, F.; Di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. miRNA in plasma exosome is stable under different storage conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef] [PubMed]
- Lange, T.; Stracke, S.; Rettig, R.; Lendeckel, U.; Kuhn, J.; Schlüter, R.; Rippe, V.; Endlich, K.; Endlich, N. Identification of miR-16 as an endogenous reference gene for the normalization of urinary exosomal miRNA expression data from CKD patients. PLoS ONE 2017, 12, e0183435. [Google Scholar] [CrossRef] [PubMed]
- Gouin, K.; Peck, K.; Antes, T.; Johnson, J.L.; Li, C.; Vaturi, S.D.; Middleton, R.; de Couto, G.; Walravens, A.S.; Rodriguez-Borlado, L.; et al. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1347019. [Google Scholar] [CrossRef] [PubMed]
- Santovito, D.; De Nardis, V.; Marcantonio, P.; Mandolini, C.; Paganelli, C.; Vitale, E.; Buttitta, F.; Bucci, M.; Mezzetti, A.; Consoli, A.; et al. Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in diabetes: Effect of glycemic control. J. Clin. Endocrinol. Metab. 2014, 99, E1681–E1685. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Yang, T. Effective enrichment of urinary exosomes by polyethylene glycol for RNA detection. Biomed. Res. 2018, 29. [Google Scholar] [CrossRef]
- Baglio, S.R.; Rooijers, K.; Koppers-Lalic, D.; Verweij, F.J.; Pérez Lanzón, M.; Zini, N.; Naaijkens, B.; Perut, F.; Niessen, H.W.; Baldini, N.; et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Domenis, R.; Cifù, A.; Quaglia, S.; Pistis, C.; Moretti, M.; Vicario, A.; Parodi, P.C.; Fabris, M.; Niazi, K.R.; Soon-Shiong, P.; et al. Pro inflammatory stimuli enhance the immunosuppressive functions of adipose mesenchymal stem cells-derived exosomes. Sci. Rep. 2018, 8, 13325. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Wu, K.C.; Harn, H.J.; Lin, S.Z.; Ding, D.C. Exosomes and Stem Cells in Degenerative Disease Diagnosis and Therapy. Cell Transplant. 2018, 27, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Suo, C.; Salim, A.; Chia, K.S.; Pawitan, Y.; Calza, S. Modified least-variant set normalization for miRNA microarray. RNA 2010, 16, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Coenen-Stass, A.M.; Wood, M.J. Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS ONE 2014, 9, e89237. [Google Scholar] [CrossRef] [PubMed]
- Sewer, A.; Gubian, S.; Kogel, U.; Veljkovic, E.; Han, W.; Hengstermann, A.; Peitsch, M.C.; Hoeng, J. Assessment of a novel multi-array normalization method based on spike-in control probes suitable for microRNA datasets with global decreases in expression. BMC Res. Notes 2014, 7, 302. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.U.; Pfaffl, M.W.; Ulbrich, S.E. Normalization strategies for microRNA profiling experiments: A ‘normal’ way to a hidden layer of complexity? Biotechnol. Lett. 2010, 32, 1777–1788. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Gray, W.D.; French, K.M.; Ghosh-Choudhary, S.; Maxwell, J.T.; Brown, M.E.; Platt, M.O.; Searles, C.D.; Davis, M.E. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ. Res. 2015, 116, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Lombaert, I.M.; Hauser, B.R.; Patel, V.N.; Hoffman, M.P. Exosomal MicroRNA Transport from Salivary Mesenchyme Regulates Epithelial Progenitor Expansion during Organogenesis. Dev. Cell 2017, 40, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Xu, C.; Zhang, Y.; Xue, C.; Yang, C.; Bi, H.; Qian, X.; Wu, M.; Ji, K.; Zhao, Y.; et al. Umbilical Cord-Derived Mesenchymal Stem Cell-Derived Exosomal MicroRNAs Suppress Myofibroblast Differentiation by Inhibiting the Transforming Growth Factor-β/SMAD2 Pathway During Wound Healing. Stem Cells Transl. Med. 2016, 5, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.W.; Wang, J.; Lee, C.J.; Liu, M.; Neelamegham, S.; Canty, J.M.; Nguyen, J. The microRNA regulatory landscape of MSC-derived exosomes: A systems view. Sci. Rep. 2018, 8, 1419. [Google Scholar] [CrossRef] [PubMed]
- Endisha, H.; Rockel, J.; Jurisica, I.; Kapoor, M. The complex landscape of microRNAs in articular cartilage: Biology, pathology, and therapeutic targets. JCI Insight 2018, 3, 121630. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Miyaki, S.; Ishitobi, H.; Matsuyama, S.; Nakasa, T.; Kamei, N.; Akimoto, T.; Higashi, Y.; Ochi, M. Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle regeneration. FEBS Lett. 2015, 589, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jia, J.; Liu, X.; Yang, S.; Ye, S.; Yang, W.; Zhang, Y. MicroRNA-16-5p Controls Development of Osteoarthritis by Targeting SMAD3 in Chondrocytes. Curr. Pharm. Des. 2015, 21, 5160–5167. [Google Scholar] [CrossRef] [PubMed]
- Kopańska, M.; Szala, D.; Czech, J.; Gabło, N.; Gargasz, K.; Trzeciak, M.; Zawlik, I.; Snela, S. MiRNA expression in the cartilage of patients with osteoarthritis. J. Orthop. Surg. Res. 2017, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gibson, G.; Kim, J.S.; Kroin, J.; Xu, S.; van Wijnen, A.J.; Im, H.J. MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene 2011, 480, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.M.; Meng, H.Y.; Yuan, X.L.; Wang, Y.; Guo, Q.Y.; Peng, J.; Wang, A.Y.; Lu, S.B. MicroRNAs’ Involvement in Osteoarthritis and the Prospects for Treatments. Evid. Based Complement. Altern. Med. 2015, 2015, 236179. [Google Scholar] [CrossRef] [PubMed]
- Essandoh, K.; Li, Y.; Huo, J.; Fan, G.C. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock 2016, 46, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Lopa, S.; Colombini, A.; Stanco, D.; de Girolamo, L.; Sansone, V.; Moretti, M. Donor-matched mesenchymal stem cells from knee infrapatellar and subcutaneous adipose tissue of osteoarthritic donors display differential chondrogenic and osteogenic commitment. Eur. Cell Mater. 2014, 27, 298–311. [Google Scholar] [PubMed]
- Tsuchida, A.I.; Beekhuizen, M.; ‘t Hart, M.C.; Radstake, T.R.; Dhert, W.J.; Saris, D.B.; van Osch, G.J.; Creemers, L.B. Cytokine profiles in the joint depend on pathology, but are different between synovial fluid, cartilage tissue and cultured chondrocytes. Arthritis Res. Ther. 2014, 16, 441. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.J.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Barilani, M.; Banfi, F.; Sironi, S.; Ragni, E.; Guillaumin, S.; Polveraccio, F.; Rosso, L.; Moro, M.; Astori, G.; Pozzobon, M.; et al. Low-affinity Nerve Growth Factor Receptor (CD271) Heterogeneous Expression in Adult and Fetal Mesenchymal Stromal Cells. Sci. Rep. 2018, 8, 9321. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Banfi, F.; Barilani, M.; Cherubini, A.; Parazzi, V.; Larghi, P.; Dolo, V.; Bollati, V.; Lazzari, L. Extracellular Vesicle-Shuttled mRNA in Mesenchymal Stem Cell Communication. Stem Cells 2017, 35, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Ramos, T.L.; Sánchez-Abarca, L.I.; Muntión, S.; Preciado, S.; Puig, N.; López-Ruano, G.; Hernández-Hernández, Á.; Redondo, A.; Ortega, R.; Rodríguez, C.; et al. MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Commun. Signal. 2016, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Kennel, P.J.; Saha, A.; Maldonado, D.A.; Givens, R.; Brunjes, D.L.; Castillero, E.; Zhang, X.; Ji, R.; Yahi, A.; George, I.; et al. Serum exosomal protein profiling for the non-invasive detection of cardiac allograft rejection. J. Heart Lung. Transplant. 2018, 37, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Cavalleri, T.; Angelici, L.; Favero, C.; Dioni, L.; Mensi, C.; Bareggi, C.; Palleschi, A.; Rimessi, A.; Consonni, D.; Bordini, L.; et al. Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos-exposed subjects suggest a 2-miRNA signature as potential biomarker of disease. PLoS ONE 2017, 12, e0176680. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034-1. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
| Accession Number | Gene Name | Target Sequence (5′–3′) | Reference |
|---|---|---|---|
| Candidate reference genes | |||
| MIMAT0000062 | let-7a-5p | UGAGGUAGUAGGUUGUAUAGUU | [27,28,29] |
| MIMAT0000069 | miR-16-5p | UAGCAGCACGUAAAUAUUGGCG | [30,31] |
| MIMAT0000078 | miR-23a-3p | AUCACAUUGCCAGGGAUUUCC | [32] |
| MIMAT0000082 | miR-26a-5p | UUCAAGUAAUCCAGGAUAGGCU | [28,32] |
| MIMAT0000099 | miR-101-3p | UACAGUACUGUGAUAACUGAA | [32] |
| MIMAT0000101 | miR-103a-3p | AGCAGCAUUGUACAGGGCUAUGA | [27] |
| MIMAT0000278 | miR-221-3p | AGCUACAUUGUCUGCUGGGUUUC | [27,28] |
| MIMAT0004748 | miR-423-5p | UGAGGGGCAGAGAGCGAGACUUU | [33] |
| MIMAT0003393 | miR-425-5p | AAUGACACGAUCACUCCCGUUGA | [33] |
| NR_004394.1 | U6 snRNA | GUGCUCGCUUCGGCAGCACAUAUACUAAAAU UGGAACGATACAGAGAAGAUUAGCAUGGCCC CUGCGCAAGGAUGACACGCAAAUUCGUGAAG CGUUCCAUAUUUU | [34] |
| miRNA target | |||
| MIMAT0000449 | miR-146a-5p | UGAGAACUGAAUUCCAUGGGUU | [35] |
| Gene Name | GeNorm M-value | NormFinder Stability | BestKeeper SD ± CP | ΔCt Mean | Geomean | Ranking Order |
|---|---|---|---|---|---|---|
| ASCs | ||||||
| miR-23a-3p | 0.044 (1) | 0.060 (2) | 0.579 (6) | 0.526 (1) | 1.86 | 1 |
| miR-16-5p | 0.044 (1) | 0.076 (3) | 0.554 (5) | 0.530 (2) | 2.34 | 2 |
| miR-101-3p | 0.165 (3) | 0.014 (1) | 0.641 (7) | 0.563 (3) | 2.82 | 3 |
| miR-423-5p | 0.369 (5) | 0.327 (6) | 0.232 (1) | 0.672 (5) | 3.50 | 4 |
| U6 snRNA | 0.315 (4) | 0.229 (4) | 0.490 (3) | 0.628 (4) | 3.72 | 5 |
| miR-26a-5p | 0.427 (6) | 0.260 (5) | 0.533 (4) | 0.679 (6) | 5.18 | 6 |
| miR-221-3p | 0.481 (7) | 0.478 (8) | 0.451 (2) | 0.810 (7) | 5.29 | 7 |
| miR-425-5p | 0.538 (8) | 0.505 (9) | 0.747 (8) | 0.850 (8) | 8.24 | 8 |
| miR-103a-3p | 0.612 (9) | 0.474 (7) | 0.885 (9) | 0.868 (9) | 8.45 | 9 |
| let-7a-5p | 0.735 (10) | 0.812 (10) | 1.071 (10) | 1.227 (10) | 10.00 | 10 |
| OA-Like Inflamed-ASCs | ||||||
| miR-23a-3p | 0.338 (4) | 0.099 (1) | 0.477 (4) | 0.645 (1) | 2.00 | 1 |
| miR-221-3p | 0.209 (1) | 0.257 (4) | 0.187 (1) | 0.695 (4) | 2.00 | 2 |
| miR-423-5p | 0.209 (1) | 0.158 (3) | 0.263 (2) | 0.652 (3) | 2.06 | 3 |
| miR-16-5p | 0.289 (3) | 0.144 (2) | 0.428 (3) | 0.647 (2) | 2.45 | 4 |
| miR-26a-5p | 0.442 (5) | 0.318 (5) | 0.506 (5) | 0.747 85) | 5.00 | 5 |
| miR-103a-3p | 0.551 (6) | 0.423 (6) | 0.709 (7) | 0.852 (6) | 6.24 | 6 |
| miR-101-3p | 0.630 (7) | 0.567 (7) | 0.722 (8) | 0.961 (7) | 7.24 | 7 |
| let-7a-5p | 0.678 (8) | 0.575 (8) | 0.862 (10) | 0.974 (8) | 8.46 | 8 |
| miR-425-5p | 0.837 (10) | 0.732 (10) | 0.545 (6) | 1.150 (10) | 8.80 | 9 |
| U6 snRNA | 0.759 (9) | 0.606 (9) | 0.808 (9) | 1.044 (9) | 9.00 | 10 |
| ALL ASCs | ||||||
| miR-16-5p | 0.212 (1) | 0.087 (2) | 0.491 (3) | 0.616 (1) | 1.57 | 1 |
| miR-23a-3p | 0.212 (1) | 0.057 (1) | 0.528 (5) | 0.618 (2) | 1.78 | 2 |
| miR-423-5p | 0.345 (3) | 0.190 (5) | 0.262 (1) | 0.687 (3) | 2.59 | 3 |
| miR-221-3p | 0.384 (4) | 0.230 (7) | 0.316 (2) | 0.781 (5) | 4.09 | 4 |
| miR-26a-5p | 0.474 (5) | 0.186 (4) | 0.525 (4) | 0.717 (4) | 4.23 | 5 |
| miR-101-3p | 0.532 (6) | 0.143 (3) | 0.739 (7) | 0.788 (6) | 5.24 | 6 |
| miR-103a-3p | 0.644 (8) | 0.229 (6) | 0.826 (8) | 0.845 (7) | 7.20 | 7 |
| U6 snRNA | 0.590 (7) | 0.242 (8) | 0.650 (6) | 0.861 (8) | 7.20 | 8 |
| let-7a-5p | 0.723 (9) | 0.328 (9) | 0.966 (10) | 1.070 (9) | 9.24 | 9 |
| miR-425-5p | 0.818 (10) | 0.350 (10) | 0.841 (9) | 1.198 (10) | 9.74 | 10 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragni, E.; Perucca Orfei, C.; De Luca, P.; Colombini, A.; Viganò, M.; Lugano, G.; Bollati, V.; de Girolamo, L. Identification of miRNA Reference Genes in Extracellular Vesicles from Adipose Derived Mesenchymal Stem Cells for Studying Osteoarthritis. Int. J. Mol. Sci. 2019, 20, 1108. https://doi.org/10.3390/ijms20051108
Ragni E, Perucca Orfei C, De Luca P, Colombini A, Viganò M, Lugano G, Bollati V, de Girolamo L. Identification of miRNA Reference Genes in Extracellular Vesicles from Adipose Derived Mesenchymal Stem Cells for Studying Osteoarthritis. International Journal of Molecular Sciences. 2019; 20(5):1108. https://doi.org/10.3390/ijms20051108
Chicago/Turabian StyleRagni, Enrico, Carlotta Perucca Orfei, Paola De Luca, Alessandra Colombini, Marco Viganò, Gaia Lugano, Valentina Bollati, and Laura de Girolamo. 2019. "Identification of miRNA Reference Genes in Extracellular Vesicles from Adipose Derived Mesenchymal Stem Cells for Studying Osteoarthritis" International Journal of Molecular Sciences 20, no. 5: 1108. https://doi.org/10.3390/ijms20051108
APA StyleRagni, E., Perucca Orfei, C., De Luca, P., Colombini, A., Viganò, M., Lugano, G., Bollati, V., & de Girolamo, L. (2019). Identification of miRNA Reference Genes in Extracellular Vesicles from Adipose Derived Mesenchymal Stem Cells for Studying Osteoarthritis. International Journal of Molecular Sciences, 20(5), 1108. https://doi.org/10.3390/ijms20051108










